Abstract
Proliferating cell nuclear antigen (PCNA), a potential anticancer target, forms a homotrimer and is required for DNA replication and numerous other cellular processes. The purpose of this study was to identify novel small molecules that modulate PCNA activity to affect tumor cell proliferation. An in silico screen of a compound library against a crystal structure of PCNA and a subsequent structural similarity search of the ZINC chemical database were carried out to derive relevant docking partners. Nine compounds, termed PCNA inhibitors (PCNA-Is), were selected for further characterization. PCNA-I1 selectively bound to PCNA trimers with a dissociation constant (Kd) of ∼0.2 to 0.4 μM. PCNA-Is promoted the formation of SDS-refractory PCNA trimers. PCNA-I1 dose- and time-dependently reduced the chromatin-associated PCNA in cells. Consistent with its effects on PCNA trimer stabilization, PCNA-I1 inhibited the growth of tumor cells of various tissue types with an IC50 of ∼0.2 μM, whereas it affected the growth of nontransformed cells at significantly higher concentrations (IC50, ∼1.6 μM). Moreover, uptake of BrdU was dose-dependently reduced in cells treated with PCNA-I1. Mechanistically the PCNA-Is mimicked the effect of PCNA knockdown by siRNA, inducing cancer cell arrest at both the S and G2/M phases. Thus, we have identified a class of compounds that can directly bind to PCNA, stabilize PCNA trimers, reduce PCNA association with chromatin, and inhibit tumor cell growth by inducing a cell cycle arrest. They are valuable tools in studying PCNA function and may be useful for future PCNA-targeted cancer therapy.
Introduction
PCNA is a ubiquitously expressed protein conserved throughout evolution. It plays crucial roles in many vital cellular processes (Moldovan et al., 2007; Naryzhny, 2008; Stoimenov and Helleday, 2009). The human PCNA monomer is a 30- to 36-kDa protein consisting of 261 amino acid residues (Almendral et al., 1987). Functional PCNA is a homotrimer forming a ring structure, in which three monomers are joined together in an antiparallel head-to-tail interaction (Kelman and O'Donnell, 1995; Gulbis et al., 1996; Naryzhny, 2008). To execute its function, PCNA needs to be loaded to DNA by replication factor C (RFC) (Waga and Stillman, 1998) and interacts with numerous protein partners, including DNA polymerases δ and ε for DNA replication; DNMT1, HDAC1, and p300 for chromatin assembly and gene regulation; DNA mismatch repair protein Msh3/Msh6 for DNA repair; p21CIP1/WAF1 for cell cycle control; and ESCO1/2 for sister-chromatid cohesion (Maga and Hubscher, 2003; Stoimenov and Helleday, 2009).
PCNA is synthesized in all stages of the cell cycle with a half-life of approximately 20 h (Bravo and Macdonald-Bravo, 1987) and is elevated in early S phase to support cell cycle progression (Bravo and Macdonald-Bravo, 1987; Naryzhny, 2008). PCNA gene deregulation and post-translational modulation are hallmarks of malignant cells. Tumor cells, regardless of their origins, express higher levels of PCNA (Celis and Olsen, 1994; Kallakury et al., 1999; Kimos et al., 2004; Malkas et al., 2006; Miyamoto et al., 2006; Naryzhny and Lee, 2007; Eltz et al., 2008; Naryzhny, 2008; Stuart-Harris et al., 2008; Zhong et al., 2008; Stoimenov and Helleday, 2009). Expression levels of PCNA correlate positively with other pathological indices in prostate cancer (Mulligan et al., 1997) and can serve as an independent prognosis marker (Miyamoto et al., 2006). Overexpression of PCNA is also a reliable biomarker for other tumor types (Kimos et al., 2004; Cappello et al., 2006; Stuart-Harris et al., 2008). These findings suggest that PCNA could be a valuable target for cancer therapy. In the present study, we have identified a series of novel compounds that directly bind to PCNA trimers, promote formation of stable PCNA trimers, reduce PCNA association with chromatin, and inhibit the growth of tumor cells of a variety of tissue origins.
Materials and Methods
Reagents.
Compounds were provided by the University of Cincinnati Drug Discovery Center (Cincinnati, OH) or purchased from Chembridge Co (San Diego, CA), ChemDiv (San Diego, CA), or Sigma-Aldrich (St Louis, MO). The recombinant His-PCNA (>95% pure) and antibody against Hus1 were purchased from Abcam (Cambridge, MA). Antibody against α-tubulin, PCNA siRNA, and scrambled siRNA control were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Lipofectamine 2000 transfection reagent and Alamar Blue were purchased from Invitrogen (Carlsbad, CA). Antibody against β-actin, propidium iodide (PI), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), concanavalin A (Con A) and protease inhibitor cocktail were purchased from Sigma-Aldrich. Antibodies against PCNA (PC10) and histone 1 and BrdU cell proliferation assay kit were purchased from Cell Signaling Technology (Danvers, MA). The enhanced chemiluminescence Western Blotting Detection System was purchased from Millipore (Billerica, MA).
Cells and Culture.
The following cell lines were used in the study:
Human cancer: LNCaP, 22Rv1, DU-145, LAPC-4, and PC-3 prostate cancer cells, MCF-7 and T47D breast cancer cells, A375 and MDA-MB435 melanoma cells.
Mouse cancer: TRAMP-C2RE3 prostate cancer cells, B16 and K1735 melanoma cells, UV2237 fibrosarcoma cells, and CT-26 colon cancer cells.
Primary human: umbilical vein endothelial cells (PromoCell, Heidelberg, Germany), bone marrow mesenchymal stem cells (PromoCell), mammary epithelial cells (Lonza Walkersville, Inc., Walkersville, MD), and primary human prostate epithelial cells (HuPrEC; Lonza Walkersville, Inc.).
Primary mouse: spleen lymphocytes (isolated in this laboratory) and bone marrow stromal cells (isolated in this laboratory).
Cells in exponential growth phase were harvested by treatment with a 0.25% trypsin and 0.02% EDTA solution, detached into RPMI 1640 medium and 10% fetal bovine serum (FBS), and resuspended in medium specific for different cells. Only suspensions of single cells with viability exceeding 95% were used.
Virtual Screening.
A three-dimensional representation of the University of Cincinnati Drug Discovery Center drug-like chemical library was screened/docked against a model of PCNA derived from an X-ray crystal structure of human PCNA (Kontopidis et al., 2005). The PCNA trimer structure (Protein Data Bank code 1VYJ) was prepared by adding missing atoms and minimizing energy of the all-atom model in explicit solvent (0.9% NaCl, pH 7.4) to remove steric clashes (Yasara Biosciences, Vienna, Austria) and verified using MolProbity (http://molprobity.biochem.duke.edu) (Hasinoff and Patel, 2009; Chen et al., 2010). The first round of screening involved individual docking of each compound structure into a rigid representation of PCNA using FRED (Openeye Scientific Software, Santa Fe, NM) under default settings. Top hits from 300,000 compounds were redocked, allowing the ligand to rotate freely within the binding site with Glide SP (Schrödinger, LLC, New York, NY). Finally, the top 2000 molecules were further docked using Glide XP (Schrödinger) at high-resolution settings as well as performing flexible-ligand, flexible-site (side chains) docking with Molegro (Molegro Bioinformatics, Aarhus, Denmark). The top 200 hits were selected for further evaluation in bioassays.
