Abstract
One of the fundamental questions in developmental biology is how the vast range of pattern and structure we observe in nature emerges from an almost uniformly homogeneous fertilized egg. In particular, the mechanisms by which biological systems maintain robustness, despite being subject to numerous sources of noise, are shrouded in mystery. Postulating plausible theoretical models of biological heterogeneity is not only difficult, but it is also further complicated by the problem of generating robustness, i.e. once we can generate a pattern, how do we ensure that this pattern is consistently reproducible in the face of perturbations to the domain, reaction time scale, boundary conditions and so forth. In this paper, not only do we review the basic properties of Turing's theory, we highlight the successes and pitfalls of using it as a model for biological systems, and discuss emerging developments in the area.
Keywords: Turing, biological pattern formation, robustness problem
1. Introduction
In 1952, Turing [1] wrote his seminal paper ‘The chemical basis of morphogenesis’ in which he proposed an ingenious new theory. Turing hypothesized that the patterns we observe during embryonic development arise in response to a spatial pre-pattern in biochemicals, which he termed morphogens. Cells would then respond to this pre-pattern by differentiating in a threshold-dependent way. Thus, Turing hypothesized that the patterns we see in nature, such as pigmentation in animals, branching in trees and skeletal structures, are reflections of inhomogeneities in underlying biochemical signalling.
To generate these pre-patterns, Turing considered the morphogens to be interacting in such a way that, in the well-mixed system, there would be a spatially uniform steady state which was stable to perturbations. He then proposed that, in the non-mixed system, this steady state would be driven unstable by diffusion. This is counterintuitive as diffusion is usually a stabilizing and a homogenizing process. In effect, what Turing showed was that from the interaction of two stabilizing processes, an instability could emerge. This is now called diffusion-driven instability (DDI) and the pattern is said to emerge or self-organize. The system Turing studied took the form
| 1.1 |
where u is a vector of chemical concentrations, D a matrix of constant diffusion coefficients (usually diagonal) and f(u) the reaction kinetics (typically nonlinear). Appropriate boundary and initial conditions, which are often periodic and perturbations of the homogeneous steady state, respectively, are applied to close the system.
We begin in §2 by demonstrating the patterning attributes of this framework with representative examples. Section 3 discusses the problem of unreliable pattern generation within Turing systems and then, in §4, we suggest that growth can in fact endow the system with robustness. Having observed that robust solutions can be generated in the deterministic case, we extend our framework in §5 to incorporate intrinsic stochastic effects, allowing us to analyse the effect of noise on Turing patterns. In §6, we change direction slightly and investigate the effects of kinetic delays on pattern formation, before summarizing our observations and placing them into the context of current research in §7.
2. Applications of the Turing model
Turing analysed the specific case of equation (1.1) when there are two interacting morphogens, u = (u,v)T. These morphogens diffuse at rates Du and Dv, respectively, and they also undergo reaction kinetics defined by f(u) = (f(u,v), g(u,v))T. By applying periodic boundary conditions to close the system description, Turing derived the following conditions for DDI:
| 2.1 |
| 2.2 |
| 2.3 |
where all the functions are evaluated at the homogeneous steady state and the subscripts are interpreted as a partial derivative of the function with respect to the subscripted variable.
The forms of the reaction kinetics depend on the problem at hand. Typical kinetics include the Gierer–Meinhardt [2] model
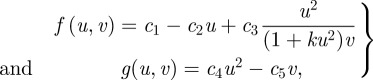 |
2.4 |
and, as a special case of one of the models proposed by Gierer & Meinhardt [2], the Schnakenberg [3] model:
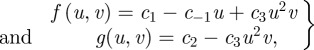 |
2.5 |
as well as the Thomas [4] reaction kinetics:
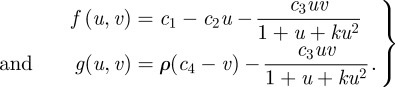 |
2.6 |
In each case, the parameters {c−1, c1, c2, …} represent the deterministic rates of the reactions, and a typical steady-state solution is shown in figure 1. The Turing model, and extensions thereof, has been applied to patterning in Hydra [5]; pigmentation patterning in fish (figure 2; [6]); shell markings, skeletal development in the limb, to name but a few examples.
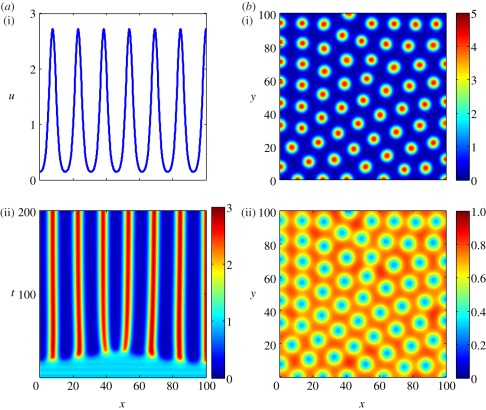
Figure 1.
Turing patterns produced through simulating the Schnakenberg kinetics, system (2.5). (a) One-dimensional simulations. (i) Final steady state of u after 1000 simulated time units. (ii) Temporal evolution. (b) Two-dimensional simulation. (i) Final steady state of u after 1000 simulated time units. (ii) Final steady state of v after 1000 simulated time units. The parameters are Du = 1, Dv = 40, c1 = 0.1, c2 = 0.9, c−1 = 1 and c3 = 1. The simulations have zero-flux boundary conditions. This means that none of the chemicals can diffuse out of the domain, i.e. the boundaries are impermeable. The initial conditions are random perturbations about the homogeneous steady state.
