Abstract
Gram-negative bacteria contain a family of outer membrane transport proteins that function in the uptake of rare nutrients, such as iron and vitamin B12. These proteins are termed TonB-dependent because transport requires an interaction with the inner membrane protein TonB. Using a combination of site-directed spin labeling and chemical denaturation, we examined the site-specific unfolding of regions of the Escherichia coli vitamin B12 transporter, BtuB. The data indicate that a portion of the N-terminal region of the protein, which occupies the lumen of the BtuB barrel, denatures prior to the unfolding of the barrel and that the free energy of folding for the N-terminus is less than that typically seen for globular proteins. Moreover, the data indicate that the N-terminal domain does not unfold in a single event, but unfolds in a series of independent steps. The unfolding of the N-terminus is reversible and removal of denaturant restores the native fold of the protein. These data are consistent with proposed transport mechanisms that involve a transient rearrangement or unfolding of the protein N-terminus, and they provide evidence for a specific protein conformation that might be an intermediate accessed during transport.
Gram-negative bacteria acquire rare nutrients by the use of high-affinity transport proteins that are localized within the outer membrane. These transporters are termed TonB-dependent because they interact with the transperiplasmic inner-membrane protein TonB and apparently extract energy for transport from the inner membrane proton potential (1–6). Crystal structures for at least 12 unique TonB-dependent transporters have been obtained. They are structurally homologous and consist of two domains: a 22-stranded β-barrel and an N-terminal domain that is folded within the interior of the barrel. At the very N-terminus is an energy coupling segment termed the Ton box, which is required for transport and directly interacts with TonB (7–9).
Shown in Fig. 1 is an X-ray crystal structure of the apo form of BtuB, the Escherichia coli TonB-dependent vitamin B12 transporter. In addition to the apo form, crystal structures of BtuB have also been obtained in ligand bound and TonB-bound forms (9–11). In these and other TonB-dependent transporter structures, there is no obvious passage for substrate through the barrel. Moreover, in the ferrichrome transporter, FhuA, molecular dynamics simulations indicate that there is no water filled channel through which substrate might pass (12). If the barrel of these proteins remains intact during transport, significant conformational rearrangements in the N-terminus would be necessary to facilitate movement of substrate through the barrel.
Figure 1.
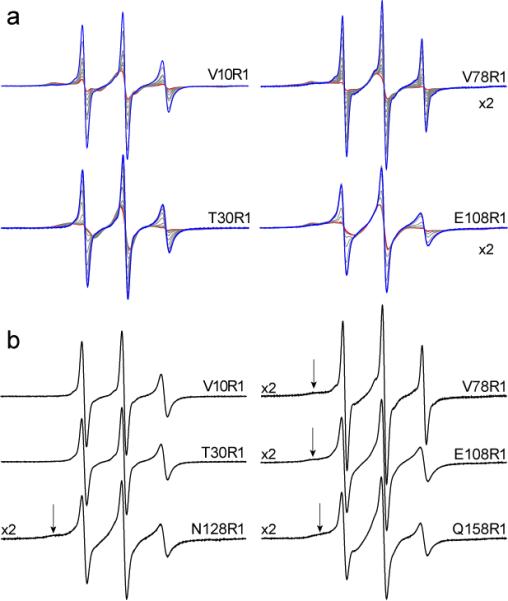
Model and EPR spectra from BtuB. In a), the high-resolution model of BtuB (PDB ID: 1NQE) where the N-terminal region is indicated in orange. The Cα carbons are indicated in green for three sites on the β-barrel that were labeled with the MTSL spin-labeled side chain R1 as well as sites in the N-terminus. In b) The N-terminal fold includes the first 136 residues of BtuB and is shown without the barrel. The N-terminal domain was spin labeled in the Ton box at position 10, as well sites on four successive β-strands at positions 30, 108, 128 and 78. The spin labeled side chain R1 is shown in c), and the EPR spectra resulting from these 8 sites is shown in d). With the exception of sites 78, 108 and 128, the EPR spectra for these sites for BtuB in POPC have been previously reported (22, 39). The dashed line indicates the position of the low-field hyperfine extrema, which is observed for sites in tertiary contact.
Transport mechanisms have been proposed that involve either the removal of the N-terminal region as an intact domain, or a series of unfolding events within the N-terminus (4, 13). As a result, the N-terminal domain is sometimes referred to as a cork, plug or hatch domain to indicate this proposed role. In the high-resolution crystal structures of BtuB, there are large numbers of waters at the interface between the N-terminal domain and the transporter β-barrel, which suggests that the N-terminal domain may be prone to conformational rearrangements or complete removal (14). In a steered molecular dynamics simulation, where the Ton box of BtuB is extracted from the periplasmic side of the β-barrel, the N-terminal core is observed to progressively unfold ultimately exposing a water-filled cavity sufficiently large for passage of the substrate (15). Denaturation experiments also suggest that the N-terminal domain and the β-barrel behave independently. For example, thermal denaturation indicates that the N-terminal domain and the β-barrel behave as two separate domains, and that the core might undergo structural rearrangements inside a static barrel (16). Similarly, in a reconstituted planar bilayer system it has been shown that a pore may be reversibly opened at moderate concentrations of urea in BtuB and related TonB-dependent transporters (17). Because these transporters retain the ability to bind ligand in the presence of urea, the data suggest that a partial and reversible unfolding of the N-terminal domain will open a channel within the transporter.
