Abstract
Hybridization among littoral cichlid species in Lake Tanganyika was inferred in several molecular phylogenetic studies. The phenomenon is generally attributed to the lake level-induced shoreline and habitat changes. These allow for allopatric divergence of geographically fragmented populations alternating with locally restricted secondary contact and introgression between incompletely isolated taxa. In contrast, the deepwater habitat is characterized by weak geographic structure and a high potential for gene flow, which may explain the lower species richness of deepwater than littoral lineages. For the same reason, divergent deepwater lineages should have evolved strong intrinsic reproductive isolation already in the incipient stages of diversification, and, consequently, hybridization among established lineages should have been less frequent than in littoral lineages. We test this hypothesis in the endemic Lake Tanganyika deepwater cichlid tribe Bathybatini by comparing phylogenetic trees of Hemibates and Bathybates species obtained with nuclear multilocus AFLP data with a phylogeny based on mitochondrial sequences. Consistent with our hypothesis, largely congruent tree topologies and negative tests for introgression provided no evidence for introgressive hybridization between the deepwater taxa. Together, the nuclear and mitochondrial data established a well-supported phylogeny and suggested ecological segregation during speciation.
1. Introduction
Cichlid fishes have undergone spectacular radiations in different parts of the world. In particular, the species flocks of the East African Great Lakes are well-known examples for rapid evolution and speciation [1–5]. Each of the three Great Lakes—Tanganyika, Malawi, and Victoria—is inhabited by hundreds of mostly endemic cichlid species [6, 7]. Notably, most of the diversity is found in the littoral habitat, whereas reduced species richness in the deep benthal and pelagial seems to be a common phenomenon in all East African Great Lakes [7–10]. At least three factors may have contributed to this pattern: (i) reduced niche diversity in the pelagic and in deepwater benthic zones, (ii) a narrow ambient light spectrum consisting only of short-wavelength blue light and hence less promotive of diversification mechanisms contingent on color perception than the shallow clear-water habitats [11–14], and (iii) the absence of strong barriers to gene flow. Indeed, deepwater cichlid species often have lake-wide distributions with very low, if any, population genetic structure over large geographic distances [10, 15, 16] (see also the Lake Tanganyika clupeid Limnothrissa miodon [17] and the centropomid Lates stappersii [18]). On the other hand, high levels of genetic differentiation, sometimes accompanied by phenotypic divergence on small geographic scales, are characteristic for the species-rich guild of stenotopic rock-dwelling cichlid species [19–31]. However, allopatric diversification in the fragmented littoral zone was not necessarily accompanied by the evolution of pre- or postzygotic isolation, so that secondary contact imposed by lake level fluctuations has often led to hybridization or introgression between previously allopatric taxa [32–35]. Even today, substrate breeders of the tribe Lamprologini can be found in mixed-species pairs [36], and interspecific fertilizations occur in communally nesting, shell-breeding lamprologines [37]. Indeed, phylogenetic analyses of predominantly littoral cichlid lineages revealed that interspecific hybridization has played, and still plays, an important role in the evolution of these fish [32, 34, 36–41]. Thus, in the majority of recent molecular studies on species relationships within littoral tribes, especially when comparing mitochondrial and nuclear phylogenies, the explanation for the tree topologies involved the claim of introgression and hybridization between established lineages in addition to incomplete lineage sorting (reviewed in [42]).
We hypothesize that this is not the case in tribes composed of deepwater species. Lacking the geographic structure introduced by littoral habitat heterogeneity, deepwater species may still be spatially separated by distance, by segregation of breeding grounds, by variable hydrological conditions [16, 43, 44], or by large-scale fragmentation of the lake basin during major droughts [45, 46]. Generally, however, the potential barriers to gene flow for deepwater species are less insurmountable than those met by stenotopic littoral cichlids. Specifically in Lake Tanganyika, the evolution of stenotopy regarding depth, bottom type, or light intensity may have been prohibited by the seasonal upwelling of anoxic waters [47]. We postulate that given the high potential for gene flow, diversification of lineages will either be curtailed (as suggested by the relative species paucity) or be attended by strong reproductive isolation right from the start. This would imply that introgression following cladogenesis occurred at much lower rates, if at all, in the deepwater species than in littoral cichlids. We test this hypothesis in the deepwater cichlid tribe Bathybatini by comparing phylogenetic trees based on mitochondrial sequence data with trees obtained with nuclear multilocus AFLP data, an approach which has previously revealed hybridization in littoral cichlids and other contexts [34, 36, 37, 39, 40, 48–54]. Moreover, in several studies, phylogenetic inference on species relationships has benefited from the use of multilocus genetic data, and we expect that the AFLP data collected in the present study will also contribute to the resolution of intergeneric relationships within the Bathybatini, which are still debated due to conflicting or ambiguous molecular and morphological evidence [46, 55–59].
1.1. The Study Species
The tribe Bathybatini (sensu Takahashi [59]) comprises 17 species in three genera: (1) the genus Bathybates comprises six large (30–40 cm), piscivorous species preying mainly on pelagic freshwater clupeids (B. fasciatus and B. leo), benthic cichlids (B. graueri, B. vittatus, and B. ferox), or undefined prey (the rare, elusive B. horni), in addition to the small (20 cm) B. minor, which is a specialized clupeid hunter. In accordance with their trophic niches, Coulter [43] distinguished three morphotypes among the Bathybates species, the fast-swimming fusiform predators B. fasciatus, B. leo, and B. horni, the generalized shape of the benthic feeders B. graueri, B. vittatus, and B. ferox, and the small clupeid-mimicking B. minor, which mingles with its prey and accompanies the diurnal clupeid migrations. Based on trawl net and gill net catches, B. minor were classified as pelagic, B. fasciatus and B. leo as chiefly bathypelagic and the remaining four species (B. graueri, B. vittatus, B. ferox, and B. horni) as chiefly benthic [43]. Except for B. minor, which was never found below 70 m, Bathybates species descend to depths of 150–200 m. (2) The member of the monotypic genus Hemibates, H. stenosoma, is an abundant benthic species on the muddy bottom of southern Lake Tanganyika feeding on fish and shrimps mainly at depths between 100 and 200 m [43]. (3) The small-bodied (<15 cm) species of the genus Trematocara (formerly assigned to the genera Trematocara and Telotrematocara, [55]) comprise nine benthic and bathypelagic species feeding on a variety on invertebrate prey, fish larvae, and phytoplankton. They are found at maximum depths of 75 to 200 m [43]. Following the upward movement of zooplankton, many Trematocara species undertake nightly migrations along slopes into the littoral.