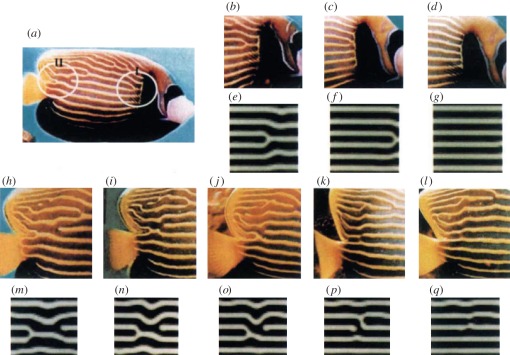
Figure 2.
Rearrangement of the stripe pattern of Pomocanthus imperator with accompanying computer simulations. Reproduced with permission from Macmillan Publishers Ltd: Kondo & Asai [6]. Copyright ©1995.
3. Properties of the model
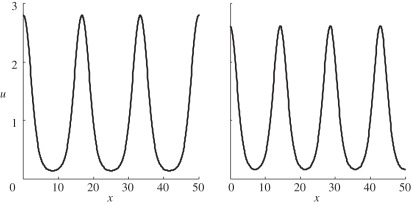
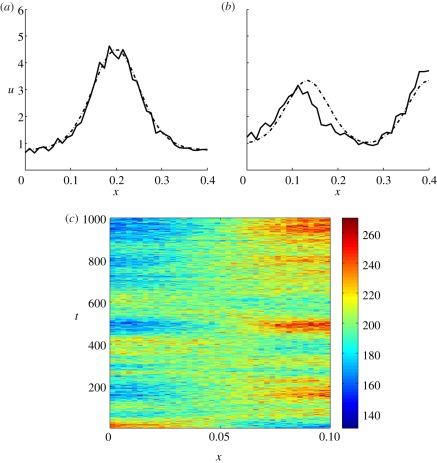
While Turing patterns have been observed in chemical systems, and morphogens identified in developmental biology, the existence of Turing patterns in biology is still an open question as concrete examples of Turing morphogens have not yet been elucidated. Thus, in light of the fact that we do not have a definitive reaction set in biology which patterns via a Turing mechanism, we can only explore hypothetical reactions, such as those mentioned above, to see if patterns can form. The parameter space for such patterns is determined by conditions (2.1)–(2.3) and it has been shown [7] that, for the standard reaction kinetic models used, this space is very restricted. Therefore, the model parameters need to be finely tuned and the patterns may therefore be non-robust in that small variations in parameters may move the system out of the Turing regime. Further, when we are in the Turing regime, different patterns can arise at the same point in parameter space, simply owing to slight variations in initial conditions ([8]; figure 3).
Figure 3.
Turing patterns produced through simulating the Schnakenberg kinetics, system (2.5), with two slightly different initial conditions. Boundary conditions and parameters are the same as in figure 1.
It has been shown that imposing different boundary conditions, such as homogeneous Dirichlet, can greatly enhance the robustness of patterns in a Turing system [9] by selecting, preferentially, certain modes at the expense of other modes which are no longer admissible. It has also been found that growth can induce robustness, and we shall now discuss this in more detail.
4. The effects of growth
Growth is an essential and readily observed process in development [10] that has been identified as an important factor in the production of spatial heterogeneity since it can fundamentally change the observed dynamics of patterning mechanisms [11]. In particular, Kondo & Asai [6] observed that, as the size of the marine angelfish Pomocanthus doubled, new stripes along the skin would develop between the old ones, thereby producing a near constant wavelength. This constant wavelength in fish pigmentation patterns is one of the defining features of a Turing pattern and is very suggestive of a Turing-like mechanism being responsible for the development of the skin pigmentation.
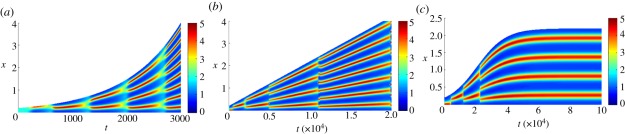
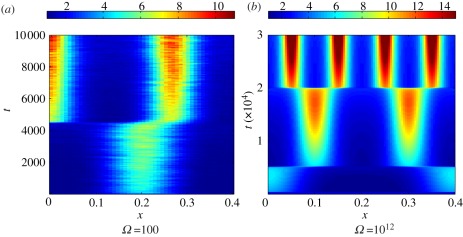
Growth was first incorporated into reaction–diffusion PDEs in an ad hoc manner. Arcuri & Murray [12] used a domain length with an explicit time dependence which treats domain growth as a reduction in strength of the diffusion rate. Their numerical simulations show a tendency to generate inconsistent pattern sequences. Owing to this apparent failure of the mechanism to generate reliable pattern sequences with domain growth, Arcuri & Murray concluded that robust biological patterns must form sequentially as any mechanism that acts over the whole domain is subject to too many sources of error for robust pattern formation. Through rigorous derivation from first principles, Crampin et al. [13] showed that uniform exponential domain growth can robustly generate certain wave forms under a persistent pattern doubling mechanism (figure 4a). Other forms of growth are also able to cause pattern doubling for a certain duration of time. However, as seen for linear growth in figure 4b, pattern doubling will eventually break down.
Figure 4.
Turing patterns on growing domains. In each image, the concentration u is visualized for the Schnakenberg kinetics, given by system (2.5). (a) Exponentially growing domain. (b) Linearly growing domain. (c) Logistically growing domain.