PCNA Binding Assay.
The his-PCNA or rabbit IgG (control) was labeled with a reactive dye NT-647 using N-hydroxy succinimide-ester chemistry using a kit (NanoTemper Technologies, Munich, Germany). PCNA-I1 at various concentrations was incubated with NT-647-labeled PCNA or IgG (100 nM) in a binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.01% Nonidet P-40 alternative). The binding of PCNA-I1 to the labeled proteins was measured by using the microscale thermophoresis technology in a Monolith NT.115 reader (NanoTemper) as detailed previously (Jerabek-Willemsen et al., 2011) using the Temperature Jump analysis. The dissociation constant (Kd) of the binding was calculated using nonlinear regression analysis (Prism 5; GraphPad Software Inc., La Jolla, CA).
Immunoblotting Analysis.
Cells were scraped into a lysis buffer (1% Triton X-100, 20 mM Tris-HCl, pH 8.0, 137 mM NaC1, 10% glycerol, 2 mM EDTA, 1 mM PMSF, and a protease inhibitor cocktail) and analyzed by immunoblotting (Zhang et al., 2002). The immunoreactive signals were revealed using the enhanced chemiluminescence methods and visualized in an IS4000MM Digital Imaging System (Carestream Health, Rochester, NY).
PCNA Trimer Stability Assay.
The assay was performed on both cell lysates (native PCNA source) and purified recombinant protein. Fifty micrograms of PC-3 cell lysate or 0.1 μg of His-PCNA was incubated with PCNA-Is or DMSO (0.1%, vehicle) in a reaction buffer (40 mM Tris-HCl, pH 7.5, 0.2 mg/ml bovine serum albumin, 10 mM MgCl2, and 10% glycerol). The reaction was stopped by the addition of the SDS-PAGE sample buffer without a reducing agent. The samples were resolved by SDS-PAGE without boiling and analyzed by immunoblotting.
Nuclear Fractionation and PCNA Chromatin Association Analysis.
Cells were lysed in buffer A (10 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, 1 mM PMSF, and protease inhibitors). Samples were pelleted by centrifugation (1500g, 2 min, 4°C) and the resulting supernatant fraction collected designated as the NP-40-extractable (NP-E) fraction. The pellet was washed in buffer B (10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM PMSF, and protease inhibitors), resuspended and digested in buffer C (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 5 mM MgCl2, 0.2 mM PMSF, and protease inhibitors) with 200 units/107 cells of DNase I for 30 min at 37°C. After centrifugation at 13,000g for 5 min at 4°C, the supernatant was collected as NP-40-resistant (NP-R) fraction.
Cell Growth Assay.
Cell growth was assessed by MTT staining as described previously (Dong et al., 1991). Growth inhibition (percentage) by the compounds was calculated using the formula: (1 − A570 of treated/A570 of control) × 100 and IC50 (the concentration that inhibited cell growth by 50%) determined. For evaluation of lymphocyte growth, the Alamar blue assay (Nakayama et al., 1997) was performed. Freshly prepared mouse spleen lymphocytes were stimulated for 72 h with 2 μg/ml concanavalin A. During the last 24 h, 20 μl/well of Alamar blue was added. The fluorescence intensity was measured at 530 nm excitation/580 nm emission in the FLUOstar Omega microplate reader (BMG Labtech, Cary, NC).
Cell Cycle Analysis.
PC-3 cells were plated onto 60-mm plates at 2 × 105 cells/plate, treated, detached by trypsinization, and resuspended in 70% ethanol in PBS on ice. The cells were resuspended in PI staining solution (PBS containing 50 μg/ml PI, 100 μg/ml RNase A, and 0.05% Triton X-100) and incubated for 45 min at 37°C, followed by washing with PBS and flow cytometry analysis in an Epics-XL-MCL system (Beckman Coulter, Fullerton, CA). The three fractions (G0/G1, G2/M, and S) were quantified by using the Synchronization Wizard of ModFit LT Flow Cytometry Cell-Cycle Analysis software (Verity Software House, Topsham, ME).
BrdU Cell Proliferation Assay.
PC-3 (2500/well) and LNCaP (5000/well) cells were seeded into 96-well plates. After an overnight incubation, the cells were treated with increasing concentrations of PCNA-I1 for 48 h. Eight hours before termination of the experiments, 10 μM BrdU was added to each well and the incorporation of BrdU into newly synthesized DNA was assessed by using an enzyme-linked Enzyme-linked immunosorbent assay (ELISA) kit following the manufacture's protocol. IC50 values were determined as described in “Cell growth inhibition assay” above.
Transfection of siRNA.
PC-3 cells were plated onto 60-mm plate at 2 × 105/plate in antibiotic-free medium and transfected with 200 pmol of PCNA-specific siRNA or control siRNA for 24 h using Lipofectamine 2000. The cells were then starved in SFM for 24 h, followed by stimulation with 5% FBS, and sampled at different times for analyses.
Statistical Analysis.
Data shown are the mean ± S.D. Differences between means were compared using the two-tailed Student's t test and were considered significantly different at the level of p < 0.05.
Results
Identification of Potential PCNA-Is.