All members of the Bathybatini are maternal mouthbrooders. Some species release their fry in shallow areas, but overall, data on bathybatine breeding behaviour is anecdotal or lacking [43, 60]. The species are sexually dimorphic, with males of Bathybates and Hemibates exhibiting species-specific patterns of dark stripes, bars and dots on a silver background and egg-spots on the anal fins, and males of the silvery Trematocara with dark dorsal fin markings. Females of all species show a uniformly silver/brown coloration. All Bathybatini have large eyes, which promote not only the detection of prey and predators but possibly also mate-recognition in the dark depths. In line with the latter, the monochromatic patterning of males may be viewed as adaptation to the short-wavelength dominated visual environment [61], in contrast to the colourful patterns of the mouthbrooding cichlids in the shallow, light-flooded littoral.
A recent phylogenetic study based on three mitochondrial genes supported the monophyly of Bathybates as well as of the species therein and indicated a polytomy of three equidistant lineages representing Bathybates, Hemibates, and Trematocara [46]. Within Bathybates, B. minor appeared ancestral to a radiation of the six large species, which showed a basal split of B. graueri and low statistical support for the branching order of the remaining species (Figure 1). The short internal branches among the large Bathybates species supported a rapid radiation at approximately 2.3–2.7 MYA, coinciding with the rapid diversification of other Lake Tanganyika cichlids [28, 45, 62]. Competition and resource partitioning as well as potential geographic isolation during an extreme low-stand of the lake were proposed as promoters of Bathybates speciation [46].
Figure 1.
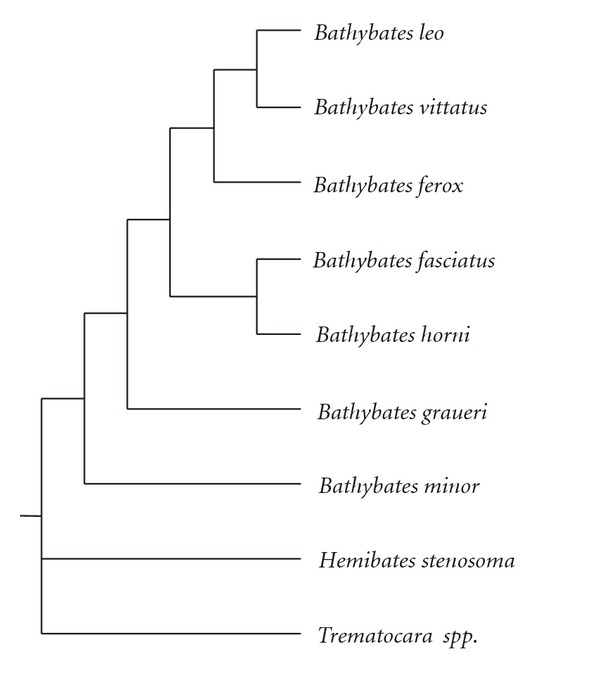
Schematic depiction of the the phylogenetic relationships within the Bathybatini as inferred from mtDNA data [46].
2. Material and Methods
2.1. Sample Collection and DNA Extraction
This study is based on a total of 38 specimens, representing all seven Bathybates species, Hemibates stenosoma as well as Trematocara unimaculata and T. macrostoma (Table 1). All specimens were obtained between 1999 and 2011 from local fishermen at Lake Tanganyika and identified to species by S. Koblmüller. Unfortunately, it was not possible to obtain a comprehensive taxon sampling for the genus Trematocara as, because of their small size and hence low market value, these fish (except for the largest species T. unimaculatum) are not caught by local fishermen.
Table 1.
List of samples with sample ID and sampling locality.
| Sample ID | Species | Sampling locality |
|---|---|---|
| 12879 | Bathybates fasciatus | Mpulungu market |
| 12885 | Bathybates fasciatus | Mpulungu market |
| 12889 | Bathybates fasciatus | Kalambo Lodge |
| 12890 | Bathybates fasciatus | Kalambo Lodge |
| 12917 | Bathybates fasciatus | Tanganyika Lodge |
| 12913 | Bathybates ferox | Lufubu estuary |
| 12877 | Bathybates graueri | Mpulungu market |
| 12878 | Bathybates graueri | Mpulungu market |
| 12883 | Bathybates graueri | Mpulungu market |
| 12893 | Bathybates graueri | Mpulungu market |
| 12897 | Bathybates graueri | Mpulungu market |
| 12901 | Bathybates graueri | Mpulungu market |
| 12902 | Bathybates graueri | Mpulungu market |
| 12911 | Bathybates graueri | Mpulungu market |
| 12912 | Bathybates graueri | Mpulungu market |
| 12919 | Bathybates graueri | North of Sumbu |
| 13101 | Bathybates horni | Mpulungu market |
| 12907 | Bathybates leo | Mpulungu market |
| 12921 | Bathybates leo | Mpulungu market |
| 12923 | Bathybates leo | Mpulungu market |
| 12925 | Bathybates leo | Mpulungu market |
| 13100 | Bathybates leo | Mpulungu market |
| 12909 | Bathybates minor | Lufubu estuary |
| 12910 | Bathybates minor | Kalambo |
| 12933 | Bathybates minor | Sumbu |
| 12882 | Bathybates vittatus | Mpulungu market |
| 12924 | Bathybates vittatus | Mpulungu market |
| 12926 | Bathybates vittatus | Mpulungu market |
| 12929 | Hemibates stenosoma | Mpulungu market |
| 12930 | Hemibates stenosoma | Mpulungu market |
| 12931 | Hemibates stenosoma | Mpulungu market |
| 12932 | Hemibates stenosoma | Mpulungu market |
| 12880 | Trematocara unimaculata | Mpulungu market |
| 12881 | Trematocara unimaculata | Mpulungu market |
| 12935 | Trematocara macrostoma | Mpulungu market |
| 12936 | Trematocara macrostoma | Mpulungu market |
Coordinates of sampling sites (if known): Kalambo, S 8°37′ E 31°12′; Kalambo Lodge, S 8°37′ E 31°37′; Lufubu estuary, S 8°32′ E 30°44′; Sumbu, S 8°31′ E 30°29′; Tanganyika Lodge, S 8°47′ E 31°05′.
Note that fish obtained at the fishmarket in Mpulungu might have been caught anywhere in southern Lake Tanganyika.
Fin clips or white muscle tissue were preserved in ethanol and DNA was isolated using a proteinase K digestion/high salt precipitation method [63]. DNA concentrations were measured using a NanoPhotometer (IMPLEN).