In the present study, we use site-directed spin labeling (SDSL1) to determine whether the N-terminal region of BtuB can be reversibly unfolded while retaining the protein β-barrel structure. This spin labeling approach was used previously to examine the site-specific denaturation of an extracellular loop and transmembrane β-strand in a related TonB-dependent transporter, FepA (18, 19). In the present case, our data indicate that the urea-induced unfolding of the BtuB N-terminus does not occur in a single cooperative step, but occurs in a series of steps. A region of the N-terminal domain, which includes the first β-strand within the core of BtuB, unfolds prior to the remainder of the core and the β-barrel. In this intermediate state, a channel is likely to be opened within the interior of BtuB that may be sufficiently large to accommodate substrate. The results demonstrate that the N-terminal domain of BtuB is capable of being reversibly unfolded and the data suggest a model for an intermediate structural or excited protein conformational state that might be accessed during transport.
Material and Methods
Mutagenesis, expression, purification, and spin labeling
BtuB mutants were engineered using the Agilent Technologies Quick Change Site Directed Mutagenesis Kit (Santa Clara, CA). The mutants were overexpressed in the E. coli strain RK5016 (metE) (20). Cells were grown in minimal “A” media containing 100 μg/mL ampicillin and supplemented with 0.24% w/v glucose, 150 μM thiamine, 3 mM MgSO4, 300 μM CaCl2, and 0.01% w/v of Met and Arg.
Outer membranes were prepared, solubilized and spin labeled with 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate (MTSL, Toronto Research Chemicals, North York, ON, Canada) to introduce the spin-labeled side chain, R1, following a previous protocol (21). The protein was then purified and reconstituted into POPC vesicles as described previously (22), and the proteoliposomes were concentrated using a Beckman Airfuge (Brea, CA).
Sample preparation and EPR measurements
Samples were titrated with urea to achieve final urea concentrations ranging from 0 to 12 M. Stock solutions of urea were dried in 0.5 ml tubes overnight using a vacuum concentrator (Savant SpeedVac, Asheville, NC), and accurate final concentrations of the denaturant were obtained by empirically determining the volume occupied by urea in solution. Reconstituted BtuB was added to the dried urea and the samples were allowed to sit for one hour at room temperature. The BtuB-urea samples then underwent four freeze-thaw cycles using liquid nitrogen and mixed after each cycle to achieve uniform urea accessibility throughout the proteoliposomes. To refold the protein from a urea denatured state, the sample was diluted 10 to 20 fold into the reconstitution buffer (10mM HEPES, 130 mM NaCl, pH 6.5), and concentrated by ultracentrifugation using a Beckman Airfuge.
EPR measurements were performed on 5 μL of sample loaded into glass capillaries with 0.6 mm i.d. × 0.84 mm o.d. (VitroCom, Mountain Lakes, NJ). These samples had protein concentrations of 100 to 200 μM. EPR spectra were obtained on a modified Varian E-line 102 Century series X-band spectrometer with a loop gap resonator (Molecular Specialties, Milwaukee, WI). LabVIEW software, provided by Dr. Christian Altenbach (University of California, Los Angeles, CA), was used for digital collection and analysis of data. All spectra were taken using 2 mW incident microwave power and 1 G p-p modulation amplitude. The scan range was 100 G, and the spectra were normalized to equivalent spin numbers by double integration of the first derivative EPR spectra.
