Abstract
Balancer chromosomes are critical tools for Drosophila genetics. Many useful transgenes are inserted onto balancers using a random and inefficient process. Here we describe balancer chromosomes that can be directly targeted with transgenes of interest via recombinase-mediated cassette exchange (RMCE).
Keywords: RMCE, targeted transgenesis, phiC31, Drosophila, balancer
In Drosophila, balancer chromosomes bearing multiple inversions are routinely used in genetic manipulations and in the maintenance of sterile or lethal mutations as balanced heterozygotes. Balancer chromosomes typically carry dominant markers, the most common of which affect adult structures only. However, using transgenic approaches, many new markers and functions have been assigned to balancers in efforts to improve their utility. For example, transgenic insertions have been created to facilitate the identification of balanced progeny at different stages of development, including balancers that carry histological or fluorescent markers driven by embryonic enhancers (including so-called “blue” and “green” balancers) (Casso et al. 2000; Halfon et al. 2002; Le et al. 2006; Panzer et al. 1993; Rudolph et al. 1999). Balancers carrying transgenic insertions of GAL80, a repressor of the UAS/GAL4 system, function similarly in cross schemes involving transgenes driven by UAS (Vef et al. 2006). More recently, the cloning of the gene responsible for Tubby1, a convenient marker that is visible during larval development and is carried on the third chromosome balancer TM6B, has led to the creation of Tubby1 transgenes inserted onto X and second chromosome balancers (Guan et al. 2006; Lattao et al. 2011; Pina and Pignoni 2012). In addition to novel markers for the identification of balanced progeny, others have created transgenic insertions on balancer chromosomes for the convenient delivery of key enzymes into genetic schemes; these include transposases for P (Lindsley and Zimm 1992) and Minos (Metaxakis et al. 2005) transposon systems, and Cre (Siegal and Hartl 1996) and FLP (Chou and Perrimon 1992) recombinases. Furthermore, autosomal balancers have been engineered to carry the cell death promoter hid in an effort to simplify fly sorting during gene replacement by homologous recombination (Huang et al. 2008). Thus, a pattern exists in which the development of new genetic technologies consistently leads researchers to target new transgenes to Drosophila balancer chromosomes.
For each of the examples listed above, transgenic insertions were incorporated onto balancers using P-element–mediated transgenesis (Rubin and Spradling 1982). P-element insertion occurs in an untargeted manner; thus, obtaining transgenes on a balancer requires one to create many independent insertions, and then to screen for those that happened to insert onto the balancer chromosome. This approach typically involves several generations of crosses and requires many lines to be discarded, representing wasted effort and resources. Therefore, we sought to create balancer chromosomes that could be directly targeted with transgenes of interest using phiC31-mediated RMCE (Bateman et al. 2006). This approach makes use of a “target cassette,” which consists of a dominant marker gene flanked by attP recognition sites for phiC31 integrase, that is first integrated into the genome. Once the target is established, a “donor cassette” carrying a transgene of interest flanked by phiC31 attB sites can be directly incorporated at the precise genomic position of the target cassette. As described below, our strategy was to use traditional P-element–mediated transgenesis to incorporate dominantly marked RMCE target cassettes onto balancer chromosomes. Once established, these targets can be used to directly incorporate transgenes of interest onto balancer chromosomes.
We previously created a P element that carries an RMCE target cassette consisting of a mini-white gene flanked by phiC31 attP sites (Bateman and Wu 2008). Using a Δ2-3 transposase source, we remobilized existing insertions of this P element and, via three different cross schemes, screened for new insertions onto the X-chromosome balancer FM7h (Heitzler 1997), the second chromosome balancer CyO, and the third chromosome balancer TM3 (see supporting information, File S1 for details of remobilization). For each balancer, we isolated three independent insertions and used inverse PCR and sequence analysis to map the precise genomic positions of the P elements (Table 1). The majority of these insertions mapped to euchromatic regions in or near broadly expressed genes and within chromatin environments expected to facilitate gene expression (Filion et al. 2010; Graveley et al. 2011; Kharchenko et al. 2011) (Table 1, Table S1). One exception, the insertion in line CyOJ01, was mapped to a Doc element that we did not precisely locate.