For logistic growth, figure 4c, not only is the system able to reliably tend to a unique mode, but it also does so even with small variations in the final domain length. This is particularly significant as it allows regulation without the need for parameter fine-tuning. This insensitivity, whereby a specific number of pattern elements are laid down despite significant variation in the domain size, is desirable in many biological systems. This is important in the context of biological pattern formation, as not only do patterns need to form, but, also, in many cases, it is imperative that the transition sequences are reliably reproducible.
However, even this mechanism is not without its problems as robust doubling is only realized within a specific (but large) region of growth rates. Outside of this region, the transition sequences once again break down. During the jump from one stable mode to another, the solution trajectory moves through a window where multiple other patterns may also be stable [14]. Thus, if the growth is too slow, period doubling may break down as even small system perturbations give rise to asymmetric peak splitting and period-doubling failure. Alternatively, if growth is too fast, period doubling breaks down because the system does not reside for sufficiently long in a specific range of stable wave modes to allow the intended pattern to become established.
5. The effects of noise
As discussed in §1, mathematical modelling is of fundamental importance in developmental biology owing to its ability to suggest and test mechanisms by which complex biological patterns can arise [15]. In all the cases discussed so far, deterministic systems of PDEs have been relied upon to capture the relevant dynamics. However, biological systems are frequently subject to noisy environments, inputs and signalling. These stochastic perturbations are important when considering the ability of such models to reproduce results consistently. In particular, owing to the Turing mechanism being very sensitive to perturbations, stochasticity is able to affect evolving patterns in ways not seen in deterministic simulations [16].
Previous research in the area of stochastic reaction–diffusion systems has focused on analysing and simulating the effects of external noise [17]. External noise is modelled as an additional effect that is controlled by the user; thus its properties are known a priori. Here, we focus on the role of intrinsic stochasticity, which stems from prescribing a probability that each reaction occurs, rather than an exact rate. For each stochastic system, a chemical master equation (CME) can be generated as an exact description of the evolution of the system [18]. Motivated by the central limit theorem, the stochastic populations, U and V, can then be linked to the concentrations, u and v, through the approximations
| 5.1 |
where Ω is the system size and links the population to the concentration scale, and ηu, ηv are random variables, assumed to be of order one. This is consistent with the observation that, as the population increases, stochastic effects are proportional to the inverse of the square root of the size of the population. By expanding the CME in terms of Ω, which is large on approaching the weak noise limit, we find that the first-order terms are exactly the deterministic equations. The second-order terms then lead to a Fokker–Plank equation that characterizes the properties of the noise. This Fokker–Plank equation can, in turn, be converted into a set of ordinary differential equations that defines the covariances of the noise [19]. Recently developed spatial Fourier transform techniques [20–22] can also be applied in order to gain an insight into the potential spatial dynamics possible in such systems.
Using these techniques, we can explore which pattern modes are stochastically excited inside and outside of the deterministic Turing domain. Within the parameter region that allows deterministic systems to realize Turing patterns, we have analytically shown that the stochastically excited wave modes correspond exactly to their deterministic analogues (figure 5; [23]). It is found that these stochastically excited wave modes grow exponentially with time and, since the noise perturbs the populations away from the uniform steady state, patterning is able to form much quicker in a stochastic system than its deterministic counterpart. Outside of this region, deterministic systems are unable to sustain viable patterns, whereas the noise inherent in a stochastic system is able to produce a state which is consistently far removed from the homogeneous steady state.
Figure 5.
(a,b) Deterministic (dashed-dotted line) and stochastic (solid line) simulations of the Schnakenberg kinetics, system (2.5), with zero-flux boundary conditions. Both systems are simulated with the same parameter values but with different initial conditions. The simulated population U has been rescaled in order to be comparable with the concentration u. Note that for the chosen parameters both one- and one-and-a-half-peak solutions are possible. (c) For the chosen parameters, the deterministic analysis predicts there are no patterns when the system is simulated on a domain of length L = 0.1. Here, we see stochastic effects are able to produce weak patterns that are able to change polarity. The parameters used are Du = 1 × 10−4, Dv = 1 × 10−2, c1 = 0.1, c−1 = 0.2, c2 = 0.3, c3 = 0.1, and the domain was discretized into K = 40 compartments. Reproduced with permission from Woolley et al. [21,23]. Copyright © 2011 American Physical Society.
Having considered the stationary domain case, we then consider the effects of growth on such stochastic systems. Since robustness in a deterministic Turing system can be achieved through domain growth, we may naïvely suspect that robustness will exist in the stochastic formulation. To achieve this, we use a domain transformation similar to that of Crampin et al. [13] in order to map the growing domain back to a stationary domain. In figure 6, we illustrate the effects of simulating stochastic kinetics on a uniformly, linearly growing domain for two different sizes of the stochastic population. For relatively small population levels (figure 6a), it is quickly realized that stochasticity destroys pattern-doubling robustness. For larger population levels (figure 6b), although pattern doubling is regained, stochasticity will eventually cause a breakdown in the mechanism since the transition from one pattern to the next becomes more delicate.
Figure 6.
Stochastic simulations of the Schnakenberg kinetics, system (2.5), with zero-flux boundary conditions and different population sizes, as governed by Ω which is listed beneath each image. In each case, the domain is growing uniformly linearly, but has been rescaled to occupy the stationary domain. The simulated population data U has been scaled by Ω; thus, each figure illustrates the evolution of concentration u. Reproduced with permission from Woolley et al. [21,23]. Copyright © 2011 American Physical Society.