Data analysis
The free energy of unfolding of a protein is known to vary linearly with the concentration of denaturant (23) as given by:
| Eq. 1 |
Where is the standard state free energy for unfolding in the absence of denaturant and C is the concentration of urea. The parameter m is related to the solvent accessible protein surface area exposed to solution upon denaturation. In the present case, we use EPR spectroscopy to follow the site-specific denaturation of the protein as described previously (18, 19). The EPR lineshapes of the protein attached nitroxide, R1 (Fig. 1c), are defined by the motion of the label on the ns time-scale, which is strongly dependent upon the local environment at the labeled site, including the tertiary contact of the label within the protein and the flexibility of the protein backbone to which the label is attached (24). In the analysis of the data obtained here, we assume that the EPR spectra reflect the protein, or protein domain, in one of two states: a folded form and an unfolded or urea denatured form. In this case, the EPR spectra at differing concentrations of urea should be a linear combination of the EPR spectra in the two states, and the peak-to-peak amplitude of the normalized first derivative EPR spectrum, A, will then be given by:
| Eq. 2 |
where fF represents the fraction of protein in the folded state, fU the fraction in the unfolded state, and Af and Au are the normalized amplitudes of the EPR spectra in the folded and urea denatured states, respectively. An inspection of Eq. 2 indicates that the peak-to-peak amplitude, A, is linearly related to the fraction of unfolded protein as given by: fU = (A−Af)/(Au−Af), and plots of A versus urea concentration should have an identical shape to those of the fraction of unfolded protein versus urea. Peak-to-peak amplitudes of EPR spectra were measured at each urea concentration for each mutant. The values for the normalized amplitudes as a function of concentration were then plotted and fit as described previously (25) using OriginPro (Origin Lab, Northhampton, MA) to an equation of the form:
| Eq. 3 |
Here, C0 is the value of C, the urea concentration, at which A=(Af+Au)/2, and dC is a measure of the width in C corresponding to the most significant change in A-values. The parameter m was calculated from m=RT/dC, where R and T have their usual meanings. Using this method of fitting, the free energy of unfolding in the standard state is given by: .
We also determined the parameters for urea denaturation using a complementary approach by finding the fractions of folded and unfolded protein from each experimental spectrum. In this case, we fit the experimental spectrum to a linear combination of the spectra that represent the folded and unfolded protein. The spectrum for the folded protein was taken as that in POPC in the absence of denaturant, and the spectrum of the unfolded protein was taken as that at the highest limiting concentrations of denaturant used. Using a least-squares approach, the best fit yielded the fraction of folded protein, fF, and the energy for unfolding at each intermediate urea concentration was then be calculated as . These free energies were plotted as a function of urea concentration, and the data fit with Eq. 1 to obtain values for m and .
Results
EPR spectra within BtuB indicate that the core or hatch region of the protein is well-folded
Shown in Fig. 1 are locations for eight sites in BtuB into which the spin labeled side chain, R1, was incorporated as well as their corresponding EPR spectra. Five of these labeled sites are located in the N-terminal core region of BtuB, and three are localized at sites in the β-barrel on strands 2 and 12. The labeled mutant proteins appear to be correctly folded. Each of the labeled mutants exhibited a behavior similar to that of wild-type protein upon purification and reconstitution into POPC bilayers. In addition, crystal structures were previously obtained for several of these spin labeled mutants and indicate that there is minimal perturbation of the protein structure upon incorporation of the label (26, 27).
The label at position 10 within the BtuB Ton box, V10R1, has been previously examined in some detail, and the crystal structure for this mutant indicates that the label lies at the base of a pocket on the periplasmic surface where it is in strong tertiary contact within the N-terminal domain of the protein (26). This is consistent with the EPR spectrum from V10R1, which indicates that the label is restricted in its motion (Fig. 1d). Positions 30, 108, 128 and 78 are located on each of four β-strands that form a sheet directed towards the extracellular surface (Fig. 1b). The EPR spectrum from T30R1 is the most mobile of the EPR spectra within the N-terminus, and it corresponds to a label having a correlation time of approximately 4 to 5 ns. This is consistent with its position, which places the label at a site of aqueous exposure. The EPR spectra at positions 108, 128 and 78 indicate that these nitroxides are restricted in their motion due to tertiary contact. Interestingly, when the spectra from these four β-strands are compared, the labels become slightly less mobile, as judged by the normalized amplitudes, when the spin label is positioned further into the protein towards the extracellular surface. These spectra indicate that these labels are folded into the protein interior, and that any unfolded population of protein N-terminal domain (which would have shown up as a mobile component in the EPR spectrum) is small.
Three labels were placed into the β-barrel of BtuB at sites 151 and 158, on strand 2 and site 371 on strand 12. Site 151 faces the interior of the protein, and it yields an EPR spectrum near the rigid motional limit, indicating that the label is present in a sterically restrained environment. The other two sites are located at the hydrocarbon surface of the β-barrel. These spectra are complex, but are consistent with R1 at sites facing the hydrocarbon surface of a β-barrel. The origins of lineshapes at these hydrocarbon-facing sites of β-barrel proteins have been described in detail elsewhere (27, 28).
EPR spectra reveal residual protein structure at the highest urea concentrations tested
Shown in Fig. 2a are normalized EPR spectra taken at increasing urea concentrations for spin labels at positions 10, 30, 78 and 108. The changes in these EPR spectra after mixing with urea were complete within an hour, and spectra that were taken a day or more after the addition of denaturant exhibited no further change. These spectra are typical of those seen at other sites, and they exhibit a dramatic increase in normalized amplitudes and a narrowing of the EPR lineshapes upon urea addition. As indicated previously, the changes in lineshape with increasing denaturant concentration are due to a loss of secondary and tertiary structure and an increase in backbone dynamics (18, 19).
