Abstract
Disrupted-In-Schizophrenia 1 (DISC1), a strong genetic candidate for psychiatric illness, encodes a multicompartmentalized molecular scaffold that regulates interacting proteins with key roles in neurodevelopment and plasticity. Missense DISC1 variants are associated with the risk of mental illness and with brain abnormalities in healthy carriers, but the underlying mechanisms are unclear. We examined the effect of rare and common DISC1 amino acid substitutions on subcellular targeting. We report that both the rare putatively causal variant 37W and the common variant 607F independently disrupt DISC1 nuclear targeting in a dominant-negative fashion, predicting that DISC1 nuclear expression is impaired in 37W and 607F carriers. In the nucleus, DISC1 interacts with the transcription factor Activating Transcription Factor 4 (ATF4), which is involved in the regulation of cellular stress responses, emotional behaviour and memory consolidation. At basal cAMP levels, wild-type DISC1 inhibits the transcriptional activity of ATF4, an effect that is weakened by both 37W and 607F independently, most likely as a consequence of their defective nuclear targeting. The common variant 607F additionally reduces DISC1/ATF4 interaction, which likely contributes to its weakened inhibitory effect. We also demonstrate that DISC1 modulates transcriptional responses to endoplasmic reticulum stress, and that this modulatory effect is ablated by 37W and 607F. By showing that DISC1 amino acid substitutions associated with psychiatric illness affect its regulatory function in ATF4-mediated transcription, our study highlights a potential mechanism by which these variants may impact on transcriptional events mediating cognition, emotional reactivity and stress responses, all processes of direct relevance to psychiatric illness.
INTRODUCTION
Disrupted-In-Schizophrenia 1 (DISC1) is a risk factor for brain disorders ranging from depression to schizophrenia (1). DISC1 encodes a multifunctional, multicompartmentalized scaffold protein with well-established roles in several aspects of neuronal physiology, including neural progenitor proliferation, migration and differentiation, as well as neurotransmission (2).
In the nucleus, DISC1 partially co-localizes with promyelocytic leukaemia nuclear bodies, which identify sites of active transcription (3), suggesting that DISC1 might be involved in transcriptional regulation. In support of this, DISC1 can interact with two highly related stress-responsive transcription factors, Activating Transcription Factor 4 (ATF4) and Activating Transcription Factor 5 (ATF5) (3–6), as well as the transcriptional repressor nuclear receptor co-repressor (N-CoR) (3). The first direct evidence for the involvement of nuclear DISC1 in transcriptional regulation was provided in a study by Sawamura et al. (3), who demonstrated that DISC1 can modulate cAMP-dependent cAMP-response element (CRE)-mediated transcription by interacting with ATF4.
ATF4 belongs to the activating transcription factor/CRE binding protein (ATF/CREB) family of basic region-leucine zipper (bZIP) transcription factors, which share the ability to bind to the CRE (7). Under basal conditions, ATF4 is expressed at very low levels, but its transcription and translation are rapidly upregulated in response to a range of different stressors (8). ATF4 can function both as a transcriptional activator and a transcriptional repressor (7). Genes whose expression is activated by ATF4 include pro- and anti-apoptotic factors (9,10), as well as genes involved in amino acid metabolism, regulation of the cell's redox balance and mitochondrial function (11). Additionally, ATF4 regulates emotional behaviour, synaptic plasticity and behavioural learning (12,13). Indeed, because of its ability to repress the CREB-mediated late phase of long-term potentiation (LTP) and long-term memory (LTM) (13), ATF4 has been referred to as a ‘memory suppressor gene’ (14,15). It is therefore possible that through its interaction with ATF4, DISC1 might contribute to the regulation of the transcriptional response to cellular stress as well as to emotional and LTP-inducing stimuli.