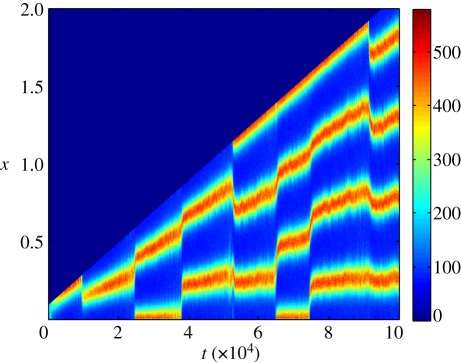
Although we lose period doubling as a mechanism of robustness, we have found that stochastic systems tend to excite consecutive patterns as the size of the domain increases. Hence, to generate robustness in a stochastic environment, we suggest apical growth as a plausible mechanism that can robustly support consecutive wave mode increasing pattern sequences (figure 7). Apical growth occurs when the growth is localized to one of the boundaries. Thus, if apical domain growth and wavenumber were connected in some form of feedback loop then, once the desired wavenumber is reached, growth would stop, leaving a stable pattern of exactly the desired wave mode.
Figure 7.
Stochastic Schnakenberg kinetics, system (2.5), simulated on a deterministic linearly growing domain (at the apex only). Only the population U is visualized. Reproduced with permission from Woolley et al. [21,23]. Copyright © 2011 American Physical Society.
6. The effects of delays
6.1. Gene-expression time delays
Modern experimental interrogations of suspected morphogen-based pattern formation systems, such as Nodal and Lefty zebrafish mesendodermal induction, use in situ hybridization [24–26]. This explicitly highlights local concentrations of specific mRNA transcripts and thus provides an indication of the rates of transcription of target genes, and emphasizes the role of gene expression in morphogenesis. Signals instigating changes in gene expression are transduced via tightly regulated biochemical cascades which ultimately result in protein production. This latter event proceeds via the transcription of nuclear DNA, forming mRNA which, in turn, is translated into protein within the cytosol. Proteins impact intercellular signalling and thus ultimately a feedback loop is created. However, signal transduction, gene transcription and mRNA translation take time; consequently, the feedback is delayed.
Empirical estimates for the magnitudes of transcriptional and translational time scales are dependent on the size of the genomic sequence and estimated to be from 10 min to several hours [27,28], which is sufficient to influence theoretical models of self-organization on developmental length- and time scales as illustrated below. However, the extent of these feedback delays is not seen in numerous theoretical patterning mechanisms that use Turing's DDI in regulating long-range cellular behaviour without gene expression, for instance, chemotactic cellular aggregation. Such systems may readily fall outside the scope of the following analyses as any feedback delays are much smaller than considered below and thus their impact, if any, is likely to be much more subtle though the subject of retarded dynamics is underexplored in these contexts.
Here, we simply focus on delays associated with the expression of genes for protein production, and these can be readily incorporated into the reaction diffusion systems considered thus far. For example, in the Gierer and Meinhardt kinetics, given by equation (2.4), the activator production proceeds by an autocatalytic interaction between two activator molecules, the rate of which is downregulated by the inhibitor, while inhibitor synthesis proceeds via activator cross catalysis. A biochemical interpretation of the form of the activator and inhibitor production can be summarized by
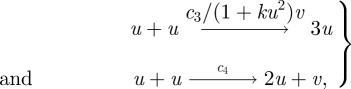 |
where c3/(1 + ku2)v and c4 are reaction rates; the v-dependent reaction rate crudely models the inhibitor's tendency to downregulate activator production.
We typically adopt two models for describing two extreme forms of gene-expression delays. In the first model, signal transduction for the production of morphogen is solely induced by reversible ligand binding at the cell surface. For the second model, all such signal transduction is assumed to proceed via internalized ligand. Additional models are briefly considered with combinations of these mechanisms of signal transduction. The first case is described by
and
where us, vs denote the activator and inhibitor proteins, respectively, at time s. Thus, morphogen-induced protein production at time t is dictated by interactions at time t – τi (i = 1,2), giving the equations
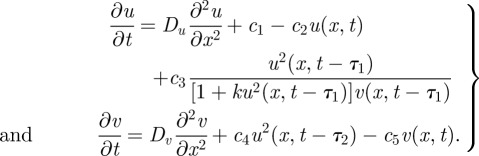 |
6.1 |
In contrast, the second case is described by
and
which yields the equations
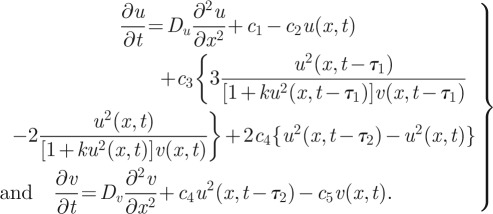 |
6.2 |
Hence, gene-expression time delays can be readily incorporated into Turing models on stationary domains for Gierer–Meinhardt kinetics and this is readily generalized for both growing domains and other Turing systems, as further motivated and illustrated in Gaffney & Monk [29], Seirin-Lee et al. [30] and Seirin-Lee & Gaffney [31]. Thus, our results are not limited to a special choice of kinetics or representation of the gene-expression dynamics and are likely to be generic.