Figure 2.
a) EPR spectra obtained from R1 at four sites in BtuB as a function of added urea concentration. Sequential spectra are shown for V10R1 from 0 to 4 M urea and T30R1 from 0 to 5M urea as urea is increased in 0.5M steps. Sequential spectra are shown for V78R1 from 0 to 9M in 1M steps and for E108R1 from 0 to 12M in 2M steps. Spectra obtained from BtuB in POPC in the absence of urea are shown in red and EPR spectra obtained at the highest urea concentrations are shown in blue. b) EPR spectra for positions 10, 30, 78, 108, 121 and 158, at the highest urea concentrations used. These concentrations were 6, 5, and 11 M urea for sites 10, 30 and 78, and 12 M urea for the remaining sites, respectively. The arrows indicate the position of a second motional component due to a more compact conformation in the urea-denatured state.
Shown in Fig. 2b are EPR spectra for a number of sites at the highest urea concentrations tested. In most cases, these are urea concentrations where the unfolding transition appears to be complete (see Fig. 3). The EPR spectra in Fig. 2b from sites 10 and 30 result from R1 having a single motional component, corresponding to a label with a correlation time of approximately 0.6 ns. This is consistent with the N-terminal segment assuming a fully unstructured random coil configuration in the presence of urea. However, at all the other sites tested, the spectra are multicomponent and they result from at least two motional components of R1: one which is isotropic and rapidly diffusing on the EPR time scale, and a second resulting from the label undergoing more restricted motion (arrows in Fig. 2b). In these cases, subtraction of the more mobile component and double integration indicates that the population of label undergoing more restricted motion is approximately 40 to 50% of the total spin signal. The mobile components from labels at sites 78, 108, 121 and 158 are isotropic, but they exhibit different correlation times from those seen at positions 10 and 30. At high urea concentrations, the label correlation time for V78R1 is approximately 0.3 ns, roughly double the diffusional rate of labels at positions 10 and 30. In contrast, the mobile isotropic signals from sites E108R1, N128R1 and Q158R1 have longer correlation times of approximately 1.0 to 1.1 ns. These differences in motion appear as a difference in linewidth and a difference in the ratio of amplitudes between the high-field and central nitroxide resonances.
Figure 3.
Plots of the normalized amplitudes of the EPR spectra from five core mutants and one of the barrel mutants as a function of urea concentration. As indicated in Methods, the normalized amplitude is linearly related to the fraction of unfolded protein. The lines represent the best fits to the data using Eq. 3 (see Methods). This fit assumes a two state model where the protein exists either in a folded or unfolded conformation.
The existence of broader components in these EPR spectra at high denaturant concentrations has been observed previously for FepA (18), and these features provide a strong indication that at the highest urea concentrations reached regions of the N-terminal core of BtuB, beyond the first β-strand, are in an equilibrium between populations of highly-unstructured (largely random coil) and more compact protein conformations. The fact that the motionally mobile components vary in lineshape and exhibit significantly different correlation times indicates that the unstructured state is not uniform, and that the protein backbone dynamics in the most unfolded state varies with position on the ns time scale. These observations are consistent with the finding that native-like protein structure can persist even in the presence of high concentrations of denaturants (29), and with the view that denatured states of proteins are more complex and more compact than a truly random coil (30).
Sites in the BtuB N-terminal domain unfold at different urea concentrations and have differing solvent exposures
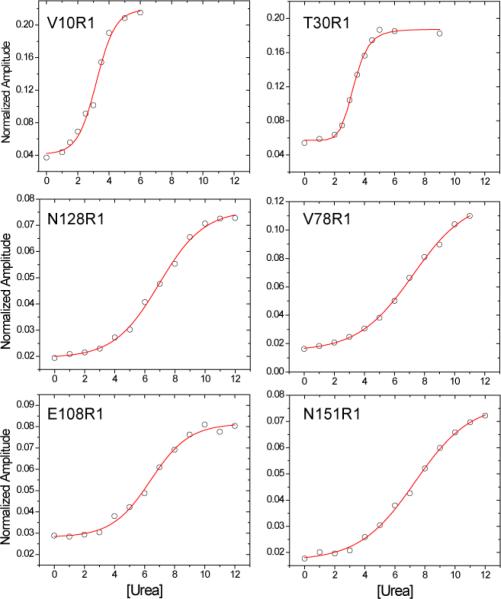
Shown in Fig. 3 are plots of the normalized EPR amplitudes (obtained from spectra such as those shown in Fig. 2a) as a function of urea concentration for a number of sites in BtuB. Also shown are fits to these data using Eq. 3. A comparison of the denaturation curves for labels in the N-terminus, particularly sites 10 and 30, indicates that they undergo an unfolding transition at lower urea concentrations than do sites deeper into the N-terminal core or in the BtuB barrel. The data were also fit as described in Methods by directly calculating the fraction of unfolded protein and performing a linear extrapolation to the data for the free energy versus urea concentration according to Eq. 1. Several of these plots are shown in Fig. 4, and a summary of the data obtained for the eight mutants is shown in Table 1. It should be noted that each experimental spectrum could be fit with a linear combination of the spectra that represent the folded and unfolded protein, consistent with the assumption that the spectra report two structural states.
