Abstract
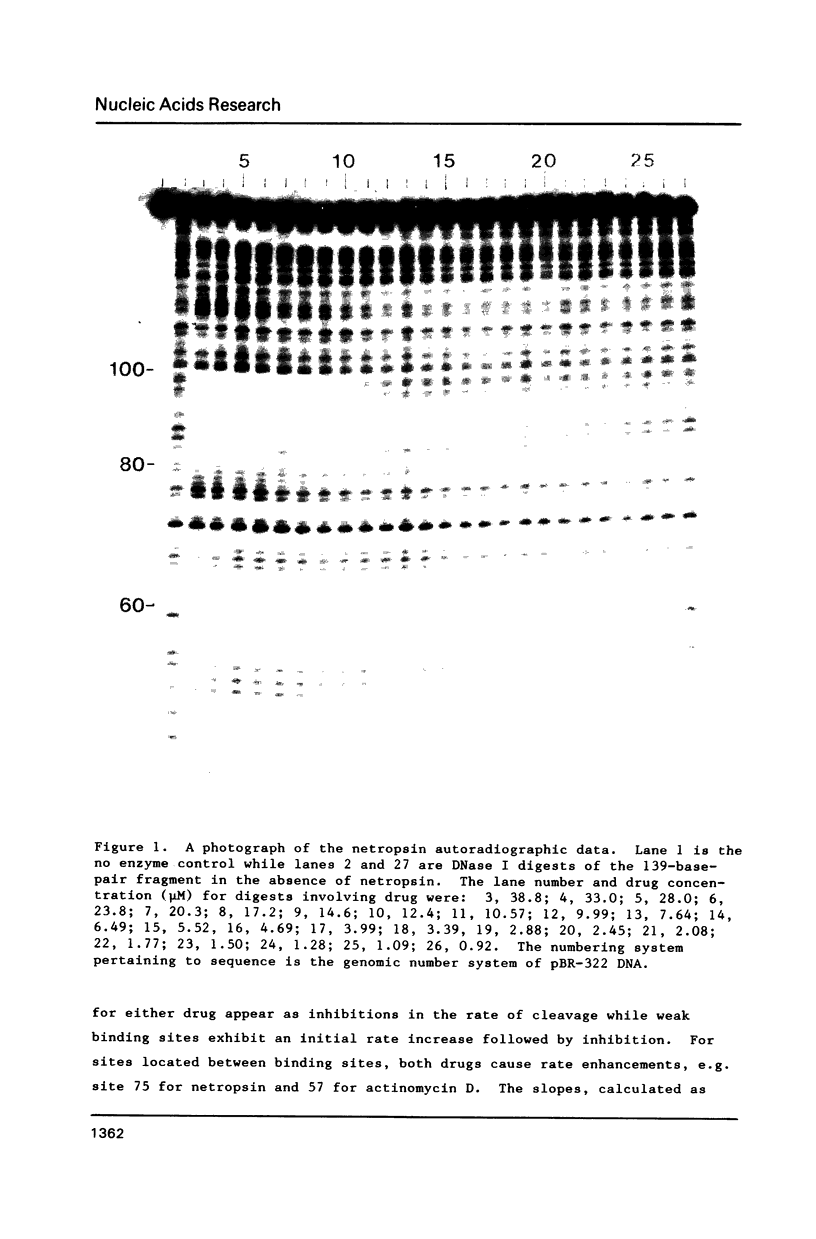
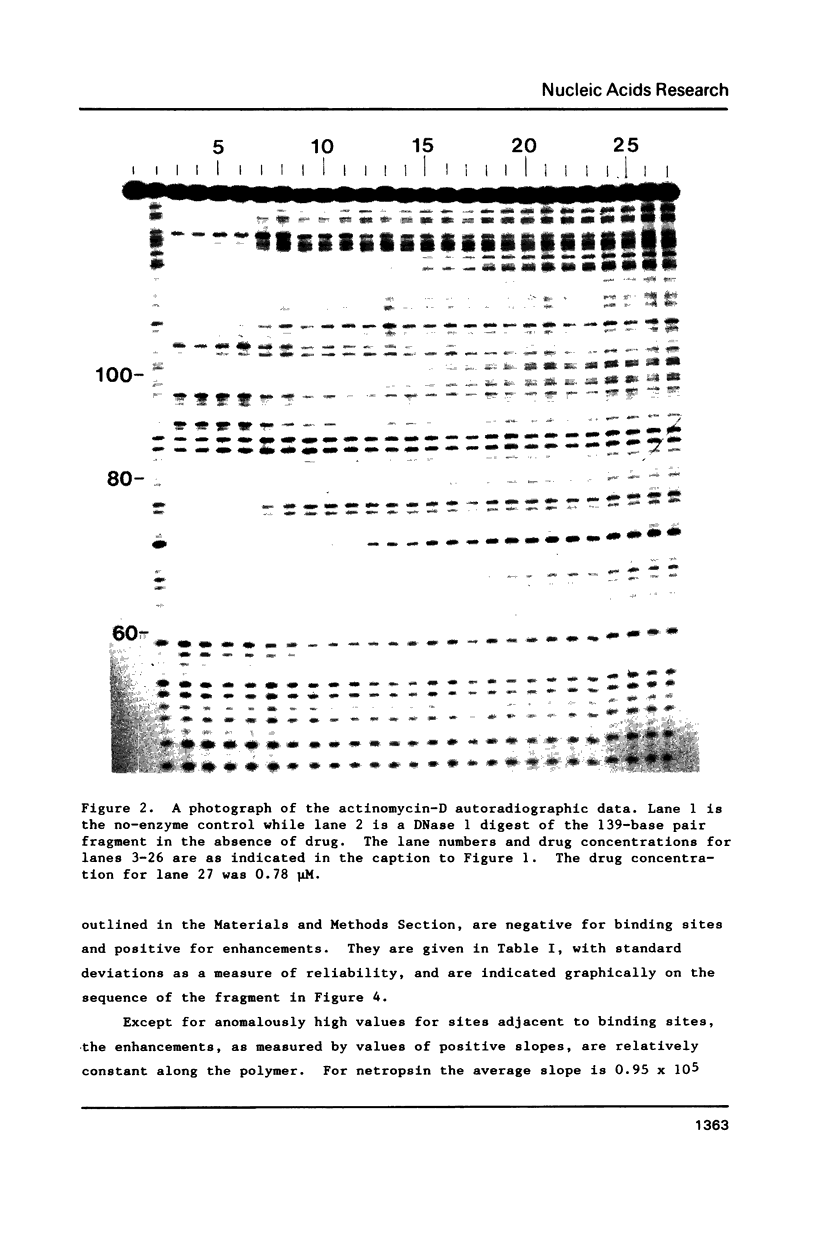
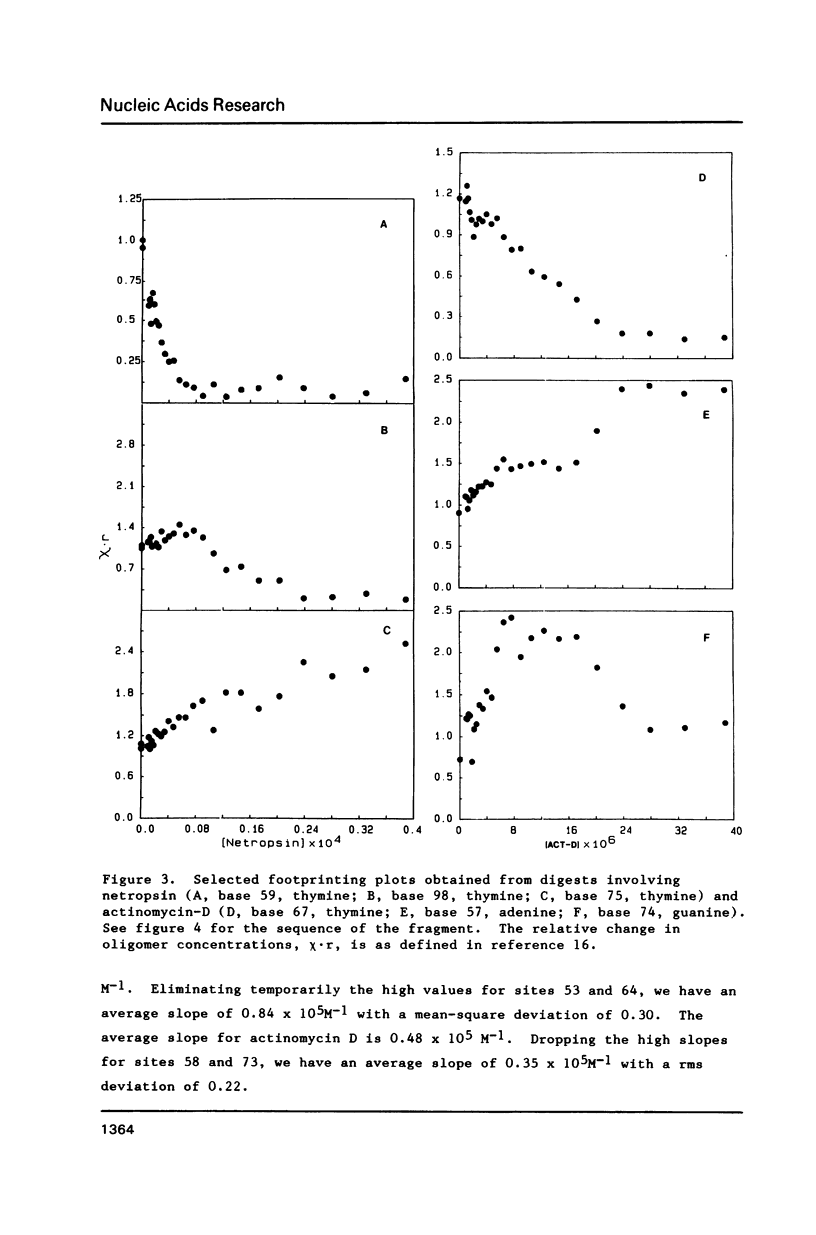
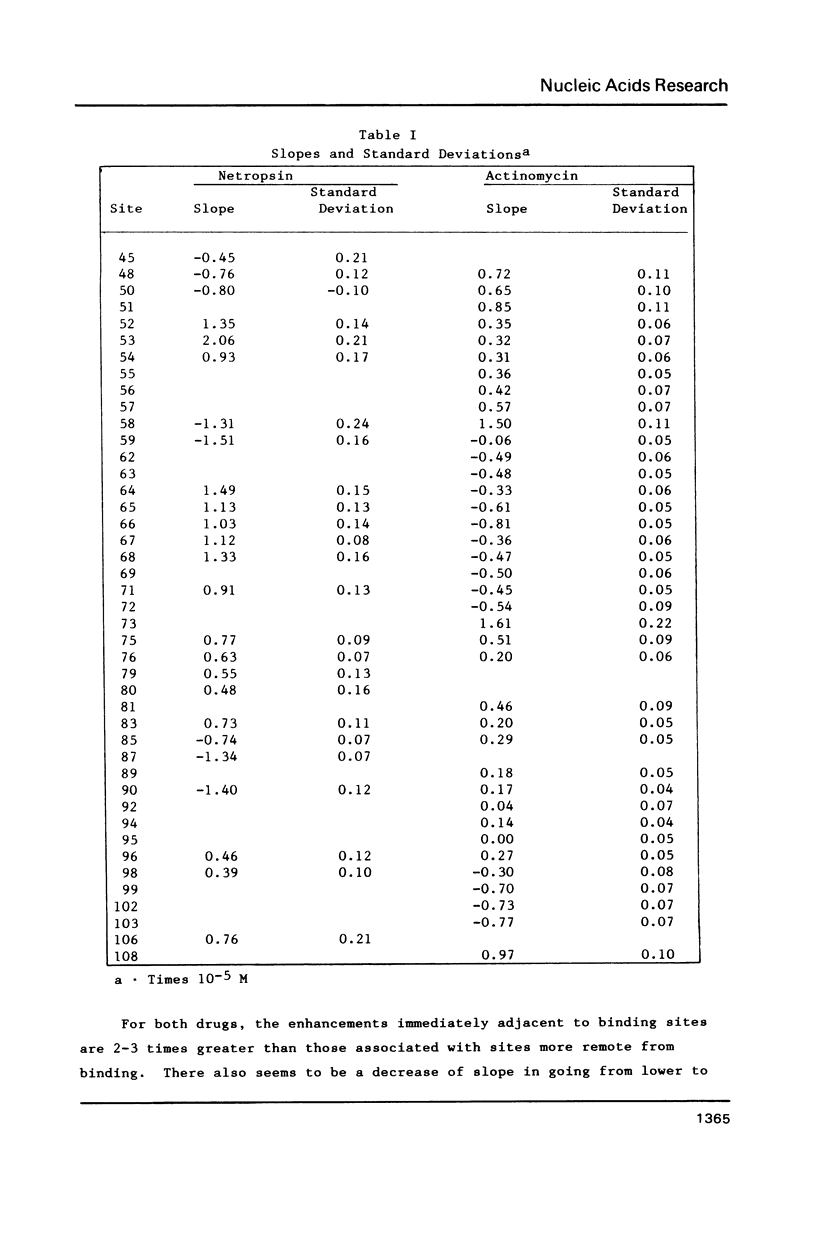
Footprinting experiments for DNase I digests of a 139-base-pair segment of pBR-322 DNA in the presence of either netropsin or actinomycin D were carried out. Plots of oligonucleotide concentration as a function of drug concentration were analyzed to study the enhancement in cleavage rates at approximately 30 sites, accompanying drug binding at other sites. The pattern of enhancements is not consistent with drug-induced DNA structural changes, but agrees with a redistribution mechanism involving DNase I. Since the total number of enzyme molecules per fragment remains unchanged, drug binding at some sites increases the enzyme concentration at other sites, giving rise to increased cleavage. The consequences of the redistribution mechanism for analysis of footprinting experiments are indicated.