6.1.1. The sensitivity of pattern structures, oscillating patterns and the breakdown of patterning
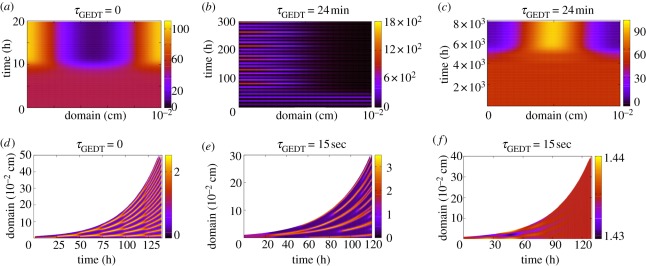
Critical effects of gene-expression time delays include the appearance of oscillating patterns with reversible ligand binding dynamics and patterning loss for both reversible ligand binding and ligand internalization dynamics (figure 8b,f). Once temporal oscillations occur, there is often an extraordinary loss of control of activator production, with an effective model breakdown, and predictions of extreme activator concentrations (figure 8b). Such pathological dynamics occurs consistently regardless of domain growth profiles. Furthermore, if the time delay is increased even slightly, the emergent pattern can fundamentally change, as shown in figure 8a–f. Such aberrant behaviour is observed even more extensively when the delay time scales differ in the representation of gene-expression dynamics [32]. Thus, the temporal sensitivity associated with gene-expression time-delay dynamics critically hinders the initiation and stabilization of Turing patterns, at least for the simple interaction models considered here.
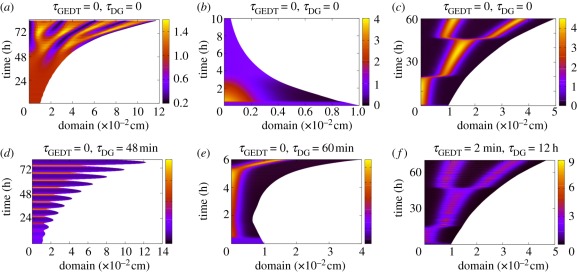
Figure 8.
Selected simulation results for delayed Gierer–Meinhardt models, (2.4), (6.1) and (6.2) on stationary and growing domains, with colour bars depicting activator concentration. (a–c) Gierer–Meinhardt delay models on stationary domains with non-saturating kinetics (k = 0 in equations (6.1) and (6.2)). (d–f) Gierer–Meinhardt delay models on uniformly exponentially growing domains with saturating kinetics (k > 0 in equations (6.1) and (6.2)). Here, the gene-expression time delays are all equal and denoted by τGEDT; thus τ1 = τ2 = τGEDT in equations (6.1) and (6.2). (b) Corresponds to reversible ligand-binding kinetics, while (c) and (e–f) represent ligand internalization. Initial conditions are given by a small perturbation from the homogeneous steady state. Only the initial conditions differ between plots (e) and (f); all other parameters are identical.
6.1.2. The aberrant patterning lag
A striking feature that can be observed on incorporating a gene-expression time delay within reaction–diffusion models is the dramatic and temporally sensitive increase in the patterning lag, as shown in figure 8a–c, especially for the ligand internalization models. In addition, gene-expression time delays sensitize the timing of the onset of patterning to fluctuations in the initial conditions for ligand internalization models on growing domains [31]. Such delays in the onset and/or stabilization of pattern impose potentially severe constraints on the applicability of reaction–diffusion models that rely on regulated gene expression, regardless of domain growth. These difficulties are particularly acute in the context of embryonic development, where the time scale of pattern establishment can be only a few hours and cells have limited time windows in which they are competent to respond to patterning cues [26].
6.1.3. Increase of the sensitivity to initial conditions
Introducing gene-expression time delays into the Gierer–Meinhardt model increases the sensitivity of the final pattern to fluctuations, such as those present in the initial conditions. This sensitivity is frequently undesirable in development, leading some authors to question Turing patterning as a feasible biological mechanism [8]. It has since been demonstrated that other mechanisms, such as domain growth, can greatly improve model robustness [13]. However, the Turing system with gene-expression time delays still exhibits sensitivity to the small perturbation of the initial conditions, for both stationary and growing domains, as illustrated by comparing, figure 8e–f.
6.2. Reactant-controlled domain growth and feedback delay dynamics
The coupling of pattern formation and the domain dynamics need not be solely one-directional as morphogens can also regulate tissue growth [33,34]. However, the response delay between cell proliferation signalling and its subsequent effect upon growth are governed by the time scales of the cell cycle, i.e. of the order of a day or more, and such an extensive retardation also has the potential to profoundly affect the Turing instability in the same manner as gene expression [35].
In fact, when tissue growth is near-uniform and the influence of morphogen concentrations on tissue growth is small, domain growth response delays do not noticeably affect pattern formation. In contrast, when the morphogens directly control tissue growth, response delays can have a critical effect on pattern formation and tissue growth. While the Schnakenberg and Gierer–Meinhardt models exhibit highly contrasting dynamics, numerous aberrant behaviours are manifested, especially domain shrinkage and oscillations, which are, in addition, highly sensitive to the size of the domain growth delay (figure 9). Thus, the dynamics of morphogen-controlled, rather than morphogen-perturbed, domain growth is difficult to reconcile with robust pattern formation via Turing morphogens, once growth response delays are incorporated. Furthermore, this mechanism does not appear to compensate for the difficulties observed once gene-expression time delays are considered within Turing's mechanism.
Figure 9.