2.2. AFLP Data Collection
Amplified fragment length polymorphism (AFLP) genotyping followed the protocol described in [34]. The ten primer combinations used for selective amplification were EcoRI-ACA/MseI-CAA, EcoRI-ACA/MseI-CAG, EcoRI-ACA/MseI-CAC, EcoRI-ACA/MseI-CAT, EcoRI-ACT/MseI-CAT, EcoRI-ACT/MseI-CAA, EcoRI-ACT/MseI-CAG, EcoRI-ACT/MseI-CAC, EcoRI-ACC/MseI-CAA, and EcoRI-ACC/MseI-CAC. Selective amplification products were visualized using an ABI 3130xl automated sequencer (Applied Biosystems) along with an internal size standard (Genescan-500 ROX; Applied Biosystems). Polymorphic positions were initially identified using GeneMapper 3.7 software (Applied Biosystems) in a range of 50–500 bp. In order to adjust misaligned bins and avoid size homoplasy, bin positions were set manually. Bins containing ambiguous low intensity peaks in a large proportion of the samples and entire profiles with short read-lengths or very low peak heights were deleted. These preprocessed, un-normalized peak-height data were analyzed with AFLPScore 1.4a [64], which optimizes thresholds for locus retention and phenotype calling based on estimated error rates. Phenotype calling thresholds were set as absolute or relative depending on the number of retained loci and the achieved error rates obtained with each option. 20 replicate samples were included to calculate the mismatch error rate for all unique loci.
2.3. Phylogenetic Inference
A neighbour joining (NJ) tree based on Nei and Li's distances [65] was constructed in PAUP 4.0b5 [66]. Bootstrap values from 1000 pseudoreplicates were used as standard measure of confidence in the inferred tree topology.
Although accurate models for Bayesian tree construction using AFLP datasets do exist [67]), the high demands for processing power make them unfeasible to use [67, 68]. Thus, Bayesian phylogenetic inference (BI) was conducted in MrBayes 3.1.2 [69], employing the restriction site model with the “noabsencesites” coding bias correction [68, 70]. The Dirichlet prior for the state frequencies was set to (2.44, 1.00) matching the actual 0/1 frequencies in the dataset. Posterior probabilities were obtained from Metropolis-coupled Markov chain Monte Carlo simulations (2 independent runs; 10 chains with 8,000,000 generations each; chain temperature: 0.2; sample frequency: 1,000; burn-in: 4,000,000 generations). Chain stationarity and run parameter convergence were checked in Tracer 1.5 [71].
To test for homoplasy excess introduced by hybridization, we conducted a tree-based method as outlined by Seehausen [74] by removing single species from the dataset and observing the change in bootstrap values in the NJ tree (see also [34, 40]). In theory, the inclusion of a hybrid taxon in a multilocus phylogeny introduces homoplasy with clades that contain its parental taxa. Hybrid taxa should be intermediate to the parental taxa since they carry a mosaic of parental characteristics. Thus, decreasing the amount of homoplasy in the dataset by removing the hybrid taxon should increase the bootstrap support for the clades that include the parental taxa or their descendants, whereas removing nonhybrid taxa should have no effect on the statistical support of other nodes (Figure 2).
Figure 2.
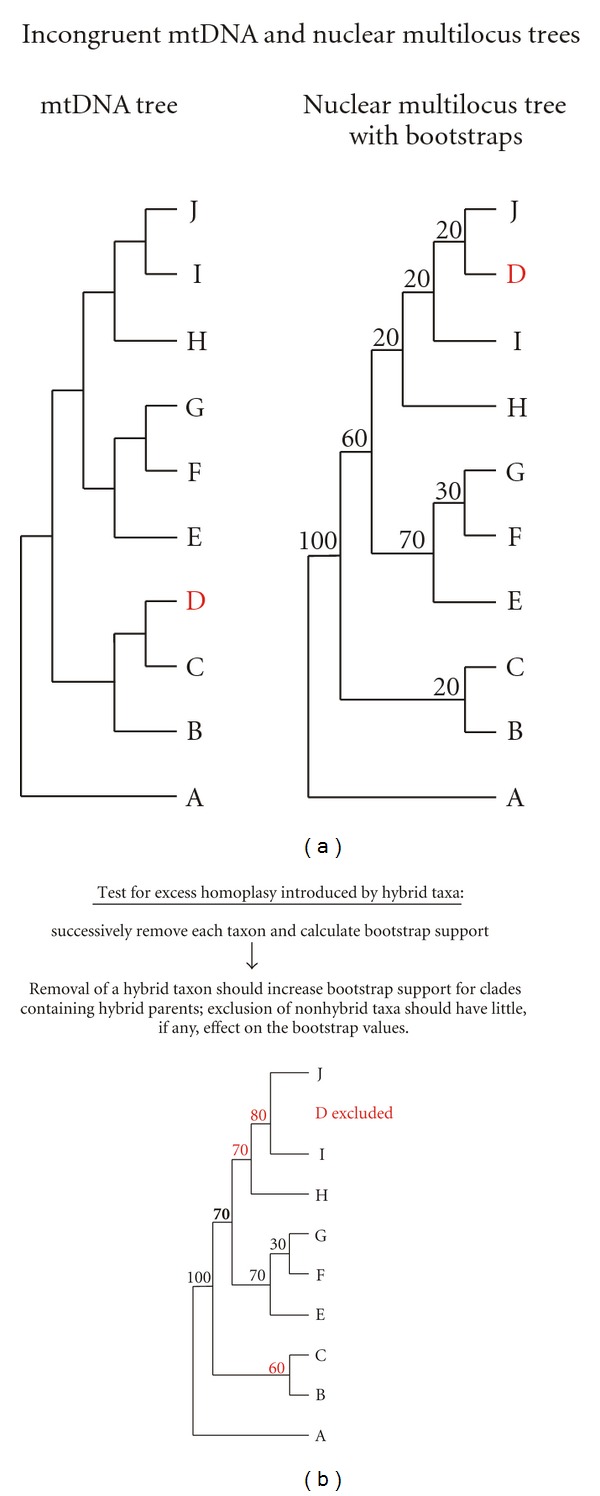
Incongruency between mtDNA and nuclear multilocus trees (e.g., AFLPs) and a test for hybridization in a multilocus phylogeny. (a) Incongruency between mtDNA and nuclear multilocus trees with regard to the placement of taxon D can result from ancient incomplete lineage sorting [72, 73] or the hybrid origin of taxon D. (b) As hybrid taxa combine nuclear alleles from both parental taxa, they introduce homoplasy into a multilocus phylogenetic tree and hence reduce bootstrap support of the nodes containing their parents [74]. Removal of the hybrid taxon from the phylogeny increases the bootstrap support of the parental clades (bold values in (b)). Conversely, removal of nonhybrid taxa should not or only slightly affect the bootstrap support of other nodes. To distinguish between informative (red values in (b)) and uninformative changes in bootstrap values, one taxon at a time is removed from the data and the resulting distribution of bootstrap values for each node is recorded. If removal of a certain taxon produces an outlier in these distributions, the removed taxon is considered a hybrid (or strongly introgressed taxon) and the clades for which support was raised are considered to contain the parental taxa (see, e.g., [34, 40, 48]). In the present example, taxon D is a hybrid between taxa C and J.