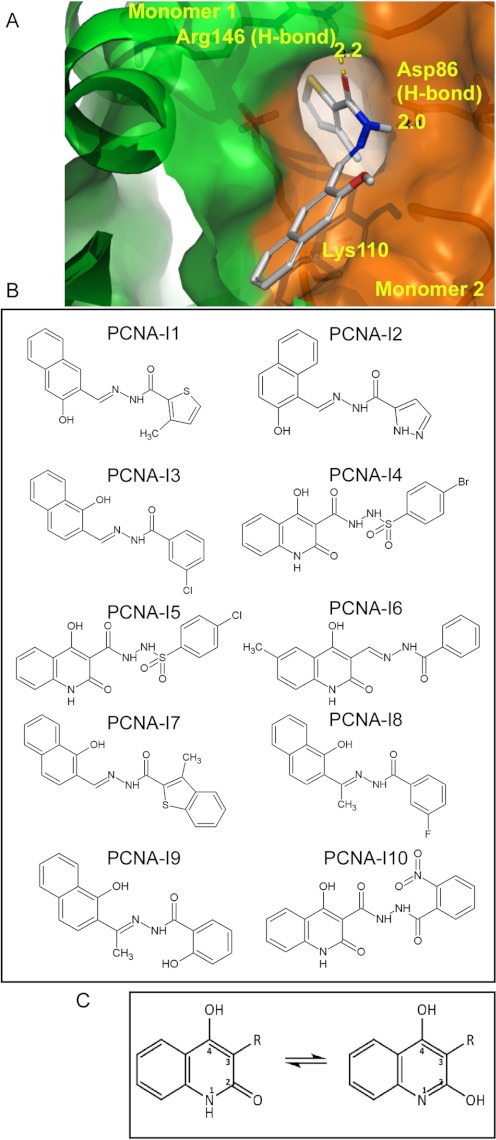
The in silico docking computation using Glide and Molegro was performed to identify compounds that potentially bind to the interfaces between PCNA monomers (Fig. 1A). Two hundred compounds with the highest combined-docking scores were tested for inhibitory effects on cell growth. Two structurally similar compounds (N′-[(1-hydroxy-2-naphthyl) methylene]-3-methyl-2-thiophenecarbo-hydrazide and N′-[(2-hydroxy-1-naphthyl) methylene]-1H-pyrazole-5-carbohydrazide, designated PCNA-I1 and -2, respectively) for which the IC50 was in the nanomolar (PCNA-I1) or nanomolar to low micromolar (PCNA-I2) range were identified (detailed below in Table 1). The docking analysis suggested that PCNA-I1 binds to Arg146 through an O-N hydrogen bond (H-bond, 2.2 Å distance) of one PCNA monomer and to Asp86 through a N-O H-bond (2.0 Å distance) of the adjacent monomer. A strong nonpolar interaction is also predicted between the lipophilic aroyl hydrazone of a naphthol on PCNA-I1 and carbons dominated by Lys110 of the adjacent monomer (Fig. 1A). To further explore the structure-activity relationships with the goal of improved intrinsic inhibitory activity and/or permeability, a structure similarity search was performed with PCNA-I1 as the template against the ZINC database (http://zinc.docking.org/). Eight additional compounds, PCNA-I3 through PCNA-I10 (Fig. 1B), were identified and selected for further characterization detailed below.
Fig. 1.
Chemical structures of PCNA-Is. A, an in silico docking image of the binding of PCNA-I1 to PCNA at the interface of two monomers. B, structures of PCNA-I1 through PCNA-I10. C, the predicted pharmacophore of PCNA-Is.
TABLE 1.
IC50 values of PCNA-I1 and PCNA-I2 (μM)
Cells were plated into 96-well plates at 1000 to 5000 cells/well. After an overnight incubation, the cells were treated for 4 days with various concentrations (up to 10 μM) of PCNA-I1 or PCNA-I1. The live cells were stained with MTT and counted as described under Materials and Methods. IC50 values are derived from growth inhibition curves. Data shown for tumor cells are mean of five to seven experiments and for primary cells are mean of three experiments. The means of IC50 for tumor cells and normal cells are 0.17 ± 0.07 and 1.60 ± 0.36, respectively (P < 0.0001).
| Species, Tissue Origin, and Cell line | PCNA-I1 | PCNA-I2 |
|---|---|---|
| μM | ||
| Tumor cell lines | ||
| Human | ||
| Breast | ||
| MCF-7 | 0.15 | 1.01 |
| T47D | 0.15 | N.D. |
| Prostate | ||
| PC-3 | 0.24 | 0.97 |
| DU145 | 0.16 | 1.19 |
| 22Rv1 | 0.18 | N.D. |
| LAPC-4 | 0.30 | 1.79 |
| LNCaP | 0.14 | 0.56 |
| Melanoma | ||
| A375 | 0.16 | 3.75 |
| MDA-MB435 | 0.29 | N.D. |
| Mouse | ||
| Prostate | ||
| TRAMP-C2RE3 | 0.20 | N.D. |
| Melanoma | ||
| B16 | 0.14 | N.D. |
| K1735 | 0.05 | N.D. |
| Fibrosarcoma | ||
| UV2237 | 0.25 | N.D. |
| Colon cancer | ||
| CT26-P | 0.13 | N.D. |
| CT26-R100 | 0.08 | N.D. |
| CT26-R500 | 0.07 | N.D. |
| Mean ± SD | 0.17 ± 0.07 | N.D. |
| Normal cells | ||
| Human | ||
| Blood vessel | ||
| HUVEC | 1.54 | N.D. |
| Bone marrow | ||
| Mesenchymal | 0.99 | N.D. |
| Stem cells | ||
| Breast | ||
| Epithelial cells | 1.67 | N.D. |
| Prostate | ||
| Epithelial cells | 2.00 | N.D. |
| Mouse | ||
| Bone marrow | ||
| Stroma cells | 1.90 | N.D. |
| Spleen | ||
| Lymphocytes | 1.50 | N.D. |
| Mean ± SD | 1.60 ± 0.36 | N.D. |
N.D., not determined.
The Binding of PCNA-Is to PCNA.
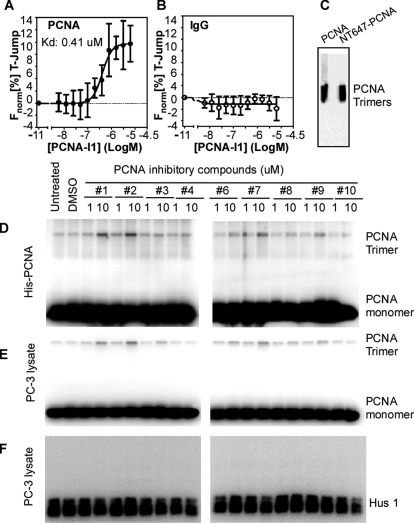
To establish that PCNA-Is bind directly to PCNA and determine the affinity of the binding, microscale thermophoresis technology analysis was performed. PCNA-I1 bound to NT-647-labeled recombinant PCNA with a Kd of 0.41 ± 0.17 μM (Fig. 2A) but not rabbit IgG (Fig. 2B), indicating that PCNA-I1 selectively bound to PCNA. The native gel electrophoresis analysis, followed by immunoblotting, revealed that NT-647-labeld PCNA was in a trimer form (Fig. 2C), indicating that PCNA-I1 bound directly to PCNA trimers. The binding of PCNA-I1 to PCNA was validated by using surface plasmon resonance (SPR) technology. The kinetic binding constants were analyzed using the BIAevaluation software (BIAcore AB, Uppsala, Sweden) in the iterative model of the best fit for the interaction parameters in a Langmuir 1:1 binding, which resulted in Kd values of 0.14 μM for PCNA-I1 (Supplemental Fig. 1A). Therefore, PCNA-I1 bound to PCNA with a Kd of 0.14 to 0.41 μM. In addition, the SPR analysis showed that PCNA-I3 bound to PCNA with a Kd of 0.17 μM (Supplemental Fig. 1B).
Fig. 2.