A number of DISC1 missense variants have been associated with the increased risk of psychiatric illness, altered brain morphology or cognitive deficits (1,2), but the molecular link between structural changes in DISC1 and clinical outcome has yet to be established. Several risk-conferring missense variants within DISC1 have the potential to modify the structure, biochemical properties and subcellular targeting of the protein, lending support to their putative pathogenic role (16). In this study, we examined the effect of a spectrum of common and rare amino acid substitutions of DISC1 associated with psychiatric illness on the nuclear targeting of the protein. We found that both the rare putatively causal variant 37W and the common 607F substitution impair nuclear targeting of DISC1, exerting a dominant-negative effect on the nuclear distribution of wild-type DISC1. Furthermore, the defective nuclear targeting of DISC1 variants 37W and 607F is reflected in their decreased ability to inhibit ATF4-dependent transcription. Recently, 607F was shown to impact on neural development by abrogating DISC1-mediated activation of wnt-dependent transcription (17). Our findings add to the evidence for a functional role of 607F in transcriptional regulation, and provide a direct link between risk-conferring genetic variants and aberrant targeting and function of nuclear DISC1.
RESULTS
Variants 37W and 607F decrease the nuclear abundance of DISC1
Several non-synonymous DISC1 variants have been associated with psychiatric illness and structural brain changes, and some have been shown to impact on specific aspects of DISC1 biology (1,2,17). Because DISC1 is a multicompartmentalized protein, we first assessed the impact of a panel of such disease-associated amino acid substitutions upon its subcellular distribution. We generated expression constructs (n = 20) carrying 37W, 432L or 603I (rare/ultra rare) or 607F (common) variants (1,2,18) in all possible combinations with the common polymorphisms R264Q and S704C (1,2). With the exception of R264Q and P432L, all of these DISC1 variants are at highly conserved positions, and all have the potential to influence the subcellular distribution of DISC1, either because they are predicted to disrupt critical structural motifs, or because they occur in regions of DISC1 that mediate binding to key partner proteins (16).
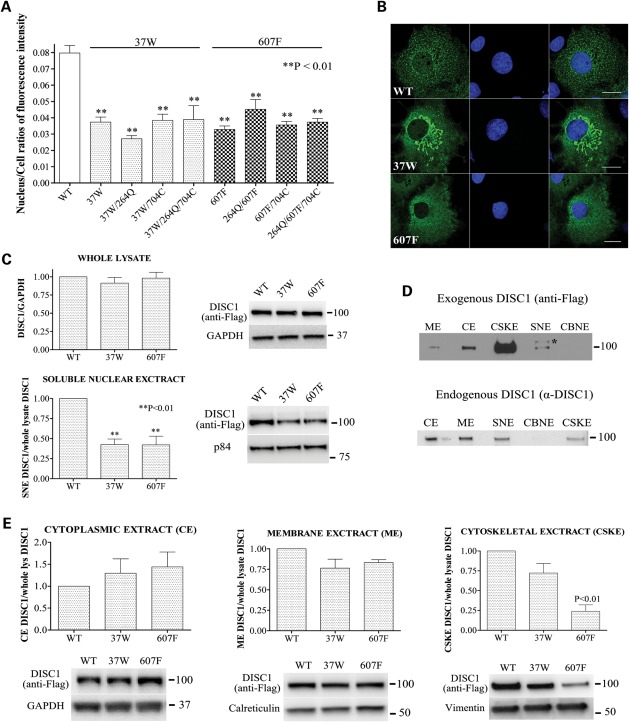
In a pilot experiment, we used immunocytochemistry to quantify the relative nuclear abundance of exogenous DISC1 in COS7 cells transfected with either one of the 20 DISC1 expression constructs. We found no evidence for an effect of substitutions at positions 264, 432, 603 or 704 on the nuclear targeting of DISC1 (Supplementary Material, Fig. S1), nor for gross alteration of the overall subcellular distribution of DISC1 (not shown). In contrast, both the R37W and the L607F substitutions result in depletion of exogenous DISC1 from the nucleus (Supplementary Material, Fig. S1). Additional variation at positions 264 and 704 does not modify the effect of 37W or 607F on the nuclear abundance of DISC1 (Supplementary Material, Fig. S1). These preliminary observations were confirmed by further immunocytochemical analysis on larger samples of cells (Fig. 1A and B). Both sequence variants reduce nuclear expression of DISC1 by ∼50% (P < 0.01). The observed decrease in nuclear expression of DISC1-37W or DISC1-607F is not due to decreased overall expression of these DISC1 variants (Supplementary Material, Fig. S2), which confirms that DISC1 carrying R or L at positions 37 and 607, respectively, is targeted to the nucleus more efficiently. DISC1-37W also induces formation of perinuclear mitochondrial clusters (Fig. 1B), which are the subject of a separate study (F. Ogawa, unpublished data). In addition, when compared with wild-type DISC1, DISC1-607F assumes a more diffuse distribution in the cytoplasm (Fig. 1B and Supplementary Material, Fig. S3). Since we found no evidence for an effect of amino acid variation at positions 264 and 704 on the subcellular distribution of DISC1, we performed all the subsequent experiments using DISC1 constructs encoding the common variants at these positions (264R and 704S). The common full-length DISC1 variant, to which all other variants analysed here are compared, will henceforth be referred to in the text as ‘wild-type (WT) DISC1’.