Selected simulation results for Schnakenberg models on reactant-controlled growing domains with gene-expression time delays and domain growth delays. The Schnakenberg model with a gene-expression time delay, denoted τGEDT, is given by exchanging u(x,t)2v(x,t) with u(x,t–τGEDT)2v(x,t − τGEDT in (2.5). (a–e) Results for the Schnakenberg model with a domain growth response delay, denoted τDG. (c,f) Results for the Schnakenberg model with both domain growth response delay and gene-expression time delays. Here, the colour bar depicts the concentration of the activator, and the initial conditions are given by a small perturbation from the homogeneous steady state. The functional dependence of the domain growth rate functions on the morphogens are the same for each plot in the pair (a,d), and similarly for the pair (b,e) and the pair (c,f); however, this functional dependence is different for inter-pair comparisons.
7. Discussion
This paper has discussed the main properties of, and issues with, the use of Turing's theory of morphogenesis to describe developmental pattern formation via the initiation and amplification of spatial heterogeneity. In particular, we have illustrated that there is still the problem of robustness. This highlights the need to investigate the effects of including stochasticity and additional biological complexity, such as detailed gene-expression dynamics, within the context of Turing's mechanism, with particular regard to the sensitivity of the model outputs.
Although only one-dimensional intervals and two-dimensional squares have been discussed in this paper, the ideas are generalizable to higher dimensions [36] and different geometries [37]. However, with increasing dimension, the number of possible patterns and arrangements of these patterns also increases. For example, in two dimensions, we can produce either spots or stripes [38], with the spots having the ability to be arranged in square, diamond, rhombic or hexagonal patterns [39]. In three-dimensions lamellae, prisms and various other cubic structures, all exist making prediction even more difficult [40,41].
We have seen that such a simple addition as growth enables the models to produce much richer dynamics than would otherwise be possible. In particular, we can generate a frequency-doubling sequence of patterns, providing the speed of growth is not too fast, nor too slow. Importantly, this sequence is not sensitive to the initial conditions, contrary to the results of Saunders & Ho [42] which are based on the numerical analyses of a model by Arcuri & Murray [12]. Further, for logistic growth, not only is the system able to reliably tend to a unique mode, it does so even with variations of the final domain length; a desirable property in many biological systems.
We then discussed the effects of intrinsic noise, first considering the specific example of Schnakenberg kinetics on a stationary domain. Here it was seen (and it can be shown analytically) that, within the deterministic Turing instability regime, the uniform steady state of the stochastic system is linearly unstable to exactly the same wave modes as the deterministic system. Similar to the standard Turing analysis, although we are able to suggest which wave modes were excited, we are unable to predict which mode the final pattern will adopt. This is due to the linearization process, which suggests that all excited modes tend to grow exponentially and ignores the higher order terms that become important as the solution evolves. However, if the number of excited modes is small, then the power spectrum suggests which mode grows most quickly. This, in turn, suggests the asymptotic behaviour of the system. Finally, parameters outside the deterministic Turing unstable region were considered and we showed that, owing to stochastic effects, not only are patterns possible, polarity switching also occurs (figure 5c).
Having investigated the stationary domain, we then moved on to consider uniformly growing domains. We saw that stochastic effects destroy any hope of robust peak-doubling sequences when species numbers are low. However, because the stochastic system excites exactly the same wave modes as the deterministic system, we suggest that the reason that the peak-doubling sequences break down is that the simulations simply switch to the mode number that is most rapidly growing, which increases consecutively. Finally, using this information, we demonstrated that apical growth can robustly generate any wave mode, owing to this consecutive increase in patterning mode.
Finally, we discussed the effects of delays, specifically those owing to gene expression and, subsequently, the response delays of tissue growth regulation. Until recently, the influence of such retardation has typically been neglected in theoretical studies of Turing models and yet, as we have seen, it may critically influence these systems. In general, typical Turing models lose robustness with the inclusion of delays associated with the time scales of gene expression or the cell cycle, regardless of the inclusion of customary stabilizing features such as tissue growth. This emphasizes the need for genuine multi-scale modelling coupling sub-cellular dynamics with large-scale self-organization within developmental biology. Numerous questions emerge, for instance, whether Turing-like behaviours are contingent on alternative pattern formation mechanisms, such as chemotaxis or haptotaxis, whose robustness to detailed sub-cellular temporal dynamics is underexplored. Alternatively, secondary stabilization mechanisms may emerge to regulate and control DDIs subject to substantial feedback delays given that even relatively simple patterning systems in cellular and developmental biology nonetheless operate within the complex framework of interacting gene networks and cross-talking signal transduction pathways. Thus, despite the difficulties associated with its sensitivity and the elusiveness of an experimental validation in cellular systems, Turing's mechanism is still being vigorously debated and explored in developmental biology, both theoretically and also experimentally, over half a century after its inception: a befitting testament to the genius behind the hypothesis.
Acknowledgements
P.K.M. was partially supported by a Royal Society Wolfson Research Merit Award. T.E.W. would like to thank the EPSRC for support. S.S.L. acknowledges funding from The Japan Society for The Promotion of Science.