Table 1. Balancer chromosomes carrying RMCE target cassettes.
| Balancer Line | Insert Cytology | Insert Position | Insert Strand | Nearest Gene | Relative Position of Insertion |
|---|---|---|---|---|---|
| FM7hFS2 | 10B6 | X:11266889 | Top | Dlg1 | Genic |
| FM7hFS4 | 18A3-4 | X:19047768 | Top | RhoGAP18B | Genic |
| FM7hFS5 | 6F3 | X:6969300 | Top | Sxl | Intergenic |
| CyOJ01 | ND | ND | ND | Doc Element | (Repeat sequence) |
| CyOJ04 | 37B8 | 2L:18987255 | Top | CG10641 | Genic |
| CyOJ08 | 37F2 | 2L:19572635 | Bottom | Spi | Genic |
| TM3FS10 | 79A2 | 3L:21872640 | Top | Mub | Genic |
| TM3FS11 | 85A5 | 3R:4502640 | Top | CG8043 | Genic |
| TM3FS18 | 100D1 | 3R:27550495 | Bottom | ttk | Genic |
Insertions were mapped by comparing sequences of inverse PCR products to release 5.3 of the Drosophila melanogaster genome sequence. The insertion in the CyOJ01 line was found in a Doc element and was not precisely mapped (ND, not determined). See Table S1 for further information on genome annotations near each insertion.
To assess the potential utility of these lines, we first confirmed that RMCE was supported at appreciable levels using at least one representative target-bearing line for each of the three balancer chromosomes, namely FM7hFS5, CyOJ01, CyOJ08, and TM3FS18. In the presence of a genomic source of the phiC31 integrase (Bischof et al. 2007), we injected donor constructs carrying attB sites flanking either an intronless yellow gene or a fluorescent marker driven by the eye-specific enhancer GMR (Moses and Rubin 1991) (Figure 1 and Table 2). Although we found experimental variation in transformation efficiencies, the lines that we tested supported RMCE at rates up to 41%, consistent with our rates of transgenesis for other genomic targets using this method and our current injection apparatus (data not shown; Bateman and Wu 2008).
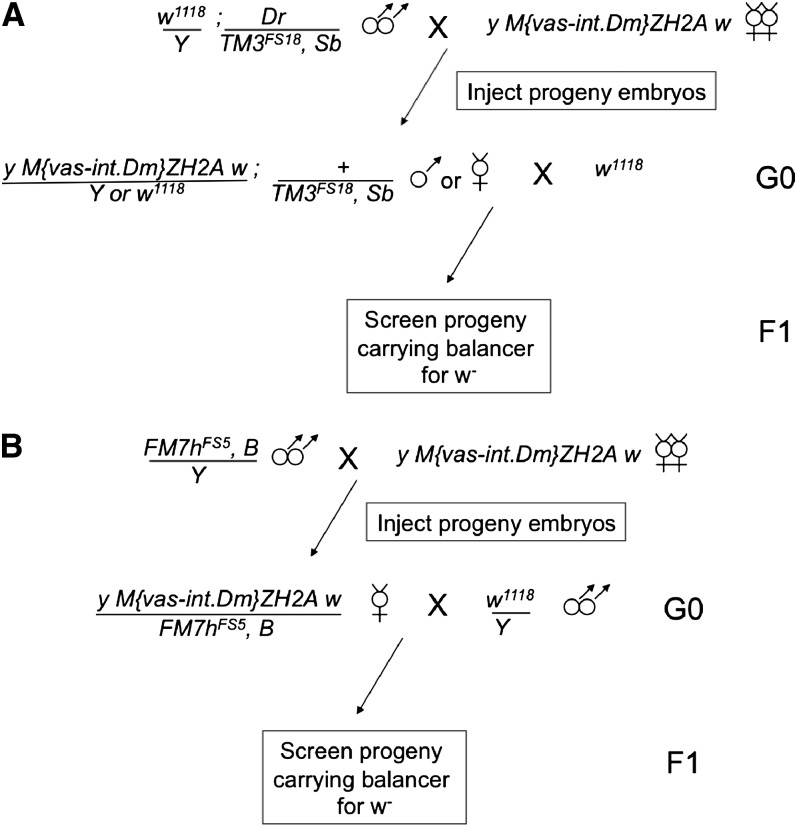
Figure 1.