Figure 4.
Linear extrapolation of free energies as a function of urea concentration to obtain the free-energy of unfolding (Eq. 1). The free energies were determined from the populations of folded and unfolded protein. These populations were estimated by determining the linear combination of folded and unfolded spectra that provide the best fit for each spectrum in the presence of urea (see Methods).
Table 1.
Thermodynamic data from urea denaturation of BtuB
| Mutant in POPC | ΔG°U (kcal/mol)a | ΔG°U (kcal/mol)b | m (kcal/mol/M) | C0 (M) |
|---|---|---|---|---|
| V10R1 | 2.77 ± 0.21 | 3.01 ± 0.24 | 0.93 ± 0.06 | 3.25 ± 0.13 |
| T30R1 | 3.23 ± 0.11 | 3.33 ± 0.12 | 1.00 ± 0.04 | 3.32 ± 0.04 |
| V78R1 | 2.22 ± 0.02 | 2.12 ± 0.05 | 0.29 ± 0.00 | 7.23 ± 0.13 |
| E108R1 | 2.99 ± 0.11 | 3.04 ± 0.13 | 0.47 ± 0.02 | 6.40 ± 0.14 |
| N128R1 | 2.39 ± 0.05 | 2.53 ± 0.08 | 0.36 ± 0.01 | 6.95 ± 0.12 |
| N151R1 | 2.42 ± 0.09 | 2.63 ± 0.11 | 0.36 ± 0.01 | 7.34 ± 0.14 |
| Q158R1 | 2.89 ± 0.18 | 2.69 ± 0.42 | 0.39 ± 0.06 | 6.91 ± 0.22 |
| W371R1 | 2.90 ± 0.07 | 3.88 ± 0.50 | 0.55 ± 0.07 | 7.05 ± 0.08 |
Free energy of unfolding calculated by linear extrapolation (y-intercept).
Free energy of unfolding calculated by mC0, where C0 is the midpoint of the unfolding transition calculated by fitting a sigmoidal curve to [urea] vs. peak-to-peak amplitude (see Methods).
The data shown in Table 1 indicate that the thermodynamic stabilities measured at sites within the N-terminal domain are similar, with the values of varying from about 2.2 to 3.2 kcal/mole. These values are smaller than those typically obtained for the denaturation of globular proteins (23, 31, 32), and they are smaller than those previously seen using SDSL for the barrel of a homologous TonB-dependent transporter, FepA (18). The values of obtained by SDSL should be viewed with some caution, because the spin label side chain, R1, is known to modulate particularly when placed at deeply buried sites or at sites in strong tertiary contact (24). Nonetheless, if these values are perturbed by the spin labels, they are remarkably similar. And this includes the value of obtained from site 30, which should not be highly perturbed due to the aqueous exposure at this site.
Urea denaturation indicates that the N-terminal domain does not unfold in a single step
Unlike the values of , which are remarkably consistent, the values of m vary substantially. As shown in Table 1, there is an approximately threefold variation in the values of m for sites in the N-terminus, with sites 10 and 30 yielding the highest values of m and 78 and 128 yielding the lowest. The values of C0 (the urea concentration at the midpoint of the unfolding) is also substantially different for sites 10 and 30, which is expected since the product of m and C0 yields the free energy of unfolding. Interestingly, the values of m become progressively smaller the further the spin label is moved towards the extracelluar surface of BtuB within the N-terminal domain. The parameter m strongly correlates with the amount of protein surface area that becomes exposed to solution upon the unfolding of the protein with urea (33). Since these numbers vary substantially within the N-terminus, this region must not unfold in a single concerted step, but must unfold in a number of independent steps. Sites 10 and 30 unfold at the lowest concentrations of urea with similar values of m, suggesting that the Ton box and the first β-strand C-terminal to the Ton box unfold in a single step with the largest exposure of protein surface area. Strands further into the protein interior unfold at higher urea concentrations and involve progressively smaller exposures of surface area.
The three labels within the β-barrel region unfold at urea concentrations that are much higher than those at sites 10 and 30, and they yield similar values for protein stability compared to those elsewhere in BtuB. These values are less than those observed previously for the stability of a β-strand in FepA, but the measurements made here were acquired in a different lipid mixture than that used previously. It should be noted that bilayer integrity seems to be maintained in the presence of urea, even at relatively high denaturant concentrations (17, 34, 35).