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. "Footprint" titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowiak J. C., Skorobogaty A., Rich N., Vary C. P., Vournakis J. N. Computer assisted microdensitometric analysis of footprinting autoradiographic DATA. Nucleic Acids Res. 1986 Jan 10;14(1):489–499. doi: 10.1093/nar/14.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Howarth N. R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley L. H., Needham-VanDevanter D. R., Lee C. S. Demonstration of the asymmetric effect of CC-1065 on local DNA structure using a site-directed adduct in a 117-base-pair fragment from M13mp1. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6412–6416. doi: 10.1073/pnas.84.18.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. J., Laplante S., Rehfuss R. P., Borer P. N., Cantor C. R. Actinomycin D facilitates transition of AT domains in molecules of sequence (AT)nAGCT(AT)n to a DNAse I detectable alternating structure. Nucleic Acids Res. 1987 Jan 26;15(2):839–852. doi: 10.1093/nar/15.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown J. W., Sondhi S. M., Ong C. W., Skorobogaty A., Kishikawa H., Dabrowiak J. C. Deoxyribonucleic acid cleavage specificity of a series of acridine- and acodazole-iron porphyrins as functional bleomycin models. Biochemistry. 1986 Sep 9;25(18):5111–5117. doi: 10.1021/bi00366a020. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Antibiotics which can alter the rotational orientation of nucleosome core DNA. Nucleic Acids Res. 1986 Nov 25;14(22):8735–8754. doi: 10.1093/nar/14.22.8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scamrov A. V., Beabealashvilli R. S. Binding of actinomycin D to DNA revealed by DNase I footprinting. FEBS Lett. 1983 Nov 28;164(1):97–101. doi: 10.1016/0014-5793(83)80027-1. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear D. F., Brenowitz M., Shea M. A., Ackers G. K. Energetics of cooperative protein-DNA interactions: comparison between quantitative deoxyribonuclease footprint titration and filter binding. Biochemistry. 1986 Nov 18;25(23):7344–7354. doi: 10.1021/bi00371a016. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. The stereochemistry of actinomycin binding to DNA and its implications in molecular biology. Prog Nucleic Acid Res Mol Biol. 1973;13:153–190. doi: 10.1016/s0079-6603(08)60103-8. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Dabrowiak J. C. Quantitative footprinting analysis of the netropsin-DNA interaction. J Biomol Struct Dyn. 1987 Apr;4(5):685–695. doi: 10.1080/07391102.1987.10507672. [DOI] [PubMed] [Google Scholar]