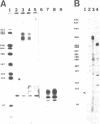
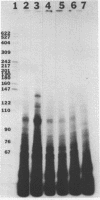
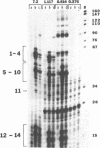
Abstract
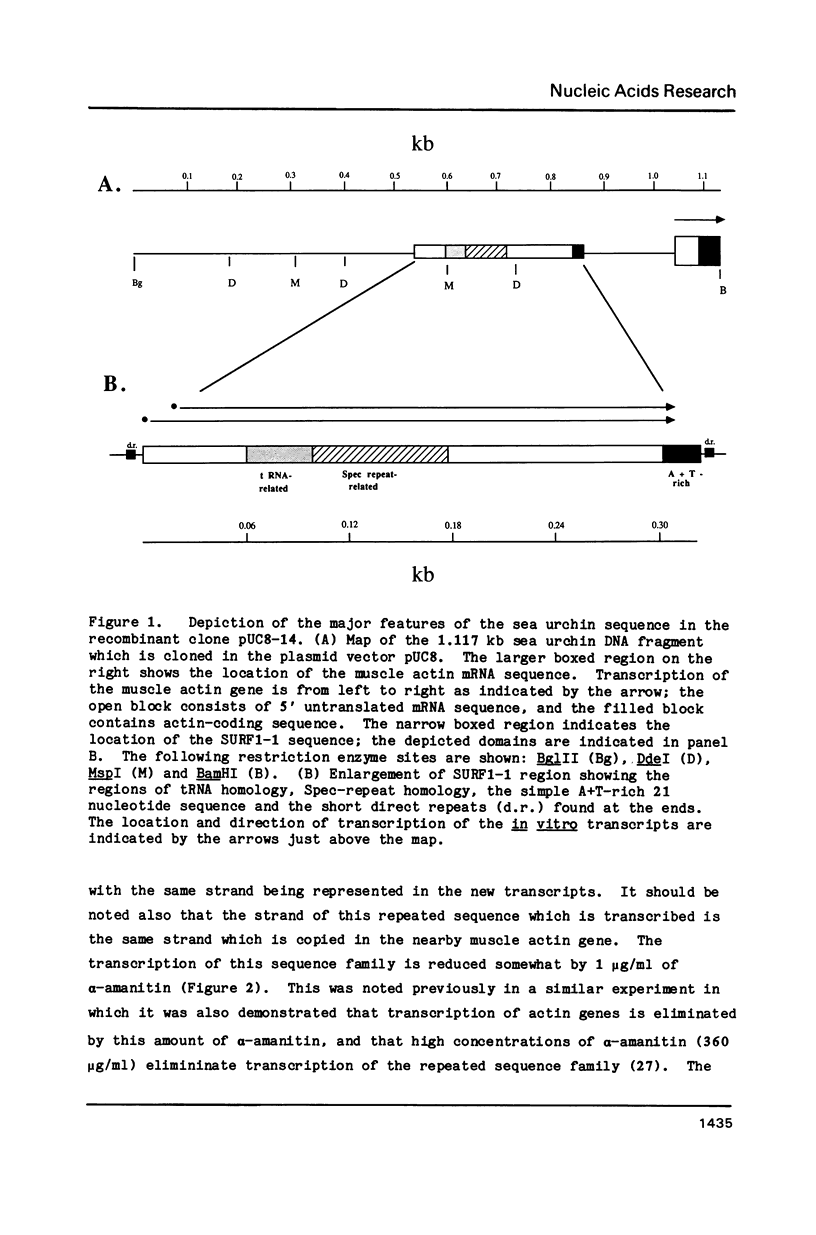
A repeated sequence element which is located about 200 nucleotides upstream from the protein-coding portion of the muscle actin gene (probably within a large 5' intron) in the genome of the sea urchin, Strongylocentrotus purpuratus has been characterized, and shown to contain the sequence features which indicate that it has been transposed by means of an RNA intermediate. This retroposon-like sequence, SURF1-1, is a member of a family which is dispersed and repeated about 800 times in the genome, referred to as SURF1 (sea urchin retroposon family 1). In vitro transcription of this sequence by RNA polymerase III defines a 300 nucleotide transcription unit which is bounded by a short direct repeated sequence. The 3' end of this unit contains a simple 21 nucleotide A+T-rich sequence characteristic of retroponons, and a consensus B box portion of an internal RNA polymerase III promotor is located 60 to 80 nucleotides downstream from the two sites of transcription initiation. This sequence also contains a 40 nucleotide region that is related to several tRNA sequences (containing the B box), and a 79 nucleotide sequence which is homologous to a repeated sequence previously shown to be present within the 3' untranslated portions of the Spec1 and Spec2 mRNAs of this species (1). Analysis of transcripts of this sequence family in RNA from several embryonic stages indicates that its expression is highest at 11 hours postfertilization (about 128 cells) and drops as development proceeds. Furthermore, most or all, transcription of this sequence family in nuclei isolated from 11 hour embryos is by RNA polymerase III, and is from the same strand which is transcribed in vitro.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. S., Eickbush T. H., Herrera R. J., Lizardi P. M. A highly reiterated family of transcribed oligo(A)-terminated, interspersed DNA elements in the genome of Bombyx mori. J Mol Biol. 1986 Feb 20;187(4):465–478. doi: 10.1016/0022-2836(86)90327-x. [DOI] [PubMed] [Google Scholar]
- Carpenter C. D., Bruskin A. M., Spain L. M., Eldon E. D., Klein W. H. The 3' untranslated regions of two related mRNAs contain an element highly repeated in the sea urchin genome. Nucleic Acids Res. 1982 Dec 11;10(23):7829–7842. doi: 10.1093/nar/10.23.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. B., Hoffman-Liebermann B., Kedes L. Structure and unusual characteristics of a new family of transposable elements in the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1985 Oct;5(10):2804–2813. doi: 10.1128/mcb.5.10.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. B., Liebermann D., Kedes L. Tsp transposons: a heterogeneous family of mobile sequences in the genome of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1985 Oct;5(10):2814–2825. doi: 10.1128/mcb.5.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain W. R., Jr, Boshar M. F., Cooper A. D., Durica D. S., Nagy A., Steffen D. The sequence of a sea urchin muscle actin gene suggests a gene conversion with a cytoskeletal actin gene. J Mol Evol. 1987;25(1):37–45. doi: 10.1007/BF02100039. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Durica D. S., Schloss J. A., Crain W. R., Jr Organization of actin gene sequences in the sea urchin: molecular cloning of an intron-containing DNA sequence coding for a cytoplasmic actin. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5683–5687. doi: 10.1073/pnas.77.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H., Okada N. Total DNA transcription in vitro: a procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci U S A. 1986 Jan;83(2):251–255. doi: 10.1073/pnas.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Deininger P. L., LaPorte P., Friedmann T., Geiduschek E. P. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981 Dec 11;9(23):6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Liebermann B., Liebermann D., Kedes L. H., Cohen S. N. TU elements: a heterogeneous family of modularly structured eucaryotic transposons. Mol Cell Biol. 1985 May;5(5):991–1001. doi: 10.1128/mcb.5.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Johnson S. A., Davidson E. H., Britten R. J. Insertion of a short repetitive sequence (D88I) in a sea urchin gene: a typical interspersed repeat? J Mol Evol. 1984;20(3-4):195–201. doi: 10.1007/BF02104726. [DOI] [PubMed] [Google Scholar]
- Klein W. H., Thomas T. L., Lai C., Scheller R. H., Britten R. J., Davidson E. H. Characteristics of individual repetitive sequence families in the sea urchin genome studied with cloned repeats. Cell. 1978 Aug;14(4):889–900. doi: 10.1016/0092-8674(78)90344-6. [DOI] [PubMed] [Google Scholar]
- Lawrence C. B., McDonnell D. P., Ramsey W. J. Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res. 1985 Jun 25;13(12):4239–4252. doi: 10.1093/nar/13.12.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann D., Hoffman-Liebermann B., Weinthal J., Childs G., Maxson R., Mauron A., Cohen S. N., Kedes L. An unusual transposon with long terminal inverted repeats in the sea urchin Strongylocentrotus purpuratus. Nature. 1983 Nov 24;306(5941):342–347. doi: 10.1038/306342a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Murakami K., Okada N. Gene for lysine tRNA1 may be a progenitor of the highly repetitive and transcribable sequences present in the salmon genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3156–3160. doi: 10.1073/pnas.83.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murphy D., Brickell P. M., Latchman D. S., Willison K., Rigby P. W. Transcripts regulated during normal embryonic development and oncogenic transformation share a repetitive element. Cell. 1983 Dec;35(3 Pt 2):865–871. doi: 10.1016/0092-8674(83)90119-8. [DOI] [PubMed] [Google Scholar]
- Perez-Stable C., Ayres T. M., Shen C. K. Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5291–5295. doi: 10.1073/pnas.81.17.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posakony J. W., Scheller R. H., Anderson D. M., Britten R. J., Davidson E. H. Repetitive sequences of the sea urchin genome. III. Nucleotide sequences of cloned repeat elements. J Mol Biol. 1981 Jun 15;149(1):41–67. doi: 10.1016/0022-2836(81)90259-x. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Okada N. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol. 1985;22(2):134–140. doi: 10.1007/BF02101691. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Singh K., Carey M., Saragosti S., Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985 Apr 11;314(6011):553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Yager L. N., Kaumeyer J. F., Lee I., Weinberg E. S. Insertion of an intermediate repetitive sequence into a sea urchin histone-gene spacer. J Mol Evol. 1987;24(4):346–356. doi: 10.1007/BF02134133. [DOI] [PubMed] [Google Scholar]