2.4. Evaluating Alternative Tree Topologies
Testing for consistency between mtDNA- and AFLP-based tree topologies employed two different strategies. In a first test we evaluated whether our AFLP-NJ-topology can be explained by the mtDNA data of Koblmüller et al. [46]. Using the mitochondrial sequences, we inferred the log likelihood of the AFLP-NJ-topology data by constraining maximum likelihood (ML) tree search to a topology identical to the species tree suggested by the NJ analyses of our AFLP data, applying the substitution model used in this previous study (HKY+I+G). To test for significant differences between the unconstrained [46] and constrained mtDNA topology we performed an ML-based Shimodaira-Hasegawa (SH) test [75] (full optimization, 1,000 bootstrap replicates) in PAUP. In a second test we evaluated by means of a Bayes factors approach [76] whether the mtDNA tree [46] can be explained by our AFLP data, and whether the AFLP-NJ and BI trees differ significantly from each other. We performed BI searches constraining the topology to that of the mtDNA-topology [46] and the interspecific relationships implied by the AFLP-NJ-tree in MrBayes 3.1.2 employing the same settings as above. Bayes factor comparison—using the harmonic means of the likelihood throughout different runs [77, 78]—among the three alternative phylogenatic hypotheses was performed in Tracer 1.5. Values of 2 × ln BF (two times the difference between the harmonic means of the two models) >10 are considered strong evidence for support of one model over another [76].
3. Results
The final AFLP dataset consisted of 659 unique loci with a mismatch error rate of roughly 3%, which falls within the acceptable limit for mismatch error rates as defined by [64]. Both the NJ and BI analysis yielded largely congruent and well-supported topologies with only minor differences between them (Figures 3(a) and 3(b)). Whereas all species were monophyletic in the NJ tree, Bathybates graueri was not resolved as a monophylum in the BI tree, but as paraphylum including the well-supported clade of the other large Bathybates species (Figure 3(b)). Despite this minor topological difference, Bayes factor comparison strongly supports the BI tree over the NJ tree (2 × ln BF = 26.828; Figure 3(c)). We note, however, that the two-state model implemented in MrBayes does not fully cover the complex genetic process of AFLP evolution and thus provides accurate phylogenetic inference less likely than distance methods [67, 79, 80]. Hence, the observed differences between AFLP and NJ tree topologies might be attributed to this problem. Both the NJ and BI analyses support the monophyly of all three genera with the genus Trematocara representing the most ancestral branch (Figures 3(a) and 3(b)). Within the genus Bathybates, the small and morphologically most distinct member of the genus, B. minor, was sister taxon to the remaining large Bathybates species. Branch lengths among the large Bathybates species are rather short and some received rather low statistical support, indicating a period of rapid cladogenesis. Nevertheless, both NJ and BI analyses revealed a largely consistent phylogenetic pattern within the large Bathybates species. Both SH-test and Bayes factor comparison revealed significant differences between mtDNA and AFLP phylogenies (SH-test: ln L of −9505.385 versus −9585.142 for mtDNA versus AFLP-NJ-topology-constraint, P < 0.001; Bayes factors: 2 × ln BF of 88.576 between mtDNA and AFLP-BI-topology and 61.748 between mtDNA and AFLP-NJ-topology, Figure 3(c)). The homoplasy excess test provided no evidence for introgressive hybridization (data not shown).
Figure 3.
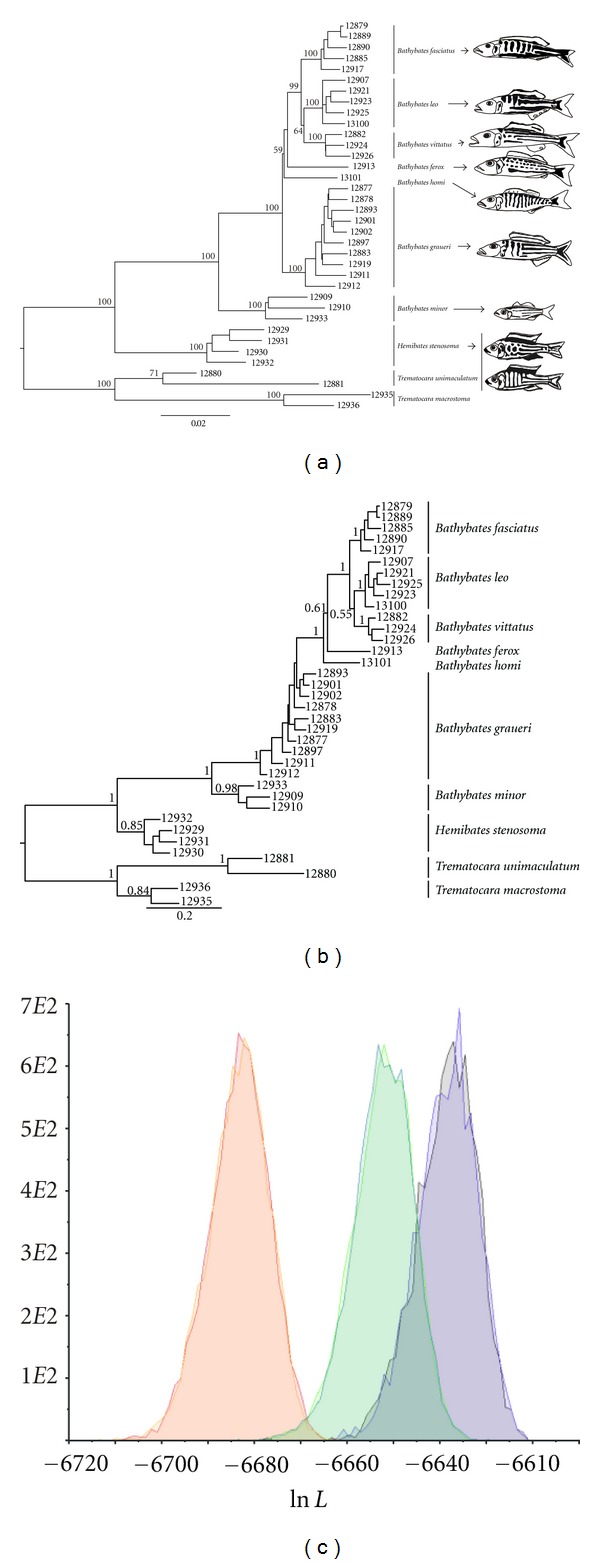
Phylogenetic relationships of the Bathybatini based on 659 polymorphic AFLP loci. (a) NJ tree (employing Nei and Li distances [65]) and (b) BI tree are shown. Only bootstrap values >50 and posterior probabilities >0.5 are shown. Branches are labelled with the samples' lab ID numbers. (c) The marginal density of posterior distribution of likelihood (Ln L) for each of the two MrBayes runs of the unconstrained AFLP data (violet, grey), the AFLP-NJ-topology-constraint (blue, green), and the mtDNA-topology-constraint (orange, red). Fish were drawn after photographs in [60] to demonstrate the interspecific differences in male nuptial patterns.