Unfolding of the N-terminal core region is reversible
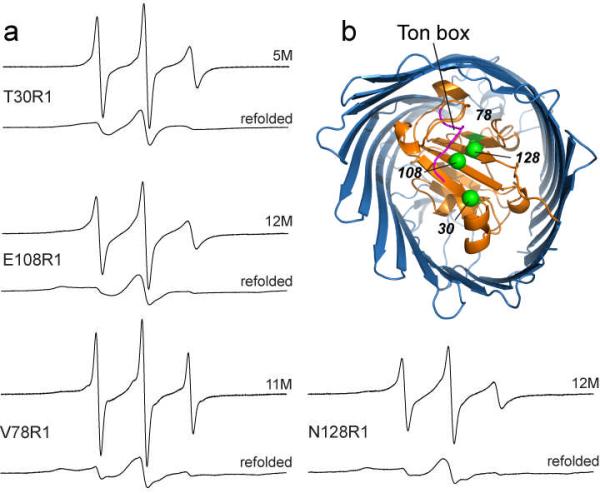
Once denatured, the N-terminal domain of BtuB can be refolded by removal of the urea. For the data shown here, urea was removed by dilution of the lipid-denaturant mixture (see Methods) and centrifugation to concentrate the sample. Shown in Fig. 5 are EPR spectra from sites T30R1, E108R1, V78R1 and N128R1 at the highest concentration of urea indicated in Fig. 3 and following the removal of the urea by dilution. The EPR spectra taken after dilution of urea are identical to those taken after reconstitution into POPC and indicate that the protein refolds under these conditions to restore local structure around the labeled site.
Figure 5.
a) EPR spectra for four R1 mutants within the core of BtuB following the removal of urea by dilution. Spectra for each site in the presence of high urea concentrations and following the removal of urea are shown. At each site, the EPR spectrum following removal of urea closely resembles the spectrum before denaturation, indicating that the local protein fold is restored at each of these sites. b) Position of these four sites within the core region of BtuB. In this model, the N-terminal domain is shown in orange and the Ton box is shown in magenta.
Discussion
The N-terminal domain that occludes the β-barrel of TonB-dependent transport proteins is generally thought to undergo a significant conformational rearrangement during transport. In one proposed transport mechanism, the N-terminal domain reversibly unfolds, where the unfolding is driven by an interaction between the transporter's Ton box and TonB (4, 13). In the present study, site-directed spin labeling was used to determine the stability of the N-terminal domain, and to characterize intermediates that could be accessed using reversible urea denaturation. The fact that the unfolding is reversible at every site examined suggests that the intermediates observed may be intermediates that are sampled during the folding of the protein (36). Moreover, these intermediates might be sampled during transport that would be driven by a TonB-dependent unfolding.
The denaturation data obtained here provide strong evidence that the N-terminal domain of BtuB unfolds in stepwise and localized fashion. The denaturation data shown in Figs. 3 and 4 can be fit by a simple two state mechanism; however, the apparent surface areas that are exposed during denaturation (as judged by the m values in Table 1) vary significantly, as do the concentrations of urea required to produce denaturation. As a result, the urea-induced unfolding of the N-terminal domain must not take place in a single cooperative step. The behavior of sites 10 and 30 to urea denaturation is different than at other sites in BtuB. First, this region of the protein unfolds at significantly lower urea concentrations than do other sites in BtuB. Second, the EPR spectra in the denatured state are similar, and they reflect a single isotropic motional component of the R1 side-chain. Unlike other sites in BtuB, there is no evidence for residual compact structure in this segment of the protein at the completion of the urea titration. Third, the value of m, which reflects the protein surface area exposed to solvent during denaturation, is identical within experimental error at sites 10 and 30, and larger than at other sites examined. Taken together, the data suggests that at modest urea concentrations (4 M) a significant segment of the N-terminus is unfolded. This includes sites 10 and 30 and may include regions further towards the C-terminus, but not extending to site 78.
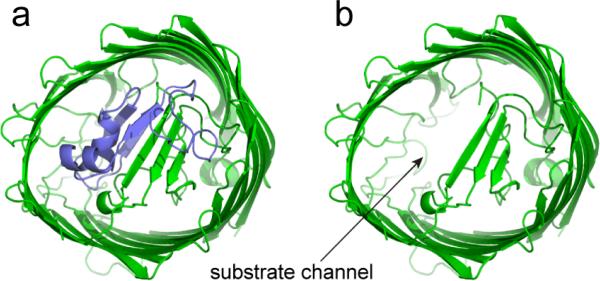
What do these data tell us about a model for transport involving a transient unfolding of the N-terminus? First, the data suggest that regions of the N-terminus are substantially less stable than typical globular proteins (perhaps by a factor of two or three) and that the N-terminus might be relatively easy to unfold. This finding is consistent with previous NMR measurements, which indicate that the N-terminal domain of FepA is not well-folded when expressed as an isolated domain (37). Second, the data indicate that the N-terminus of BtuB can be refolded from this denatured state, and that refolding from a TonB-driven unfolding event should be spontaneous. Finally, if a segment including residues 10 and 30 and extending up to residues 60 or 65 is unfolded from the interior of BtuB, a cavity is opened within the interior of BtuB, as shown in Fig. 6. This unfolded segment includes several substrate contact sites that are conserved in TonB-dependent transporters (14). A similar cavity to that shown in Fig. 6 was observed in steered molecular dynamics simulations where extraction of the Ton box of BtuB eventually opened a channel sufficiently large for substrate passage (15). Moreover, a cavity such as that shown in Fig. 6 may be the source of the ion conduction observed for reconstituted BtuB under similar denaturing conditions (17). As a result, the opening of this cavity, for example by an interaction of the Ton box with TonB, might release bound vitamin B12 from its extracellular binding pocket and allow passage of the substrate into the periplasm. It should be noted that although these denaturation experiments indicate the types of conformational transition states that might be sampled during a directed unfolding of the BtuB N-terminus, there are no data to indicate that such a unfolding takes place during transport. Further work under conditions where active transport is taking place or where an intermediate state is trapped would be required to verify such a model.