References
- 1.Turing M. A. 1952. The chemical basis of morphogenesis. Phil. Trans. R. Soc. B 237, 37–72 [Google Scholar]
- 2.Gierer A., Meinhard H. 1972. A theory of biological pattern formation. Biol. Cybern. 12, 30–39 10.1007/BF00289234 (doi:10.1007/BF00289234) [DOI] [PubMed] [Google Scholar]
- 3.Schnakenberg J. 1979. Simple chemical reaction systems with limit cycle behaviour. J. Theor. Biol. 81, 389–400 10.1016/0022-5193(79)90042-0 (doi:10.1016/0022-5193(79)90042-0) [DOI] [PubMed] [Google Scholar]
- 4.Thomas D. 1975. Artifical enzyme membranes, transport, memory, and oscillatoryphenomena. In Analysis and control of immobilized enzyme systems (Thomas D., Kernevez J. P.), pp. 115–150. New York, NY: Springer Verlag. [Google Scholar]
- 5.Meinhardt H. 1982. Models of biological pattern formation, vol. 6 London, UK: Academic Press [Google Scholar]
- 6.Kondo S., Asai R. 1995. A reaction–diffusion wave on the skin of the marine angelfish Pomacanthus. Nature 376, 765–768 10.1038/376765a0 (doi:10.1038/376765a0) [DOI] [PubMed] [Google Scholar]
- 7.Murray J. D. 1982. Parameter space for Turing instability in reaction diffusion mechanisms: a comparison of models. J. Theor. Biol. 98, 143. 10.1016/0022-5193(82)90063-7 (doi:10.1016/0022-5193(82)90063-7) [DOI] [PubMed] [Google Scholar]
- 8.Bard J., Lauder I. 1974. How well does Turing's theory of morphogenesis work? J. Theor. Biol. 45, 501–531 10.1016/0022-5193(74)90128-3 (doi:10.1016/0022-5193(74)90128-3) [DOI] [PubMed] [Google Scholar]
- 9.Dillon R., Maini P. K., Othmer H. G. 1994. Pattern formation in generalized Turing systems. J. Math. Biol. 32, 345–393 10.1007/BF00160165 (doi:10.1007/BF00160165) [DOI] [Google Scholar]
- 10.Gicquel C., Le Bouc Y. 2006. Hormonal regulation of fetal growth. Horm. Res. Paediatr. 65, 28–33 [DOI] [PubMed] [Google Scholar]
- 11.Maini P. K., Crampin E. J., Madzvamuse A., Wathen A. J., Thomas R. D. K. 2003. Implications of domain growth in morphogenesis. In Mathematical Modelling & Computing in Biology and Medicine: 5th ESMTB Conf. 2002, vol. 1, 67–73 [Google Scholar]
- 12.Arcuri P., Murray J. D. 1986. Pattern sensitivity to boundary and initial conditions in reaction–diffusion models. J. Math. Biol. 24, 141–165 10.1007/BF00275996 (doi:10.1007/BF00275996) [DOI] [PubMed] [Google Scholar]
- 13.Crampin E. J., Gaffney E. A., Maini P. K. 1999. Reaction and diffusion on growing domains: scenarios for robust pattern formation. Bull. Math. Biol. 61, 1093–1120 10.1006/bulm.1999.0131 (doi:10.1006/bulm.1999.0131) [DOI] [PubMed] [Google Scholar]
- 14.Barrass I., Crampin E. J., Maini P. K. 2006. Mode transitions in a model reaction-diffusion system driven by domain growth and noise. Bull. Math. Biol. 68, 981–995 10.1007/s11538-006-9106-8 (doi:10.1007/s11538-006-9106-8) [DOI] [PubMed] [Google Scholar]
- 15.Maini P. K., Baker R. E., Chuong C. M. 2006. The Turing model comes of molecular age. Science 314, 1397–1398 10.1126/science.1136396 (doi:10.1126/science.1136396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biancalani T., Fanelli D., Di Patti F. 2010. Stochastic Turing patterns in the Brusselator model. Phys. Rev. E 81, 046215. 10.1103/PhysRevE.81.046215 (doi:10.1103/PhysRevE.81.046215) [DOI] [PubMed] [Google Scholar]
- 17.García-Ojalvo J., Sancho J. M. 1999. Noise in spatially extended systems. Berlin, Germany: Springer; 10.1007/978-1-4612-1536-3 (doi:10.1007/978-1-4612-1536-3) [DOI] [PubMed] [Google Scholar]
- 18.Gardiner C. W. 1985. Handbook of stochastic methods. Berlin, Germany: Springer [Google Scholar]
- 19.van Kampen N. G. 2007. Stochastic processes in physics and chemistry, 3rd edn. Amsterdam, The Netherlands: North Holland [Google Scholar]
- 20.Lugo C. A., McKane A. J. 2008. Quasicycles in a spatial predator-prey model. Phys. Rev. E 78, 051911. 10.1103/PhysRevE.78.051911 (doi:10.1103/PhysRevE.78.051911) [DOI] [PubMed] [Google Scholar]
- 21.Woolley T. E., Baker R. E., Gaffney E. A., Maini P. K. 2011. Influence of stochastic domain growth on pattern nucleation for diffusive systems with internal noise. Phys. Rev. E 84, 041905. 10.1103/PhysRevE.84.041905 (doi:10.1103/PhysRevE.84.041905) [DOI] [PubMed] [Google Scholar]
- 22.Woolley T. E., Baker R. E., Gaffney E. A., Maini P. K. 2011. Power spectra methods for a stochastic description of diffusion on deterministically growing domains. Phys. Rev. E 84, 021915. 10.1103/PhysRevE.84.021915 (doi:10.1103/PhysRevE.84.021915) [DOI] [PubMed] [Google Scholar]