4. Discussion
In the original classification of Lake Tanganyika cichlid tribes by Poll [55], Bathybates and Hemibates were included in a tribe Bathybatini as sister group to the Trematocarini (equivalent to the genus Trematocara), a hypothesis supported both by lepidological [58] as well as allozyme data [57]. In contrast, based on morphological characteristics, Stiassny [56] and Takahashi [59] proposed a sister group relationship of Bathybates and Trematocara. Currently, all three genera, Bathybates, Hemibates, and Trematocara, are united in the tribe Bathybatini [59]. A previous mtDNA phylogenetic study remained equivocal with regard to the two competing morphological classifications and suggested that Bathybates, Hemibates, and Trematocara diverged rapidly from their common ancestor [46]. In the present AFLP phylogeny, the length differences between the most ancestral branches favour Poll's original classification with Trematocara as sister group to Hemibates + Bathybates [55, 57, 58]. Consistent with the mitochondrial phylogeny [46], the AFLP data confirm the split between Bathybates minor and the larger members of the genus Bathybates with B. graueri as their most basal representative and identifies a period of rapid cladogenesis at the onset of the diversification of the large Bathybates species. However, mtDNA and AFLP phylogenies differ significantly with respect to the branching pattern among the remaining large Bathybates species. Introgressive hybridization (including the possibility of complete mtDNA replacement [41]) and ancient incomplete lineage sorting are two alternative sources of topological disagreement between nuclear and mitochondrial trees [40, 41, 81–83], resulting in similar phylogenetic patterns that are difficult to resolve by strict hypothesis testing [84]. Circumstantial inference can be based on the fact that lineage sorting is expected to lag behind rapid cladogenetic events, such that the rapid radiation of the large Bathybates species predisposes this clade to mitonuclear phylogenetic incompatibilities without implying postcladogenetic introgression [72, 73]. Likewise, monophyly of species in both mitochondrial and nuclear trees (excepting the paraphyly of B. graueri in Bayesian AFLP tree) and negative tests for homoplasy excess in the AFLP data do not indicate the presence of hybrid taxa in the genus Bathybates. These findings support our hypothesis that deepwater species are less prone to introgressive hybridization and hybrid speciation than littoral species, but reservations arise on the one hand from the possibility that events of introgression were not detected in our samples and data and given the power of our analyses, and on the other hand from the small number of species in the phylogeny. Principally, rates of interspecific introgression may not differ between littoral and deepwater cichlids, but will nonetheless lead to higher incidences of introgression in the species-rich groups than in a less speciose clade. If this was the case, the lack of a signal of introgression in Bathybates and Hemibates would be fully explained by the low diversification rate of the lineage and hence limited opportunity for interspecific hybridization, without implying the evolution of complete reproductive isolation early on during diversification.
The branching order of Hemibates and the basal Bathybates species, which is reconstructed congruently by mtDNA and AFLP markers, suggests repeated transitions between benthic and bathypelagic feeding mode (ecological data from [43]). The basal H. stenosoma represents a benthic generalist feeding on shrimps and various species of fish. The next split led to the specialized pelagic clupeid hunter B. minor, which mimics its prey in size and coloration and stages surprise attacks from within the sardine shoals. Then, B. graueri took a step back to the benthic habitat, specialized on cichlid prey and evolved a large body size. The chronology of the following radiation of the large bathypelagic clupeid hunters and benthic cichlid hunters remains unresolved, but involved at least one transition from benthic to bathypelagic habitat preferences. Depth preferences may vary between these species [43], such that speciation may have involved both niche and spatial segregation. The apparent ecological differentiation among lineages may have reduced the fitness of hybrids [85–87] and may have promoted the evolution of a mate recognition system, perhaps based on the species-specific melanic patterns of male Hemibates and Bathybates. The efficacy of monochromatic black, silvery, and white body and fin patterns in mediating assortative mating in the dark, short-wavelength dominated environment has recently been demonstrated for deepwater cichlid species of Lake Malawi. These sympatric and morphologically similar species differ primarily in male nuptial patterns and their reproductive isolation is corroborated by genetic differentiation estimates [61]. However, there is increasing evidence that color pattern is not the only cue for mate recognition in cichlids fish and it is possible and likely that auditory [88] and olfactory cues [89] play a role in mediating assortative mating in deepwater species, too.
5. Summary and Conclusions
In concert with previous mitochondrial data, the present study provides an informative phylogeny of the species in the deepwater genera Hemibates and Bathybates. As far as the branching pattern can be resolved, it suggests ecological segregation during speciation. The rapid radiation within Bathybates mirrors a burst of speciation observed in several other cichlid tribes of Lake Tanganyika and reveals a congruent cladogenetic pattern across vastly different habitats, which suggests some kind of synchronization by environmental factors [27, 45, 46, 62]. Consistent with the hypothesis that lineages evolving in the weakly structured deepwater habitat would develop stronger reproductive isolation than the allopatric lineages of the fragmented littoral, our data provided no evidence for the presence of hybrid taxa in the deepwater dwelling genus Bathybates. In further support of the hypothesis, introgressive hybridization is also not indicated by the mitochondrial and AFLP phylogenies of the Lake Tanganyika cichlid genus Xenotilapia [90], which includes several deepwater-dwelling species comprising the prey of the benthic Bathybates and Hemibates. An increased sample size to evaluate this pattern will be attained by the analyses of additional open-water and deepwater species, for example, the tribe Limnochromini and the genus Trematocara.
Acknowledgments
The authors acknowledge financial support by the Theodor Körner Foundation (to S. Koblmüller) and the Austrian Science Fund (Grant P20883 to K. M. Sefc).