Injection scheme for RMCE using targets on (A) autosomal or (B) X chromosomal balancers. Germline-targeted phiC31 integrase is supplied from the X-chromosomal ZH2A insertion (Bischof et al. 2007). In the G0 generation, single males or females (A) or females only (B) that carry the RMCE cassette-bearing balancer (50% of progeny expected) are mated singly to flies with a w− genotype, and the F1 generation is screened for balanced progeny in which the mini-white eye color of the target cassette is lost. Insertions onto CyO are obtained through a scheme analogous to (A). See Figure S1 for an alternate strategy using stocks carrying the integrase source and the target balancer concurrently.
Table 2. Target cassettes on balancer chromosomes support RMCE.
| Balancer Line | Donor Cassette Gene | Embryos Injected | Vials with RMCE | % Vials with RMCE |
|---|---|---|---|---|
| FM7hFS5 | yellow | 175 | 3/12 | 25% |
| FM7hFS5 | GMR-GFP | 200 | 1/17 | 5.9% |
| TM3FS18 | yellow | 165 | 2/13 | 15% |
| TM3FS18 | GMR-GFP | 243 | 7/17 | 41% |
| CyOJ01 | yellow | 150 | 4/26 | 15.4% |
| CyOJ01 | GMR-GFP | 150 | 2/28 | 7.1% |
| CyOJ08 | GMR-mCherry | 250 | 2/19 | 10.5% |
Donor constructs in sterile water were injected according to the scheme in Figure 1. DNA concentrations were 250 ng/µl (yellow), 325 ng/µl (GMR-GFP), and 115 ng/µl (GMR-mCherry).
Finally, we verified that transgene expression was supported in the transformants that we generated. First, we assessed adult body pigmentation of transformants carrying an insertion of the intronless yellow gene in an otherwise yellow mutant background, and we found that all (9/9) transformed lines produced fully penetrant levels of yellow pigmentation indistinguishable from wild-type flies (Figure 2 and data not shown). To address gene expression at earlier points of development, we assessed the expression of donor cassettes carrying GMR-GFP or GMR-mcherry in whole-mounted late-stage embryos and in eye imaginal discs from wandering third instar larvae (Moses and Rubin 1991). Insertions into FM7hFS5, CyOJ01, CyOJ08, and TM3FS18 produced robust tissue-specific fluorescence (Figure 2, Figure S2), demonstrating that our modified balancers can support gene expression at multiple stages of development.
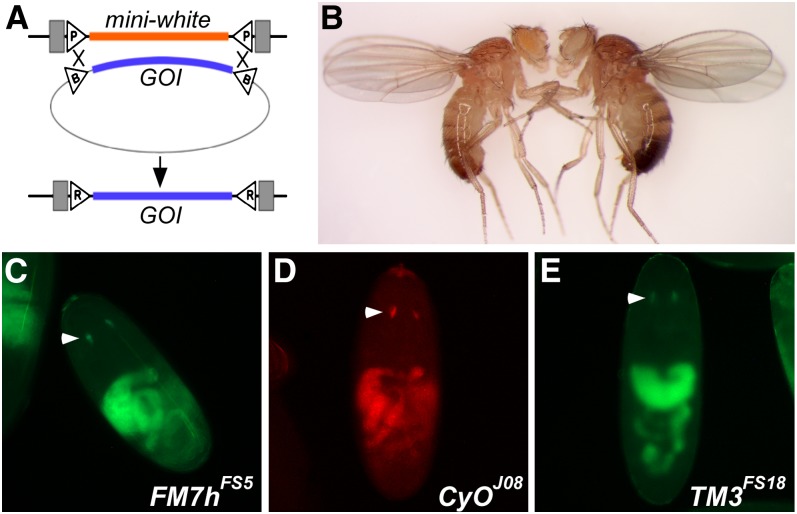
Figure 2.