Biochemical analyses of PCNA-I binding to PCNA. A, PCNA-I1 binds to purified human PCNA protein with a Kd of ∼407 ± 168 nM (−6.390 ± 0.174 Log M). B, PCNA-I1 does not bind to the negative control purified rabbit IgG protein. Data in A and B are baseline-subtracted and analyzed as described under Materials and Methods. ● (A) and ○ (B) represent the mean ± S.E.M. measurements from three separate single point experiments. C, NT-647-PCNA was resolved in a native gel electrophoresis and analyzed by immunoblotting. D and E, elevation of PCNA trimer in recombinant PCNA (D) and cell lysate (E) treated with PCNA-Is. The recombinant His-PCNA (0.1 μg/reaction) or PC-3 cell lysates (50 μg/reaction) was incubated at room temperature for 3 h with dimethyl sulfoxide (DMSO; 0.1%, a vehicle control) or PCNA-Is (1 or 10 μM). The reactions were stopped by addition of SDS-PAGE sample buffer, resolved by SDS-PAGE without the sample boiling pretreatment, and analyzed by immunoblotting using the PC10 PCNA antibody. F, a duplicate set of samples of PCNA-Is-treated PC-3 cell lysate was analyzed by immunoblotting using an antibody to Hus1.
As an approach to help to define potential effects of the binding of PCNA-Is on stability of PCNA trimer structure, we used SDS-PAGE separation analysis of PCNA pretreated with PCNA-Is and analyzed by immunoblotting. The His-tagged recombinant PCNA was incubated with dimethyl sulfoxide (vehicle) or PCNA-Is (1 or 10 μM). PCNA-I5 has a structure almost identical to that of PCNA-I4 and hence was not included. As shown in Fig. 2D, approximately 2 to 5% of PCNA was present as the trimer form under the experimental conditions. After incubation with PCNA-Is, PCNA-I1 and PCNA-I2 in particular, the amount of PCNA in the trimer form was significantly elevated. Likewise, the treatment with PCNA-Is enhanced the amount of PCNA trimers by natural PCNA present in a lysate of PC-3 cells (Fig. 2E). To determine specificity of this effect of PCNA-Is, the trimer stability assay was performed to examine trimer formation by the 9-1-1 proteins as a control. The 9-1-1 protein complex (approximately 110 kDa) is another member of the clamp family proteins and a heterotrimer formed by Rad9, Rad1, and Hus1. The 9-1-1 protein complex also encircles DNA and is involved in DNA repair (Song et al., 2007, 2009; Park et al., 2009; Xu et al., 2009). PC-3 cell lysate was subjected to the treatment with 1 or 10 μM PCNA-Is and analyzed by immunoblotting using an antibody against Hus1. As shown in Fig. 2F, a high level of Hus1 was detected in the PC-3 cell lysate. However, the treatment with PCNA-Is did not detectably induce 9-1-1 trimer formation. Taken together, these data indicated that the PCNA-Is bind directly and selectively to PCNA trimers, which will potentially stabilize the trimer structure of PCNA.
Effects of PCNA-I1 on the Association of PCNA with Chromatin.
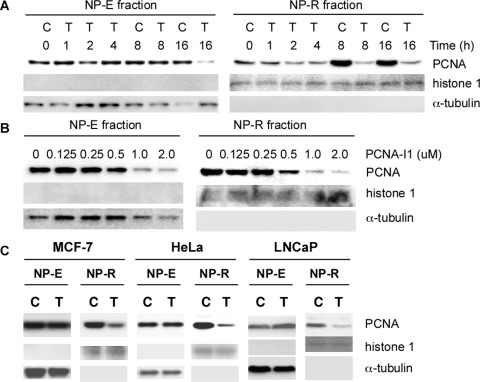
Association with chromatin is a prerequisite for PCNA to execute its functions. We therefore determined whether treatment of cells with PCNA-I1 alters the association of PCNA with chromatin. PC-3 cells were treated for various times with 1 μM PCNA-I1 and lysed in a buffer containing 0.5% NP-40 (Savio et al., 1996). PCNA in the NP-E and NP-R fractions was analyzed by immunoblotting to identify the free and chromatin-associated PCNA with α-tubulin (free form) and histone 1 (chromatin protein) as loading controls, respectively. As shown in Fig. 3A, PCNA in NP-E fraction (the free-form PCNA), was not significantly altered in cells treated by PCNA-I1 for up to 8 h but reduced in cells treated for 16 h. In contrast, the level of PCNA in NP-R fraction (the chromatin-associated PCNA) was reduced in 1 to 2 h in cells treated with PCNA-I1. A more significant reduction of the chromatin-associated PCNA was observed in cells treated with PCNA-I1 for 8 h. Therefore, the reduction of the chromatin-associated PCNA occurred much earlier than did the free-form PCNA. We next analyzed this effect of PCNA-I1 in cells treated for 8 h with increasing concentrations of PCNA-I1. Data in Fig. 3B show that the effects of PCNA-I1 on reduction of chromatin-associated PCNA were dose-dependent and could be observed in cells treated with the compounds at concentrations of 0.5 μM or higher. The same treatment did not significantly alter the level of free-form PCNA. The similar reduction of chromatin-associated PCNA by PCNA-I1 was also observed in other cell lines examined, including LNCaP, HeLa, and A375 cells (Fig. 3C).
Fig. 3.
Effects of PCNA-I on PCNA association with chromatin. PC-3 cells were treated with 1 μM PCNA-I1 for various times (A) or for 8 h with various concentrations of PCNA-I1 (B). The NP-E and NP-R fractions of protein were analyzed by immunoblotting using PC10 PCNA antibody or using an antibody to histone 1 (chromatin-associated protein control) or α-tubulin (NP-40 extractable free form PCNA control). C, MCF-7, HeLa, or LNCaP cells were treated for 8 h with 1 μM PCNA-I1. The NP-E and NP-R fractions were analyzed as described for PC-3 cells in A and B. Data shown are from one experiment that is representative of three.
Inhibitory Effects of PCNA-Is on Cell Growth.
The growth-inhibitory effects of PCNA-I1 and PCNA-I2 were tested on a panel of human and mouse tumor cell lines, as well as primary cells and immortalized cells, of various tissue origins. As summarized in Table 1, PCNA-I1 inhibited growth of all tumor cells, regardless of tissue origins, with IC50 values at nanomolar levels. CT26-R100 and CT26-R500 cells (Killion et al., 1993), which overexpress P-glycoprotein and exhibit the multidrug resistance phenotype, were more sensitive than their parental CT26 cells to PCNA-I1 (Table 1). PCNA-I2 was 3 to 5 times less potent than PCNA-I1 in suppressing growth of most cell lines examined (Table 1). PCNA-I1 also inhibited growth of primary cultures of bone marrow mesenchymal stem cells, endothelial cells, lymphocytes, mammary epithelial cells, and prostate epithelial cells (Table 1). However, the potency of PCNA-I1 on growth of all normal cells was significantly lower than that on tumor cells. As revealed in Table 1, the IC50 of PCNA-I1 on normal cells was approximately 9 times higher than that for tumor cells (p < 0.001). Next, we further determined effects of other PCNA-Is (PCNA-I3–PCNA-I10, except for PCNA-I5) on growth of PC-3 and LNCaP cells (Table 2). It was noteworthy that, with the exception of PCNA-I3, all compounds identified through the structural similarity search were less potent than PCNA-I1 and PCNA-I2 in suppressing tumor cell growth (Table 2). It is noteworthy that the potencies of growth inhibition by the compounds correlated closely with those of their effects on PCNA trimer stability (Fig. 2, D and E).