References
- 1.Schluter D. The Ecology of Adaptive Radiation. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 2.Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nature Reviews Genetics. 2004;5(4):288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 3.Turner GF. Adaptive radiation of cichlid fish. Current Biology. 2007;17(19):R827–R831. doi: 10.1016/j.cub.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 4.Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Molecular Ecology. 2009;18(2):169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- 5.Sturmbauer C, Husemann M, Danley P. Explosive speciation and adaptive radiation in East African cichlid fishes. In: Zachos FE, Habel JC, editors. Biodiversity Hotspots. Berlin, Germany: Springer; 2011. pp. 333–362. [Google Scholar]
- 6.Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL. How many species of cichlid fishes are there in African lakes? Molecular Ecology. 2001;10(3):793–806. doi: 10.1046/j.1365-294x.2001.01200.x. [DOI] [PubMed] [Google Scholar]
- 7.Koblmüller S, Sefc KM, Sturmbauer C. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia. 2008;615(1):5–20. [Google Scholar]
- 8.Turner GF. Offshore Cichlids of Lake Malawi. Lauenau, Germany: Cichlid Press; 1996. [Google Scholar]
- 9.Seehausen O, Witte F, Katunzi EF, Smits J, Bouton N. Patterns of the remnant cichlid fauna in southern Lake Victoria. Conservation Biology. 1997;11(4):890–904. [Google Scholar]
- 10.Shaw PW, Turner GF, Idid MR, Robinson RL, Carvalho GR. Genetic population structure indicates sympatric speciation of Lake Malawi pelagic cichlids. Proceedings of the Royal Society B. 2000;267(1459):2273–2280. doi: 10.1098/rspb.2000.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seehausen O, van Alphen JJM, Witte F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science. 1997;277(5333):1808–1811. [Google Scholar]
- 12.Seehausen O, van Alphen JJM, Lande R. Color polymorphism and sex ratio distortion in a cichlid fish as an incipient stage in sympatric speciation by sexual selection. Ecology Letters. 1999;2(6):367–378. [Google Scholar]
- 13.Knight ME, Turner GF. Laboratory mating trials indicate incipient speciation by sexual selection among populations of the cichlid fish Pseudotropheus zebra from Lake Malawi. Proceedings of the Royal Society B. 2004;271(1540):675–680. doi: 10.1098/rspb.2003.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maan ME, Seehausen O, Söderberg L, et al. Intraspecific sexual selection on a speciation trait, male coloration, in the Lake Victoria cichlid Pundamilia nyererei . Proceedings of the Royal Society B. 2004;271(1556):2445–2452. doi: 10.1098/rspb.2004.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genner MJ, Nichols P, Shaw PW, Carvalho GR, Robinson RL, Turner GF. Genetic homogeneity among breeding grounds and nursery areas of an exploited Lake Malawi cichlid fish. Freshwater Biology. 2008;53(9):1823–1831. [Google Scholar]
- 16.Genner MJ, Nichols P, Shaw PW, Carvalho GR, Robinson RL, Turner GF. Population structure on breeding grounds of Lake Malawi’s “twilight zone” cichlid fishes. Journal of Biogeography. 2010;37(2):258–269. [Google Scholar]
- 17.Hauser L, Carvalho GR, Pitcher TJ. Genetic population structure in the Lake Tanganyika sardine Limnothrissa miodon . Journal of Fish Biology. 1998;53(Supplement A):413–429. [Google Scholar]
- 18.Kuusipalo L. Genetic differentiation of endemic nile perch Lates stappersi (Centropomidae, Pisces) populations in Lake Tanganyika suggested by RAPD markers. Hydrobiologia. 1999;407:141–148. [Google Scholar]
- 19.van Oppen MJH, Turner GF, Rico C, et al. Unusually fine-scale genetic structuring found in rapidly speciating Malawi cichlid fishes. Proceedings of the Royal Society of London Series B. 1997;264(1289):1803–1812. [Google Scholar]
- 20.Arnegard ME, Markert JA, Danley PD, Stauffer JR, Ambali AJ, Kocher TD. Population structure and colour variation of the cichlid fish Labeotropheus fuelleborni Ahl along a recently formed archipelago of rocky habitat patches in southern Lake Malawi. Proceedings of the Royal Society B. 1999;266(1415):119–130. [Google Scholar]
- 21.Marker JA, Arnegard ME, Danley PD, Kocher TD. Biogeography and population genetics of the Lake Malawi cichlid Melanochromis auratus: habitat transience, philopatry and speciation. Molecular Ecology. 1999;8(6):1013–1026. [Google Scholar]
- 22.Danley PD, Markert JA, Arnegard ME, Kocher TD. Divergence with gene flow in the rock-dwelling cichlids of Lake Malawi. Evolution. 2000;54(5):1725–1737. doi: 10.1111/j.0014-3820.2000.tb00716.x. [DOI] [PubMed] [Google Scholar]
- 23.Taylor MI, Rüber L, Verheyen E. Microsatellites reveal high levels of population substructuring in the species-poor Eretmodine cichlid lineage from Lake Tanganyika. Proceedings of the Royal Society B. 2001;268(1469):803–808. doi: 10.1098/rspb.2000.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rico C, Turner GF. Extreme microallopatric divergence in a cichlid species from Lake Malawi. Molecular Ecology. 2002;11(8):1585–1590. doi: 10.1046/j.1365-294x.2002.01537.x. [DOI] [PubMed] [Google Scholar]
- 25.Duftner N, Sefc KM, Koblmüller S, et al. Distinct population structure in a phenotypically homogeneous rock-dwelling cichlid fish from Lake Tanganyika. Molecular Ecology. 2006;15(9):2381–2395. doi: 10.1111/j.1365-294X.2006.02949.x. [DOI] [PubMed] [Google Scholar]
- 26.Duftner N, Sefc KM, Koblmüller S, Salzburger W, Taborsky M, Sturmbauer C. Parallel evolution of facial stripe patterns in the Neolamprologus brichardi/pulcher species complex endemic to Lake Tanganyika. Molecular Phylogenetics and Evolution. 2007;45(2):706–715. doi: 10.1016/j.ympev.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Koblmüller S, Sefc KM, Duftner N, Warum M, Sturmbauer C. Genetic population structure as indirect measure of dispersal ability in a Lake Tanganyika cichlid. Genetica. 2007;130(2):121–131. doi: 10.1007/s10709-006-0027-0. [DOI] [PubMed] [Google Scholar]
- 28.Koblmüller S, Salzburger W, Obermüller B, Eigner E, Sturmbauer C, Sefc KM. Separated by sand, fused by dropping water: habitat barriers and fluctuating water levels steer the evolution of rock-dwelling cichlid populations in Lake Tanganyika. Molecular Ecology. 2011;20(11):2272–2290. doi: 10.1111/j.1365-294X.2011.05088.x. [DOI] [PubMed] [Google Scholar]
- 29.Sefc KM, Baric S, Salzburger W, Sturmbauer C. Species-specific population structure in rock-specialized sympatric cichlid species in Lake Tanganyika, East Africa. Journal of Molecular Evolution. 2007;64(1):33–49. doi: 10.1007/s00239-006-0011-4. [DOI] [PubMed] [Google Scholar]