Figure 6.
Unfolding a section of the N-terminus including the first β-strand within BtuB should open a pore that enables the passage of substrate. On the left a) is a view from the periplasmic surface of BtuB in its folded state where residues 6 to 65 are highlighted in blue. On the right is a view of the periplasmic surface b) with the first 65 residues removed from the core of the protein.
The data obtained here by SDSL on the denatured state of BtuB provide a unique view of membrane protein structure under highly denaturing conditions. It is generally believed that the urea-denatured states of proteins are not random coils and that a significant level of compact structure exists even at relatively high urea concentrations (29, 30). The EPR data obtained here support this view. Two distinct motional components are observed from most sites in BtuB, suggesting that significant populations of an extended (random coil) and a more compact protein form are present under denaturing conditions. At X-band, conversion between these states must be slower than a few tens of ns for these states to be resolved, and additional experiments examining the relaxation rates of these labels could put a time-scale on the rate of conversion between these forms (38). In addition to compact and more extended forms, the EPR spectra indicate that the backbone dynamics of the extended form varies as a function of position along the sequence. Thus, even the more open form seen for the denatured protein is not a true random coil. This mixture of states generally applies to both the N-terminal domain and to the membrane imbedded β-barrel of BtuB. As a result, the ability of the protein to refold at every site examined is perhaps not surprising, since significant residual structure is likely maintained even under strongly denaturing conditions.
In summary, active transport requires the input of energy, and in the case of TonB-dependent transport, this energy is believed to be derived from the inner-membrane proton potential through an interaction with TonB. The results presented here indicate that a region of the N-terminus of BtuB can be unfolded under moderately denaturing conditions, and that the N-terminus denatures in a series of steps. The data are consistent with a model for transport that involves a partial unfolding of the N-terminal domain of BtuB, promoted by an interaction with energized TonB. This partial unfolding would lead to a release of the substrate from its binding site and could facilitate substrate diffusion through the BtuB barrel.
Acknowledgements
We would like to thank Christian Altenbach (UCLA) for providing LabVIEW programs used to process and simulate EPR spectra. We would also like to thank Gail Fanucci and Jae Lee for carrying out preliminary urea denaturation studies on BtuB.
Footnotes
This work was supported by a grant from the National Institutes of Health, NIGMS, GM 035215.
Abbreviations used: EPR, electron paramagnetic resonance; MTSL, methanethiosulfonate spin label; POPC, palmitoyloleoylphosphatidylcholine; R1, spin-labeled side chain produced by derivatization of a cysteine with the MTSL; SDSL, site-directed spin labeling.