TABLE 2.
IC50 values of PCNA-Is
The growth-inhibitory effects of the compounds were determined by the MTT assay, and IC50 values were derived from growth-inhibitory curves as detailed in the legend to Table 1. Data shown are the mean of two independent experiments.
| PCNA-Is | PC-3 | LNCaP |
|---|---|---|
| μM | ||
| PCNA-I3 | 0.44 | 0.26 |
| PCNA-I4 | 3.00 | 0.68 |
| PCNA-I6 | >10.0 | >10.0 |
| PCNA-I7 | 3.60 | 2.20 |
| PCNA-I8 | 1.00 | 0.68 |
| PCNA-I9 | 2.20 | 0.71 |
| PCNA-I10 | >10.0 | 2.20 |
Effects of PCNA-Is on Cell-Cycle Distribution.
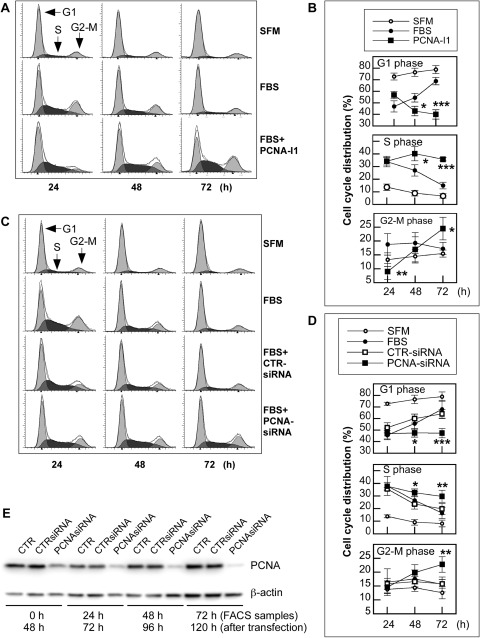
Given the pivotal roles of PCNA in cell-cycle regulation, the effects of the compounds, PCNA-I1 in particular, on cell-cycle progression were determined. PC-3 cells were starved in serum-free medium (SFM) for 24 h, which partially synchronized the cells in G1 phase, then stimulated with 5% FBS in the absence or presence of 1 μM PCNA-I1. As expected, the serum starvation led to accumulation of cells in G1 phase at all times examined. The G1 arrest was rescued upon serum stimulation, leading to a significant reduction of cells in G1 phase and an increase of cells in S and G2/M phases in the first 24 h (Fig. 4A and 4B). The cell cycle progressed smoothly over the next 48 h. By 72 h, the distribution of cells treated with serum in all phases of the cell cycle was similar to that in the culture without serum stimulation (Fig. 4, A and B). A significantly different cell distribution profile was observed in cultures treated with PCNA-I1. PCNA-I1 partially attenuated the serum-stimulated G1 decrease at 24 h but led to significant G1 reductions at 48 and 72 h. On the other hand, the number of cells in S and G2/M phases gradually increased over the 72 h, resulting in an accumulation of cells in S and G2/M phase of the cell cycle (Fig. 4, A and B). A similar S and G2/M arrest was also observed in cells treated with other PCNA-Is (Supplemental Fig. 2). Therefore, the cell cycle distribution analysis indicated that treatment of PC-3 cells with PCNA-Is led to G1 phase accumulation during the first 24 h and an S and G2/M phase arrest by 72 h.
Fig. 4.
Effects of PCNA-Is on cell cycle progression. A and B, PC-3 cells were plated onto 60-mm plates at 2 × 105/plate. After an overnight incubation, the cells were starved for 24 h in SFM. The starved cells were then cultured in fresh SFM or stimulated in the medium supplemented with 5% FBS and sampled 24, 48, or 72 h later for flow cytometry analysis. C and D, PC-3 cells were plated onto 60-mm plates at 2 × 105/plate in antibiotics-free medium. After an overnight incubation, the cells were transfected for 24 h with PCNA-specific siRNA or a scrambled siRNA control using Lipofectamine 2000. The cells were then starved for 24 h in SFM, followed by stimulation with fresh medium supplemented with 5% FBS. Cells were sampled 24, 48, or 72 h later for flow cytometry analysis. The profile files (A and C) are from one experiment that is representative of five. Data in B and D are the mean ± S.D. of five experiments. E, cells were transfected as described in C and D and sampled for Western blotting immediately after the 24-h starvation (0 h) or 24 to 72 h after FBS stimulation. *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTR, control; FACS, fluorescence-activated cell sorting.
Effects of knocking down endogenous PCNA protein on cell cycle distribution were determined to validate S and G2/M arrest induced by PCNA-Is. As shown in Fig. 4, C and D, the transfection of PC-3 cells with PCNA-specific siRNA, but not the control scrambled siRNA, led to an accumulation of cells in the S and G2/M phases of the cell cycle over the 72-h incubation, mimicking the effects of the treatment with PCNA-Is. However, the partial G1 phase accumulation induced by PCNA-Is was not observed, possibly because of the timing of the sampling (72 h after the transfection). The knocking down of PCNA protein by the siRNA was confirmed in a parallel set of samples and shown in Fig. 4E. The PCNA protein level in PCNA-siRNA–transfected cells was reduced by 50% at 0 h, 50% at 24 h (the first time point of sampling for flow cytometry analysis and 72 h after the transfection), 60% at 48 h, and 80% at 72 h (Fig. 4E). These data strongly suggest that the S and G2/M arrest induced by the PCNA-Is could be caused by their interference with PCNA function.
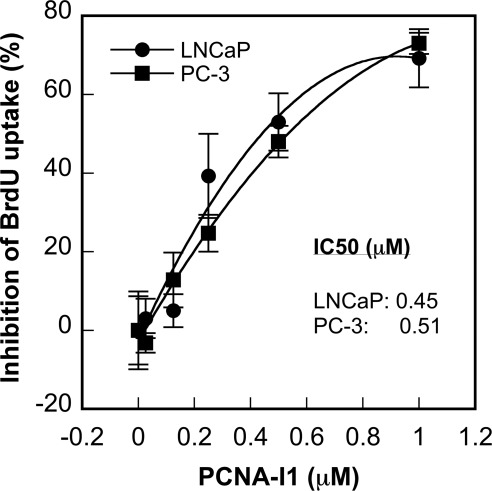
To further validate effects of PCNA-I1 on DNA replication, PC-3 and LNCP cells were treated with increasing concentrations of PCNA-I1 for 48 h and uptake of BrdU into cellular DNA was measured. Data in Fig. 5 show that incorporation of BrdU in both cell lines was dose dependently inhibited by the treatment of PCNA-I1 with IC50 values of 0.51 and 0.45 μM in PC-3 and LNCaP cells, respectively, which is consistent with their IC50 values in the growth inhibition assay (Table 1).