- 30.Wagner CE, McCune AR. Contrasting patterns of spatial genetic structure in sympatric rock-dwelling cichlid fishes. Evolution. 2009;63(5):1312–1326. doi: 10.1111/j.1558-5646.2009.00612.x. [DOI] [PubMed] [Google Scholar]
- 31.Konijnendijk N, Joyce DA, Mrosso HDJ, Egas M, Seehausen O. Community genetics reveal elevated levels of sympatric gene flow among morphologically similar but not among morphologically dissimilar species of Lake Victoria cichlid fish. International Journal of Evolutionary Biology. 2011;2011:12 pages. doi: 10.4061/2011/616320. Article ID 616320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rüber L, Meyer A, Sturmbauer C, Verheyen E. Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression. Molecular Ecology. 2001;10(5):1207–1225. doi: 10.1046/j.1365-294x.2001.01259.x. [DOI] [PubMed] [Google Scholar]
- 33.Salzburger W, Baric S, Sturmbauer C. Speciation via introgressive hybridization in East African cichlids? Molecular Ecology. 2002;11(3):619–625. doi: 10.1046/j.0962-1083.2001.01438.x. [DOI] [PubMed] [Google Scholar]
- 34.Egger B, Koblmüller S, Sturmbauer C, Sefc KM. Nuclear and mitochondrial data reveal different evolutionary processes in the Lake Tanganyika cichlid genus Tropheus . BMC Evolutionary Biology. 2007;7, article 137 doi: 10.1186/1471-2148-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nevado B, Fazalova V, Backeljau T, Hanssens M, Verheyen E. Repeated unidirectional introgression of nuclear and mitochondrial dna between four congeneric Tanganyikan cichlids. Molecular Biology and Evolution. 2011;28(8):2253–2267. doi: 10.1093/molbev/msr043. [DOI] [PubMed] [Google Scholar]
- 36.Sturmbauer C, Salzburger W, Duftner N, Schelly R, Koblmüller S. Evolutionary history of the Lake Tanganyika cichlid tribe Lamprologini (Teleostei: Perciformes) derived from mitochondrial and nuclear DNA data. Molecular Phylogenetics and Evolution. 2010;57(1):266–284. doi: 10.1016/j.ympev.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koblmüller S, Duftner N, Sefc KM, et al. Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika—the result of repeated introgressive hybridization. BMC Evolutionary Biology. 2007;7, article 7 doi: 10.1186/1471-2148-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schelly R, Salzburger W, Koblmüller S, Duftner N, Sturmbauer C. Phylogenetic relationships of the lamprologine cichlid genus Lepidiolamprologus (Teleostei: Perciformes) based on mitochondrial and nuclear sequences, suggesting introgressive hybridization. Molecular Phylogenetics and Evolution. 2006;38(2):426–438. doi: 10.1016/j.ympev.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Koblmüller S, Egger B, Sturmbauer C, Sefc KM. Evolutionary history of Lake Tanganyika’s scale-eating cichlid fishes. Molecular Phylogenetics and Evolution. 2007;44(3):1295–1305. doi: 10.1016/j.ympev.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Koblmüller S, Egger B, Sturmbauer C, Sefc KM. Rapid radiation, ancient incomplete lineage sorting and ancient hybridization in the endemic Lake Tanganyika cichlid tribe Tropheini. Molecular Phylogenetics and Evolution. 2010;55(1):318–334. doi: 10.1016/j.ympev.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 41.Nevado B, Koblmüller S, Sturmbauer C, Snoeks J, Usano-Alemany J, Verheyen E. Complete mitochondrial DNA replacement in a Lake Tanganyika cichlid fish. Molecular Ecology. 2009;18(20):4240–4255. doi: 10.1111/j.1365-294X.2009.04348.x. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi T, Koblmüller S. The adaptive radiation of cichlid fish in Lake Tanganyika: a morphological perspective. International Journal of Evolutionary Biology. 2011;2011:14 pages. doi: 10.4061/2011/620754. Article ID 620754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulter GW. Lake Tanganyika and Its Life. London, UK: Oxford University Press; 1991. [Google Scholar]
- 44.Fryer G. Evolution in ancient lakes: radiation of Tanganyikan atyid prawns and speciation of pelagic cichlid fishes in Lake Malawi. Hydrobiologia. 2006;568(1):131–142. [Google Scholar]
- 45.Duftner N, Koblmüller S, Sturmbauer C. Evolutionary relationships of the Limnochromini, a tribe of benthic deepwater cichlid fish endemic to Lake Tanganyika, East Africa. Journal of Molecular Evolution. 2005;60(3):277–289. doi: 10.1007/s00239-004-0017-8. [DOI] [PubMed] [Google Scholar]
- 46.Koblmüller S, Duftner N, Katongo C, Phiri H, Sturmbauer C. Ancient divergence in bathypelagic Lake Tanganyika deepwater cichlids: mitochondrial phylogeny of the tribe Bathybatini. Journal of Molecular Evolution. 2005;60(3):297–314. doi: 10.1007/s00239-004-0033-8. [DOI] [PubMed] [Google Scholar]
- 47.Eccles DH. Is speciation of demersal fishes in Lake Tanganyika restrained by physical limnological conditions? Biological Journal of the Linnean Society. 1986;29(2):115–122. [Google Scholar]
- 48.Schliewen UK, Klee B. Reticulate sympatric speciation in Cameroonian crater lake cichlids. Frontiers in Zoology. 2004;1, article 5 doi: 10.1186/1742-9994-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan JP, Lavoué S, Arnegard ME, Hopkins CD. AFLPs resolve phylogeny and reveal mitochondrial introgression within a species flock of African electric fish (Mormyroidea: Teleostei) Evolution. 2004;58(4):825–841. doi: 10.1111/j.0014-3820.2004.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 50.Herder F, Nolte AW, Pfaender J, Schwarzer J, Hadiaty RK, Schliewen UK. Adaptive radiation and hybridization in Wallace’s Dreamponds: evidence from sailfin silversides in the Malili Lakes of Sulawesi. Proceedings of the Royal Society B. 2006;273(1598):2209–2217. doi: 10.1098/rspb.2006.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi R, Watanabe K, Nishida M, Hori M. Evolution of feeding specialization in Tanganyikan scale-eating cichlids: a molecular phylogenetic approach. BMC Evolutionary Biology. 2007;7(1, article 195) doi: 10.1186/1471-2148-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen PA, Marchán-Rivadeneira MR, Baker RJ. Natural hybridization generates mammalian lineage with species characteristics. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(25):11447–11452. doi: 10.1073/pnas.1000133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternkopf V, Liebers-Helbig D, Ritz MS, Zhang J, Helbig AJ, de Knijff P. Introgressive hybridization and the evolutionary history of the herring gull complex revealed by mitochondrial and nuclear DNA. BMC Evolutionary Biology. 2010;10(1, article 348) doi: 10.1186/1471-2148-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joyce DA, Lunt DH, Genner MJ, Turner GF, Bills R, Seehausen O. Repeated colonization and hybridization in Lake Malawi cichlids. Current Biology. 2011;21(3):R108–R109. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 55.Poll M. Classification des Cichlidae du lac Tanganika. Tribus, genres et espèces. Academie Royale de Belgique, Mémoires de la Classe des Sciences. 1986;45:1–163. [Google Scholar]