Figure 1.
Effect of DISC1 variants on its subcellular distribution. (A) Relative abundance of DISC1 variants in the nucleus of transfected COS7 cells calculated as the ratio between the total pixel intensity of DISC1 staining in the nucleus and the total pixel intensity of DISC1 staining in the whole cell. The bars represent the mean values measured in three independent experiments in which 50 cells/variant were analysed. (B) Representative confocal images of COS7 cells expressing wild-type DISC1, DISC1-37W or DISC1-607F. DISC1 is in green, and the nuclei are stained with DAPI (blue). Scale bars are 20 μm. (C) Distribution of wild-type DISC1, DISC1-37W or DISC1-607F in whole-cell lysates and soluble nuclear extracts (SNE) prepared from transfected SH-SY5Y cells. Each bar represents the average of four independent experiments. (D) Equal amounts of sub-cellular protein extracts obtained from SH-SY5Y cells that were either untransfected (bottom) or transfected with wild-type DISC1 (top) were analysed by western blotting using antibodies to detect endogenous or exogenous DISC1, respectively. ME, membrane extract; CE, cytoplasmic extract; CSKE, cytoskeletal extract; CBNE, chromatin-bound nuclear extract. *Non-specific band. (E) The indicated sub-cellular protein extracts were obtained from SH-SY5Y cells transfected with wild-type, 37W or 607F DISC1 and analysed by western blotting to detect exogenous DISC1, followed by band densitometry. The loading controls are proteins known to be preferentially enriched in either of the different subcellular fractions analysed. The bars represent the average of four independent experiments. All the densitometry data are normalized to the relative band intensity of wild-type DISC1. The position and size (kDa) of the protein markers is indicated.
To further examine the effect of DISC1 variants 37W and 607F on the subcellular distribution of the protein, we prepared whole-cell lysates and subcellular protein fractions from transfected and untransfected SH-SY5Y neuroblastoma cells and analysed them by western blotting. While the total protein levels of exogenous wild-type, DISC1-37W and DISC1-607F were comparable, we detected a ∼50% decrease in the relative nuclear abundance of DISC1-37W and DISC1-607F (P < 0.01, Fig. 1C), and observed a similar effect by immunocytochemistry (Supplementary Material, Fig. S4). This reduction in nuclear expression in SH-SY5Y cells is equivalent to that observed in COS7 cells. Besides being clearly detectable in the soluble nuclear protein extract, the full-length 100 kDa DISC1 isoform (both endogenous and exogenous) is also present in the cytoplasmic, membrane-bound and cytoskeletal extracts, but not in the chromatin-bound protein fraction (Fig. 1D). While endogenous DISC1 is predominantly enriched in the cytoplasmic and membrane-associated fractions, the vast majority of exogenous wild-type DISC1 is present in the cytoskeletal fraction (Fig. 1D). Interestingly, the relative protein abundance of DISC1-WT, DISC1-37W and DISC1-607F is comparable in the cytoplasmic and membrane-associated fractions; however, variant 607F is strongly depleted from the cytoskeletal fraction (P < 0.01, Fig. 1E), consistent with its aberrant cytoplasmic distribution (Fig. 1B and Supplementary Material, Fig. S3).