Fig. 5.
Effects of PCNA-I1 on DNA replication in cells. PC-3 and LNCaP cells were plated into 96-well plates. After an overnight incubation, the cells were treated with PCNA-I1 for 48 h, and 10 μM BrdU were added to each well during the last 8 h. The BrdU incorporated into DNA was detected by enzyme-linked immunosorbent assay. The data shown are mean ± S.D. of three independent experiments.
Discussion
PCNA is required for DNA synthesis and repair as well as numerous other pivotal cellular activities. It is one of very few proteins universally overexpressed in all types of tumors and is therefore a potentially valuable target for cancer therapy. The present study, through a combination of the computational virtual docking screen, ligand-binding assays, biochemical assays, and bioassays on cell growth, has identified a series of small molecule compounds that directly bind PCNA trimers, reduce chromatin-associated PCNA in cells, inhibit cell growth through induction of S and G2/M arrest, and attenuate DNA replication in cells.
Structural analysis reveals that PCNA-I3 is most similar to PCNA-I1 because thiophenes are generally phenyl-like in character, differing primarily in the shift of the hydroxyl functions from the 1 to 3 position. Indeed, SPR analysis confirmed that the two compounds bound to PCNA with very similar Kd values (0.14 and 0.17 μM, respectively). PCNA-I2 reverses the acyl hydrazone and hydroxyl positions from PCNA-I3, leaving the naphthyl ring more perpendicular to the acyl hydrazone chain, resulting in an approximate 3- to 5-fold loss in activity. PCNA-I2 also differs in that it has a pyrazole ring; although relatively nonbasic, it is smaller and capable of being an H-bond donor relative to the aryl groups of PCNA-I1 and -3. The weaker cell growth-inhibitory activity of PCNA-I2 is possibly due to its lower cell permeability, which is predicted in a computer simulation analysis using QikProp software (Schrödinger LLC). PCNA-I8 is nearly identical to PCNA-I3 except for the methyl group at the imine carbon of the hydrazone and the halogen substitution. This methyl group may induce greater deplanarization of the overall structure and a consequent 2-fold loss of activity. Comparison of PCNA-I9 to PCNA-I8 suggests a loss of activity by addition of H-bonding functions in the aroyl moiety, a trend consistent with the comparison of PCNA-I7 to PCNA-I1, with the latter pair suggesting some size restrictions in this moiety also. The leap from the naphthol scaffolds in PCNA-I1, -2, -3, -7, -8, and -9 to the 4-hydroxy-2-quinolone scaffolds in PCNA-I4, -6, and -10 is suggested by the good potency observed with hydroxyl positions in PCNA-I1 and PCNA-I3 on either side of the hydrazone chain. Overall, the dominant pharmacophore is a lipophilic aroyl hydrazone of a naphthol (Fig. 1C). This template will serve as the basis for further analog selection, wherein substitutions of the naphthyl and aroyl moieties will be further explored.
The majority of PCNA is present as the trimer form in the nucleoplasm, but only the chromatin-associated PCNA trimers are functional. Loading of PCNA trimers to DNA requires the RFC complex (Waga and Stillman, 1998). Extensive interactions with RFC open the PCNA ring, and the engagement of the RFC-PCNA with the primer-template junctions of DNA results in ATP hydrolysis, closing of the ring, and release of the sliding clamp on DNA (Bowman et al., 2004). Because the PCNA-Is directly bind to PCNA trimers at the junction of the interfaces (by the docking analysis), which may stabilize the trimer structure of PCNA (implied by the SDS-PAGE analysis data), it is possible that the PCNA-I-complexed PCNA becomes relatively insensitive to the RFC loading, resulting in the reduction of chromatin-associated PCNA. On the other hand, it has been shown that Lys110 is required for chromatin association and formation of double homotrimer of PCNA through its interaction with Arg5 on the other homotrimer (Naryzhny et al., 2005; Kim and Lee, 2008). The docking analysis suggested that PCNA-Is interact with Lys110, which would potentially interfere with the formation of the double homotrimers or recognition of DNA replication and repair foci. It should be mentioned that although the effects of PCNA-Is on PCNA trimer stability correlate with their inhibitory potencies on cell growth, they might not directly correlate with the Kd values of the compound to PCNA.
In addition to the reduction of the chromatin-associated PCNA, treatment with PCNA-I1, particularly after a longer duration, also leads to a significant down-regulation of total PCNA. Because treatment with PCNA-I1 does not lead to acceleration of PCNA degradation (data not shown) and PCNA is synthesized mainly in early S phase to support cell cycle progression (Bravo and Celis, 1980; Bravo and Macdonald-Bravo, 1987; Naryzhny, 2008), it is very possible that this down-regulation of total PCNA by the compounds is due indirectly to the blockade of cell-cycle progression.
PCNA, through its interaction with many cell-cycle regulatory proteins, such as cyclin-dependent kinases, cyclins, p21, Cdc25 (Luo et al., 1995; Kawabe et al., 2002; Helt et al., 2004), plays a crucial role in cell-cycle regulation. Therefore, it was not surprising that treatment with the PCNA-Is or PCNA knock-down led to cell cycle arrest. A minor differential effect on cell cycle distribution was observed in the first 24 h in cells treated with PCNA-Is and knocking down PCNA expression. Whereas the PCNA-Is induced a partial G1 arrest during the first 24 h, this effect was not observed in experiments knocking down PCNA expression. Given that flow cytometric analysis on the siRNA-transfected cells was started at 48 h after the transfection when PCNA in the cells was already significantly reduced, it is very likely that a transient G1 arrest occurred in the first 48 h. Indeed, it has been reported by others that transfection of antisense oligodeoxynucleotide of PCNA inhibited the progression of cells through G1 to S phase (Yang et al., 2002).
One of the major dose-limiting factors for successful chemotherapy of cancer is toxicity of the therapeutic drugs. In contrast to many chemotherapeutic drugs used in clinics, the PCNA-Is show more profound inhibitory effects on tumor cells versus normal cells, which would provide a significant therapeutic window. This differential sensitivity was reported previously in studies knocking down PCNA expression by antisense oligonucleotides (Sakakura et al., 1994). The differential sensitivity is probably due to higher demands of PCNA in tumor cells. In addition to their constantly growing and requiring more PCNA, tumor cells are genetically more unstable because of accumulation of gene mutations, and they require more functional PCNA for DNA repair to survive and are therefore more vulnerable to a PCNA-targeting agent.