- 56.Stiassny MLJ. Phylogenetic versus convergent relationship between piscivorous cichlid fishes from Lakes Malawi and Tanganyika. Bulletin of the British Museum of Natural History. 1981;40:67–101. [Google Scholar]
- 57.Nishida M. Phylogenetic relationships and evolution of Tanganyikan cichlids: a molecular perspective. In: Kawanabe H, Hori M, Nagoshi M, editors. Fish Communities in Lake Tanganyika. Kyoto, Japan: Kyoto University Press; 1997. pp. 1–23. [Google Scholar]
- 58.Lippitsch E. Phylogenetic study of cichlid fishes in Lake Tanganyika: a lepidological approach. Journal of Fish Biology. 1998;53(4):752–766. [Google Scholar]
- 59.Takahashi T. Systematics of Tanganyikan cichlid fishes (Teleostei: Perciformes) Ichthyological Research. 2003;50(4):367–382. [Google Scholar]
- 60.Konings A. Tanganyika Cichlids in Their Natural Habitat. El Paso, Tex, USA: Cichlid Press; 1998. [Google Scholar]
- 61.Genner MJ, Nichols P, Carvalho GR, Robinson RL, Shaw PW, Turner GF. Reproductive isolation among deep-water cichlid fishes of Lake Malawi differing in monochromatic male breeding dress. Molecular Ecology. 2007;16(3):651–662. doi: 10.1111/j.1365-294X.2006.03173.x. [DOI] [PubMed] [Google Scholar]
- 62.Koblmüller S, Salzburger W, Sturmbauer C. Evolutionary relationships in the sand dwelling cichlid lineage of Lake Tanganyika suggest multiple colonization of rocky habitats and convergent origin of biparental mouthbrooding. Journal of Molecular Evolution. 2004;58(1):79–96. doi: 10.1007/s00239-003-2527-1. [DOI] [PubMed] [Google Scholar]
- 63.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16(3):p. 1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitlock R, Hipperson H, Mannarelli M, Butlin RK, Burke T. An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Molecular Ecology Resources. 2008;8(4):725–735. doi: 10.1111/j.1755-0998.2007.02073.x. [DOI] [PubMed] [Google Scholar]
- 65.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swofford DL. PAUP∗: Phylogenetic Analysis Using Parsimony (and Other Methods) version 4.0b2a. Sunderland, Mass, USA: Sinauer; 2000. [Google Scholar]
- 67.Luo R, Hipp AL, Larget B. A Bayesian model of AFLP marker evolution and phylogenetic inference. Statistical Applications in Genetics and Molecular Biology. 2007;6(1):1–30. doi: 10.2202/1544-6115.1152. [DOI] [PubMed] [Google Scholar]
- 68.Koopman WJM, Wissemann V, de Cock K, et al. AFLP markers as a tool to reconstruct complex relationships: a case study in Rosa (Rosaceae) American Journal of Botany. 2008;95(3):353–366. doi: 10.3732/ajb.95.3.353. [DOI] [PubMed] [Google Scholar]
- 69.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 70.Ronquist F, Huelsenbeck JP, van der Mark P. MrBayes 3.1. Manual Draft 5/26.2005. Tallahassee, Fla, USA: School of Computational Science, Florida State University; 2005. [Google Scholar]
- 71.Rambaut A, Drummond AJ. Tracer v1.5. 2009. http://tree.bio.ed.ac.uk/software/tracer/
- 72.Takahashi K, Terai Y, Nishida M, Okada N. Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in lake Tanganyika as revealed by analysis of the insertion of retroposons. Molecular Biology and Evolution. 2001;18(11):2057–2066. doi: 10.1093/oxfordjournals.molbev.a003747. [DOI] [PubMed] [Google Scholar]
- 73.McCracken KG, Sorenson MD. Is homoplasy or lineage sorting the source of incongruent mtDNA and nuclear gene trees in the stiff-tailed ducks (Nomonyx-Oxyura)? Systematic Biology. 2005;54(1):35–55. doi: 10.1080/10635150590910249. [DOI] [PubMed] [Google Scholar]
- 74.Seehausen O. Hybridization and adaptive radiation. Trends in Ecology and Evolution. 2004;19(4):198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 75.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16(8):1114–1116. [Google Scholar]
- 76.Kass RE, Raftery AE. Bayes factors. Journal of the American Statistical Association. 1995;90(430):773–795. [Google Scholar]
- 77.Newton MA, Raftery AE. Approximate Bayesian inference by weighted likelihood bootstrap. Journal of the Royal Statistical Society Series B. 1994;56(1):3–48. [Google Scholar]
- 78.Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Molecular Biology and Evolution. 2001;18(6):1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 79.Althoff DM, Gitzendanner MA, Segraves KA. The utility of amplified fragment length polymorphisms in phylogenetics: a comparison of homology within and between genomes. Systematic Biology. 2007;56(3):477–484. doi: 10.1080/10635150701427077. [DOI] [PubMed] [Google Scholar]
- 80.Holland BR, Clarke AC, Meudt HM. Optimizing automated AFLP scoring parameters to improve phylogenetic resolution. Systematic Biology. 2008;57(3):347–366. doi: 10.1080/10635150802044037. [DOI] [PubMed] [Google Scholar]
- 81.Shaw KL. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):16122–16127. doi: 10.1073/pnas.242585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. Proceedings of the Royal Society B. 2003;270(1520):1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willyard A, Cronn R, Liston A. Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Molecular Phylogenetics and Evolution. 2009;52(2):498–511. doi: 10.1016/j.ympev.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 84.Knowles LL. The burgeoning field of statistical phylogeography. Journal of Evolutionary Biology. 2004;17(1):1–10. doi: 10.1046/j.1420-9101.2003.00644.x. [DOI] [PubMed] [Google Scholar]
- 85.Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256(5054):193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- 86.Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution. 1999;53(3):866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- 87.Schluter D. Ecology and the origin of species. Trends in Ecology and Evolution. 2001;16(7):372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- 88.Verzijden MN, van Heusden J, Bouton N, Witte F, Ten Cate C, Slabbekoorn H. Sounds of male Lake Victoria cichlids vary within and between species and affect female mate preferences. Behavioral Ecology. 2010;21(3):548–555. [Google Scholar]
- 89.Plenderleith M, van Oosterhout C, Robinson RL, Turner GF. Female preference for conspecific males based on olfactory cues in a Lake Malawi cichlid fish. Biology Letters. 2005;1(4):411–414. doi: 10.1098/rsbl.2005.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kidd MR, Duftner N, Koblmüller S, Sturmbauer C, Hofmann H. Repeated parallel evolution of parental care strategies within Xenotilapia, a genus of cichlid fishes from Lake Tanganyika. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0031236. Article ID e31236. [DOI] [PMC free article] [PubMed] [Google Scholar]