Insertion of donor cassettes onto balancers supports expression of transgenes. (A) Schematic of the exchange reaction. Integrase-mediated crossovers at both ends of the aligned target and donor cassettes result in removal of mini-white and integration of the gene of interest (GOI) into the genome. Triangles, att sites; gray boxes, P-element ends. (B) FM7hFS5/Y males before (left) and after (right) RMCE integration of an intronless yellow transgene. The fly on the left retains the mini-white eye pigmentation and lacks yellow pigmentation, whereas on the right, the mini-white eye color is lost, and expression of the yellow transgene is evident in the wing and abdomen. Transgenic insertions of the intronless yellow cassette on CyOJ01 and TM3FS18 produced similar pigmentation (not shown). (C–E) Embryonic expression of GMR-GFP (A, C) or GMR-mCherry (B) insertions onto balancers. White arrowheads, GMR-specific expression (Moses and Rubin 1991); autofluorescence of the gut is also evident.
We anticipate that the modified balancer chromosomes described here will greatly facilitate future efforts at incorporating new transgenes onto balancers and may foster new approaches to balancer chromosome modifications by removing a significant barrier to obtaining balancer insertions. As balancers are developed for other genetic models (Hentges and Justice 2004), a similar scheme for simplified balancer marking may also be beneficial in those systems.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for fly stocks and for agreeing to distribute the balancer stocks described here; the Mount Desert Island Biological Laboratory DNA Sequencing Core for DNA sequencing; and Michael Palopoli and Bruce Kohorn for helpful comments. This project was supported by grants from the National Center for Research Resources (5-P20-RR-016463-12) and the National Institute of General Medical Sciences (8-P20-GM-103423-12) of the National Institutes of Health.
Footnotes
Communicating editor: K. S. McKim
Literature Cited
- Bateman J. R., Wu C. T., 2008. A simple polymerase chain reaction-based method for the construction of recombinase-mediated cassette exchange donor vectors. Genetics 180: 1763–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso D., Ramirez-Weber F., Kornberg T. B., 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91: 451–454 [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N., 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Middlebrooks B. W., Alexander S., Wasserman S. A., 2006. Mutation of TweedleD, a member of an unconventional cuticle protein family, alters body shape in Drosophila. Proc. Natl. Acad. Sci. USA 103: 16794–16799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M. S., Gisselbrecht S., Lu J., Estrada B., Keshishian H., et al. , 2002. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis 34: 135–138 [DOI] [PubMed] [Google Scholar]
- Heitzler P., 1997. New FM7 versions from Strasbourg. Drosoph. Inf. Serv. 80: 103 [Google Scholar]
- Hentges K. E., Justice M. J., 2004. Checks and balancers: balancer chromosomes to facilitate genome annotation. Trends in genetics. TIG 20: 252–259 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou W., Watson A. M., Jan Y. N., Hong Y., 2008. Efficient ends-out gene targeting in Drosophila. Genetics 180: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattao R., Bonaccorsi S., Guan X., Wasserman S. S., Gatti M., 2011. Tubby-tagged balancers for the Drosophila X and second chromosomes. Fly (Austin) 5: 369–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T., Liang Z., Patel H., Yu M. H., Sivasubramaniam G., et al. , 2006. A new family of Drosophila balancer chromosomes with a w- dfd-GMR yellow fluorescent protein marker. Genetics 174: 2255–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G., 1992. The Genome of Drosophila melanogaster. Academic Press, Inc., San Diego, CA [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A., Savakis C., 2005. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K., Rubin G. M., 1991. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 5: 583–593 [DOI] [PubMed] [Google Scholar]
- Panzer S., Fong A., Beckendorf S. K., 1993. Genetic notes: new lacZ-marked balancer. Drosoph. Inf. Serv. 72: 197 [Google Scholar]
- Pina C., Pignoni F., 2012. Tubby-RFP balancers for developmental analysis: FM7c 2xTb-RFP, CyO 2xTb-RFP, and TM3 2xTb-RFP. Genesis 50: 119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353 [DOI] [PubMed] [Google Scholar]
- Rudolph T., Lu B., Westphal T., Szidonya J., Eissenberg J., et al. , 1999. New type of CyO and TM3 green balancers. Drosoph. Inf. Serv. 82: 99–100 [Google Scholar]
- Siegal M. L., Hartl D. L., 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vef O., Cleppien D., Loffler T., Altenhein B., Technau G. M., 2006. A new strategy for efficient in vivo screening of mutagenized Drosophila embryos. Dev. Genes Evol. 216: 105–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.