References
- 1.Ferguson AD, Deisenhofer J. TonB-dependent receptors-structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- 2.Faraldo-Gomez JD, Sansom MS. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 3.Postle K, Kadner R. Touch and go: tying TonB to transport. Mol. Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 4.Wiener MC. TonB-dependent outer membrane transport: going for Baroque? Curr Opin Struct Biol. 2005;15:394–400. doi: 10.1016/j.sbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Endriss F. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals. 2007;20:219–231. doi: 10.1007/s10534-006-9072-5. [DOI] [PubMed] [Google Scholar]
- 6.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadieux N, Kadner RJ. Site-directed disulfide bonding reveals an interaction site between energy coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 9.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 10.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nature structural biology. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 11.Cherezov V, Yamashita E, Liu W, Zhalnina M, Cramer WA, Caffrey M. In meso structure of the cobalamin transporter, BtuB, at 1.95 A resolution. J Mol Biol. 2006;364:716–734. doi: 10.1016/j.jmb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faraldo-Gomez JD, Smith GR, Sansom MS. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a comparative study of the ferrichrome-free and bound states. Biophys J. 2003;85:1406–1420. doi: 10.1016/S0006-3495(03)74573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 14.Chimento DP, Kadner RJ, Wiener MC. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins. 2005;59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 15.Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonhivers M, Desmadril M, Moeck GS, Boulanger P, Colomer-Pallas A, Letellier L. Stability studies of FhuA, a two-domain outer membrane protein from Escherichia coli. Biochemistry. 2001;40:2606–2613. doi: 10.1021/bi001725i. [DOI] [PubMed] [Google Scholar]
- 17.Udho E, Jakes KS, Buchanan SK, James KJ, Jiang X, Klebba PE, Finkelstein A. Reconstitution of bacterial outer membrane TonB-dependent transporters in planar lipid bilayer membranes. Proc Natl Acad Sci U S A. 2009;106:21990–21995. doi: 10.1073/pnas.0910023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klug CS, Feix JB. Guanidine hydrochloride unfolding of a transmembrane beta-strand in FepA using site-directed spin labeling. Protein Sci. 1998;7:1469–1476. doi: 10.1002/pro.5560070624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klug CS, Su W, Liu J, Klebba PE, Feix JB. Denaturant unfolding of the ferric enterobactin receptor and ligand-induced stabilization studied by site-directed spin labeling. Biochemistry. 1995;34:14230–14236. doi: 10.1021/bi00043a030. [DOI] [PubMed] [Google Scholar]
- 20.Heller K, Kadner RJ. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. Journal of Bacteriology. 1985;161:904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coggshall KA, Cadieux N, Piedmont C, Kadner R, Cafiso DS. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry. 2001;40:13946–13971. doi: 10.1021/bi015602p. [DOI] [PubMed] [Google Scholar]
- 22.Fanucci GE, Cadieux N, Piedmont CA, Kadner RJ, Cafiso DS. Structure and dynamics of the beta-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry. 2002;41:11543–11551. doi: 10.1021/bi0259397. [DOI] [PubMed] [Google Scholar]
- 23.Yao M, Bolen DW. How valid are denaturant-induced unfolding free energy measurements? Level of conformance to common assumptions over an extended range of ribonuclease A stability. Biochemistry. 1995;34:3771–3781. doi: 10.1021/bi00011a035. [DOI] [PubMed] [Google Scholar]
- 24.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 25.Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 26.Freed DM, Horanyi PS, Wiener MC, Cafiso DS. Conformational Exchange in a Membrane Transport Protein Is Altered in Protein Crystals. Biophysical journal. 2010;99:1604–1610. doi: 10.1016/j.bpj.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freed DM, Khan AK, Horanyi PS, Cafiso DS. Molecular origin of electron paramagnetic resonance line shapes on beta-barrel membrane proteins: the local solvation environment modulates spin-label configuration. Biochemistry. 2011;50:8792–8803. doi: 10.1021/bi200971x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores Jimenez RH, Freed DM, Cafiso DS. Lipid and Membrane Mimetic Environments Modulate Spin Label Side Chain Configuration in the Outer Membrane Protein A. J Phys Chem B. 2011 doi: 10.1021/jp207420d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 30.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 31.Pace CN, Vanderburg KE. Determining globular protein stability: guanidine hydrochloride denaturation of myoglobin. Biochemistry. 1979;18:288–292. doi: 10.1021/bi00569a008. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad F, Bigelow CC. Estimation of the free energy of stabilization of ribonuclease A, lysozyme, alpha-lactalbumin, and myoglobin. J Biol Chem. 1982;257:12935–12938. [PubMed] [Google Scholar]
- 33.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastoriza-Gallego M, Oukhaled G, Mathe J, Thiebot B, Betton JM, Auvray L, Pelta J. Urea denaturation of alpha-hemolysin pore inserted in planar lipid bilayer detected by single nanopore recording: loss of structural asymmetry. FEBS Lett. 2007;581:3371–3376. doi: 10.1016/j.febslet.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 35.Feng Y, Yu ZW, Quinn PJ. Effect of urea, dimethylurea, and tetramethylurea on the phase behavior of dioleoylphosphatidylethanolamine. Chem Phys Lipids. 2002;114:149–157. doi: 10.1016/s0009-3084(01)00198-0. [DOI] [PubMed] [Google Scholar]
- 36.Religa TL, Markson JS, Mayor U, Freund SM, Fersht AR. Solution structure of a protein denatured state and folding intermediate. Nature. 2005;437:1053–1056. doi: 10.1038/nature04054. [DOI] [PubMed] [Google Scholar]
- 37.Usher KC, Ozkan E, Gardner KH, Deisenhofer J. The plug domain of FepA, a TonB-dependent transport protein from Escherichia coli, binds its siderophore in the absence of the transmembrane barrel domain. Proc Natl Acad Sci U S A. 2001;98:10676–10681. doi: 10.1073/pnas.181353398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bridges MD, Hideg K, Hubbell WL. Resolving Conformational and Rotameric Exchange in Spin-Labeled Proteins Using Saturation Recovery EPR. Applied magnetic resonance. 2010;37:363. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. Substrate-induced conformational changes of the periplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry. 2003;42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]