- 23.Woolley T. E., Baker R. E., Gaffney E. A., Maini P. K. 2011. Stochastic reaction and diffusion on growing domains: understanding the breakdown of robust pattern formation. Phys. Rev. E 84, 046216. 10.1103/PhysRevE.84.046216 (doi:10.1103/PhysRevE.84.046216) [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Schier A. F. 2002. Lefty proteins are long-range inhibitors of squint-mediated nodal signaling. Curr. Biol. 12, 2124–2128 10.1016/S0960-9822(02)01362-3 (doi:10.1016/S0960-9822(02)01362-3) [DOI] [PubMed] [Google Scholar]
- 25.Jing X. H., Zhou S. M., Wang W. Q., Chen Y. 2006. Mechanisms underlying long- and short-range nodal signaling in Zebrafish. Mech. Dev. 123, 388–394 10.1016/j.mod.2006.03.006 (doi:10.1016/j.mod.2006.03.006) [DOI] [PubMed] [Google Scholar]
- 26.Solnica-Krezel L. 2003. Vertebrate development: taming the nodal waves. Curr. Biol. 13, R7–R9 10.1016/S0960-9822(02)01378-7 (doi:10.1016/S0960-9822(02)01378-7) [DOI] [PubMed] [Google Scholar]
- 27.Lewis J. 2003. Autoinhibition with transcriptional delay: a simple mechanism for the Zebrafish somitogenesis oscillator. Curr. Biol. 13, 1398–1408 10.1016/S0960-9822(03)00534-7 (doi:10.1016/S0960-9822(03)00534-7) [DOI] [PubMed] [Google Scholar]
- 28.Tennyson C. N., Klamut H. J., Worton R. G. 1995. The human dystrophin gene requires 16 hr to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9, 184–190 10.1038/ng0295-184 (doi:10.1038/ng0295-184) [DOI] [PubMed] [Google Scholar]
- 29.Gaffney E. A., Monk N. A. M. 2006. Gene expression time delays and Turing pattern formation system. Bull. Math. Biol. 68, 99–130 10.1007/s11538-006-9066-z (doi:10.1007/s11538-006-9066-z) [DOI] [PubMed] [Google Scholar]
- 30.Seirin-Lee S., Gaffney E. A., Monk N. A. M. 2010. The influence of gene expression time delays on Gierer-Meinhardt pattern formation systems. Bull. Math. Biol. 72, 2139–2160 10.1007/s11538-010-9532-5 (doi:10.1007/s11538-010-9532-5) [DOI] [PubMed] [Google Scholar]
- 31.Seirin-Lee S., Gaffney E. A. 2010. Aberrant behaviours of reaction diffusion self-organisation models on growing domains in the presence of gene expression time delays. Bull. Math. Biol. 72, 2161–2179 10.1007/s11538-010-9533-4 (doi:10.1007/s11538-010-9533-4) [DOI] [PubMed] [Google Scholar]
- 32.Gaffney E. A., Seirin-Lee S. In press. The sensitivity of Turing self-organisation to biological feedback delays: 2D models of Zebrafish pigmentation. Bull. Math. Biol. 10.1007/s11538-011-9634-8 (doi:10.1007/s11538-011-9634-8) [DOI] [PubMed] [Google Scholar]
- 33.Rogulja D., Irvine K. D. 2005. Regulation of cell proliferation by a morphogen gradient. Cell 123, 449–461 10.1016/j.cell.2005.08.030 (doi:10.1016/j.cell.2005.08.030) [DOI] [PubMed] [Google Scholar]
- 34.Tabata T., Takei Y. 2004. Morphogens, their identification and regulation. Development 131, 703–712 10.1242/dev.01043 (doi:10.1242/dev.01043) [DOI] [PubMed] [Google Scholar]
- 35.Seirin-Lee S., Gaffney E. A., Baker R. E. 2011. The dynamics of Turing patterns for morphogen-regulated growing domains with cellular response delays. Bull. Math. Biol. 73, 2527–2551 10.1007/s11538-011-9634-8 (doi:10.1007/s11538-011-9634-8) [DOI] [PubMed] [Google Scholar]
- 36.Painter K. J., Maini P. K., Othmer H. G. 1999. Stripe formation in juvenile Pomacanthus explained by a generalized Turing mechanism with chemotaxis. Proc. Natl Acad. Sci. USA 96, 5549–5554 10.1073/pnas.96.10.5549 (doi:10.1073/pnas.96.10.5549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gjorgjieva J., Jacobsen J. 2007. Turing patterns on growing spheres: the exponential case. Dynamical systems and differential equations. In Proc. 6th AIMS Int. Conf. Disc. Cont. Dyn. Sys. Supp., Poitiers, France, September 2007, pp. 436–445 Springfield, MO: AIMSciences. [Google Scholar]
- 38.Ermentrout B. 1991. Stripes or spots? Nonlinear effects in bifurcation of reaction-diffusion equations on the square. Proc. R. Soc. Lond. A 434, 413–417 10.1098/rspa.1991.0100 (doi:10.1098/rspa.1991.0100) [DOI] [Google Scholar]
- 39.Leppänen T. 2004 Computational studies of pattern formation in turing systems. PhD thesis. Helsinki, Finland: Helsinki University of Technology Laboratory of Computational Engineering Publications. [Google Scholar]
- 40.Callahan T. K., Knobloch E. 1996. Bifurcations on the fcc lattice. Phys. Rev. E 53, 3559–3562 10.1103/PhysRevE.53.3559 (doi:10.1103/PhysRevE.53.3559) [DOI] [PubMed] [Google Scholar]
- 41.Callahan T. K., Knobloch E. 1999. Pattern formation in three-dimensional reaction-diffusion systems. Physica D 132, 339–362 10.1016/S0167-2789(99)00041-X (doi:10.1016/S0167-2789(99)00041-X) [DOI] [Google Scholar]
- 42.Saunders P. T., Ho M. W. 1995. Reliable segmentation by successive bifurcation. Bull. Math. Biol. 57, 539–556 [Google Scholar]