The PCNA-Is identified in our studies (not in the pharmacophore) show a structural similarity of iron-binding site with some iron chelators, suggesting that they might be able to bind to iron (Richardson and Milnes, 1997; Merlot et al., 2010). Iron is required for many cellular processes, and iron chelators have been shown to inhibit tumor cell replication and to induce apoptosis (Richardson and Milnes, 1997; Merlot et al., 2010). However, the findings that the inhibitory effects of PCNA-Is on cell growth and cell cycle distribution are recapitulated in cells by knocking down PCNA using siRNA and that the Kd of PCNA-I1 to PCNA correlates with its IC50 in tumor cell growth inhibition strongly suggest these PCNA-binding compounds inhibit cell growth through an attenuation of PCNA function. It is noteworthy that many chemotherapeutic compounds, such as doxorubicin (Hasinoff and Patel, 2009), cisplatin (Baliga et al., 1998) and curcumin (Jiao et al., 2009), as well as some typical iron chelators (Rao et al., 2009), are all capable of iron binding but were shown to inhibit cell growth and induce apoptosis through mechanisms independent of iron chelation.
The binding and stabilization of protein structures by small-molecule ligands is not an uncommon phenomenon. A group of compounds has been shown to bind to and stabilize transthyretin tetramers at nanomolar to micromolar Kd values (Alhamadsheh et al., 2011). The present studies have identified a series of novel compounds that bind to PCNA, promote stable PCNA trimer formation, reduce chromatin-associated PCNA, and inhibit tumor cell proliferation through induction of S and G2/M cell-cycle arrest. These unique compounds are the first in class and provide a novel pharmacological tool for studying PCNA functions and show promise as lead compounds in the development a novel PCNA-targeting cancer therapy.
Supplementary Material
Acknowledgments
We thank Dr. Ruben Papoian (Drug Discovery Center, University of Cincinnati College of Medicine, Cincinnati, OH) for evaluating this project, Mary Palascak for technical assistance in the flow cytometry analysis, and Dr. Robert Franco (Department of Medicine, University of Cincinnati College of Medicine, Cincinnati, OH) for critical reading of the manuscript.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grants R01-CA131137-01A1, R01-CA121289]; and by Millennium Scholar Funds from the University of Cincinnati Cancer Institute.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- siRNA
- small interfering RNA
- PI
- propidium iodide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- FBS
- fetal bovine serum
- PCNA-I
- PCNA inhibitor
- PMSF
- phenylmethylsulfonyl fluoride
- PAGE
- polyacrylamide gel electrophoresis
- NP-E
- NP-40-extractable
- NP-R
- NP-40-resistant
- BrdU
- bromodeoxyuridine
- PCNA-I1
- N′-[(1-hydroxy-2-naphthyl) methylene]-3-methyl-2-thiophenecarbo-hydrazide
- PCNA-I2
- N′-[(2-hydroxy-1-naphthyl) methylene]-1H-pyrazole-5-carbohydrazide)
- SPR
- surface plasmon resonance
- SFM
- serum-free medium.
Authorship Contributions
Participated in research design: Wortman, Lu, and Dong.
Conducted experiments: Tan, Wortman, Dillehay, Evelyn, Smith.
Contributed new reagents or analytic tools: Wortman, Seibel, and Dong.
Performed data analysis: Wortman, Seibel, Evelyn, Smith, Malkas, Zheng, and Dong.
Wrote or contributed to the writing of the manuscript: Wortman, Seibel, Malkas, Zheng, Lu, and Dong.
References
- Alhamadsheh MM, Connelly S, Cho A, Reixach N, Powers ET, Pan DW, Wilson IA, Kelly JW, Graef IA. (2011) Potent kinetic stabilizers that prevent transthyretin-mediated cardiomyocyte proteotoxicity. Sci Transl Med 3:97ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendral JM, Huebsch D, Blundell PA, Macdonald-Bravo H, Bravo R. (1987) Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins. Proc Natl Acad Sci USA 84:1575–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga R, Zhang Z, Baliga M, Ueda N, Shah SV. (1998) In vitro and in vivo evidence suggesting a role for iron in cisplatin-induced nephrotoxicity. Kidney Int 53:394–401 [DOI] [PubMed] [Google Scholar]
- Bowman GD, O'Donnell M, Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429:724–730 [DOI] [PubMed] [Google Scholar]
- Bravo R, Celis JE. (1980) A search for differential polypeptide synthesis throughout the cell cycle of HeLa cells. J Cell Biol 84:795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Macdonald-Bravo H. (1987) Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol 105:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello F, Ribbene A, Campanella C, Czarnecka AM, Anzalone R, Bucchieri F, Palma A, Zummo G. (2006) The value of immunohistochemical research on PCNA, p53 and heat shock proteins in prostate cancer management: a review. Eur J Histochem 50:25–34 [PubMed] [Google Scholar]
- Celis JE, Olsen E. (1994) A qualitative and quantitative protein database approach identifies individual and groups of functionally related proteins that are differentially regulated in simian virus 40 (SV40) transformed human keratinocytes: an overview of the functional changes associated with the transformed phenotype. Electrophoresis 15:309–344 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong ZY, Ward NE, Fan D, Gupta KP, O'Brian CA. (1991) In vitro model for intrinsic drug resistance: effects of protein kinase C activators on the chemosensitivity of cultured human colon cancer cells. Mol Pharmacol 39:563–569 [PubMed] [Google Scholar]
- Eltz S, Comperat E, Cussenot O, Rouprêt M. (2008) Molecular and histological markers in urothelial carcinomas of the upper urinary tract. BJU Int 102:532–535 [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297–306 [DOI] [PubMed] [Google Scholar]
- Hasinoff BB, Patel D. (2009) The iron chelator Dp44mT does not protect myocytes against doxorubicin. J Inorg Biochem 103:1093–1101 [DOI] [PubMed] [Google Scholar]
- Helt CE, Staversky RJ, Lee YJ, Bambara RA, Keng PC, O'Reilly MA. (2004) The Cdk and PCNA domains on p21Cip1 both function to inhibit G1/S progression during hyperoxia. Am J Physiol Lung Cell Mol Physiol 286:L506–L513 [DOI] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9:342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wilkinson J, 4th, Di X, Wang W, Hatcher H, Kock ND, D'Agostino R, Jr, Knovich MA, Torti FM, Torti SV. (2009) Curcumin, a cancer chemopreventive and chemotherapeutic agent, is a biologically active iron chelator. Blood 113:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallakury BV, Sheehan CE, Rhee SJ, Fisher HA, Kaufman RP, Jr, Rifkin MD, Ross JS. (1999) The prognostic significance of proliferation-associated nucleolar protein p120 expression in prostate adenocarcinoma: a comparison with cyclins A and B1, Ki-67, proliferating cell nuclear antigen, and p34cdc2. Cancer 85:1569–1576 [DOI] [PubMed] [Google Scholar]
- Kawabe T, Suganuma M, Ando T, Kimura M, Hori H, Okamoto T. (2002) Cdc25C interacts with PCNA at G2/M transition. Oncogene 21:1717–1726 [DOI] [PubMed] [Google Scholar]
- Kelman Z, O'Donnell M. (1995) Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res 23:3613–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killion JJ, Radinsky R, Dong Z, Fishbeck R, Whitworth P, Fidler IJ. (1993) The immunogenic properties of drug-resistant murine tumor cells do not correlate with expression of the MDR phenotype. Cancer Immunol Immunother 36:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Lee H. (2008) Lys-110 is essential for targeting PCNA to replication and repair foci, and the K110A mutant activates apoptosis. Biol Cell 100:675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimos MC, Wang S, Borkowski A, Yang GY, Yang CS, Perry K, Olaru A, Deacu E, Sterian A, Cottrell J, et al. (2004) Esophagin and proliferating cell nuclear antigen (PCNA) are biomarkers of human esophageal neoplastic progression. Int J Cancer 111:415–417 [DOI] [PubMed] [Google Scholar]
- Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, Fischer PM, Walkinshaw MD. (2005) Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc Natl Acad Sci USA 102:1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Hurwitz J, Massagué J. (1995) Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 375:159–161 [DOI] [PubMed] [Google Scholar]
- Maga G, Hubscher U. (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 116:3051–3060 [DOI] [PubMed] [Google Scholar]
- Malkas LH, Herbert BS, Abdel-Aziz W, Dobrolecki LE, Liu Y, Agarwal B, Hoelz D, Badve S, Schnaper L, Arnold RJ, et al. (2006) A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proc Natl Acad Sci USA 103:19472–19477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot AM, Pantarat N, Lovejoy DB, Kalinowski DS, Richardson DR. (2010) Membrane transport and intracellular sequestration of novel thiosemicarbazone chelators for the treatment of cancer. Mol Pharmacol 78:675–684 [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Ito K, Kurokawa K, Suzuki K, Suzuki K, Yamanaka H. (2006) Clinical validity of proliferating cell nuclear antigen as an objective marker for evaluating biologic features in patients with untreated prostate cancer. Int J Urol 13:767–772 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129:665–679 [DOI] [PubMed] [Google Scholar]
- Mulligan JM, Mai KT, Parks W, Gerridzen RG. (1997) Proliferating cell nuclear antigen (PCNA) and MIB 1: Markers of locally advanced and biologically aggressive prostate cancer. Can J Urol 4:422–425 [PubMed] [Google Scholar]
- Nakayama GR, Caton MC, Nova MP, Parandoosh Z. (1997) Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods 204:205–208 [DOI] [PubMed] [Google Scholar]
- Naryzhny SN. (2008) Proliferating cell nuclear antigen: a proteomics view. Cell Mol Life Sci 65:3789–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryzhny SN, Lee H. (2007) Characterization of proliferating cell nuclear antigen (PCNA) isoforms in normal and cancer cells: there is no cancer-associated form of PCNA. FEBS Lett 581:4917–4920 [DOI] [PubMed] [Google Scholar]
- Naryzhny SN, Zhao H, Lee H. (2005) Proliferating cell nuclear antigen (PCNA) may function as a double homotrimer complex in the mammalian cell. J Biol Chem 280:13888–13894 [DOI] [PubMed] [Google Scholar]
- Park MJ, Park JH, Hahm SH, Ko SI, Lee YR, Chung JH, Sohn SY, Cho Y, Kang LW, Han YS. (2009) Repair activities of human 8-oxoguanine DNA glycosylase are stimulated by the interaction with human checkpoint sensor Rad9-Rad1-Hus1 complex. DNA repair 8:1190–1200 [DOI] [PubMed] [Google Scholar]
- Rao VA, Klein SR, Agama KK, Toyoda E, Adachi N, Pommier Y, Shacter EB. (2009) The iron chelator Dp44mT causes DNA damage and selective inhibition of topoisomerase IIalpha in breast cancer cells. Cancer Res 69:948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DR, Milnes K. (1997) The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents II: the mechanism of action of ligands derived from salicylaldehyde benzoyl hydrazone and 2-hydroxy-1-naphthylaldehyde benzoyl hydrazone. Blood 89:3025–3038 [PubMed] [Google Scholar]
- Sakakura C, Hagiwara A, Tsujimoto H, Ozaki K, Sakakibara T, Oyama T, Ogaki M, Takahashi T. (1994) Inhibition of gastric cancer cell proliferation by antisense oligonucleotides targeting the messenger RNA encoding proliferating cell nuclear antigen. Br J Cancer 70:1060–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio M, Stivala LA, Scovassi AI, Bianchi L, Prosperi E. (1996) p21waf1/cip1 protein associates with the detergent-insoluble form of PCNA concomitantly with disassembly of PCNA at nucleotide excision repair sites. Oncogene 13:1591–1598 [PubMed] [Google Scholar]
- Song W, Levin DS, Varkey J, Post S, Bermudez VP, Hurwitz J, Tomkinson AE. (2007) A conserved physical and functional interaction between the cell cycle checkpoint clamp loader and DNA ligase I of eukaryotes. J Biol Chem 282:22721–22730 [DOI] [PubMed] [Google Scholar]
- Song W, Pascal JM, Ellenberger T, Tomkinson AE. (2009) The DNA binding domain of human DNA ligase I interacts with both nicked DNA and the DNA sliding clamps, PCNA and hRad9-hRad1-hHus1. DNA Repair (Amst) 8:912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoimenov I, Helleday T. (2009) PCNA on the crossroad of cancer. Biochemical Society transactions 37:605–613 [DOI] [PubMed] [Google Scholar]
- Stuart-Harris R, Caldas C, Pinder SE, Pharoah P. (2008) Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast 17:323–334 [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. (1998) The DNA replication fork in eukaryotic cells. Ann Rev Biochem 67:721–751 [DOI] [PubMed] [Google Scholar]
- Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. (2009) Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem 284:20457–20461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ye SD, Feng LG, Wang JS, Zhai W. (2002) [In vitro experiment of inhibiting cell proliferation on human esophageal squamous cell cancer T]. Tn cell line by proliferating cell nuclear antigen antisense oligodeoxynucleotide. Ai Zheng 21:276–280 [PubMed] [Google Scholar]
- Zhang F, Lu W, Dong Z. (2002) Tumor-infiltrating macrophages are involved in suppressing growth and metastasis of human prostate cancer cells by INF-beta gene therapy in nude mice. Clin Cancer Res 8:2942–2951 [PubMed] [Google Scholar]
- Zhong W, Peng J, He H, Wu D, Han Z, Bi X, Dai Q. (2008) Ki-67 and PCNA expression in prostate cancer and benign prostatic hyperplasia. Clin Invest Med 31:E8–E15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.