Dominant-negative effect of 37W and 607F upon wild-type DISC1 nuclear distribution
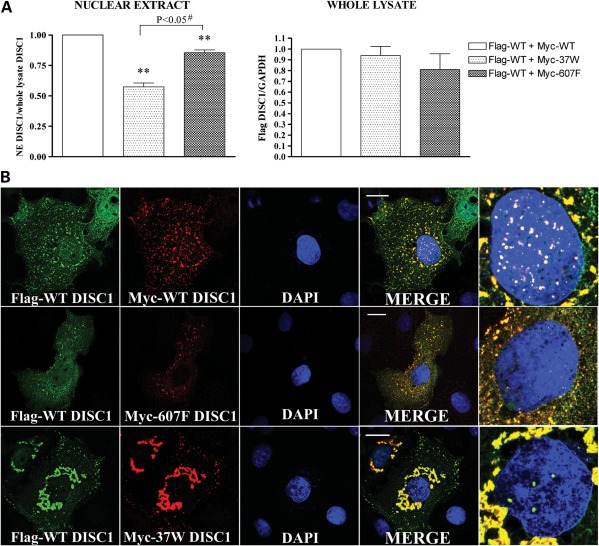
Next, due to the propensity of DISC1 to oligomerize (19–22), we asked if risk-conferring DISC1 variants 37W and 607F act in a dominant-negative fashion. Whole-cell lysates and nuclear protein extracts from SH-SY5Y cells expressing wild-type DISC1 alone or in combination with either DISC1-37W or DISC1-607F were analysed by western blotting. We verified that the different DISC1 expression constructs used in these experiments achieved comparable levels of protein expression in SH-SY5Y cells (Supplementary Material, Fig. S5). As shown in Figure 2A, co-expression of DISC1-37W or DISC1-607F results in a significant decrease in nuclear abundance of wild-type DISC1 (P < 0.01), with this effect being particularly pronounced for the 37W variant when compared with 607F (P < 0.05). To further examine this, we co-expressed DISC1-37W or DISC1-607F with wild-type DISC1 in COS7 and SH-SY5Y cells and analysed the sub-cellular distribution of each variant by immunocytochemistry, using cells transfected with wild-type DISC1 only as a control. As expected, when wild-type DISC1 is expressed alone, it translocates to the nucleus, where it is detectable as numerous bright puncta on a more diffuse background (Fig. 2B and Supplementary Material, Fig. S6). Consistent with the results of our subcellular protein fractionation experiment, both DISC1-37W and DISC1-607F reduce the formation of wild-type DISC1 puncta in the nucleus (Fig. 2B and Supplementary Material, Fig. S6). Furthermore, we noted that the cytoplasmic distribution of wild-type DISC1 appears more diffuse in the majority of cells co-expressing DISC1-607F (Fig. 2B and Supplementary Material, Fig. S6). Our results predict that 37W or 607F carriers will have substantially reduced nuclear DISC1 expression, and that 607F homozygotes will be similar to 607F heterozygotes in this particular respect.
Figure 2.
Dominant-negative effect of 37W and 607F DISC1. (A) SH-SY5Y cells were transfected with Flag-WT DISC1 and an equal amount of either Myc-DISC1-WT, Myc-DISC1-37W or Myc-DISC1-607F. The relative abundance of Flag-WT DISC1 was then quantified by western blotting in whole-cell lysates and nuclear extracts (NE) prepared from the transfected cells. The bars represent the average of three independent experiments. The data are normalized to the relative band density of Flag-DISC1 in samples expressing WT DISC1 only. (B) Sub-cellular distribution of Flag-DISC1 (green) and Myc-DISC1 (red) in representative COS7 cells expressing Flag-WT DISC1 in combination with an equal amount of Myc-DISC1-WT (top), Myc-DISC1-607F (middle) or Myc-DISC1-37W (bottom). Magnifications of the cell nuclei are shown in the far-right panels. **P < 0.01; #two-tailed paired Student's t-test. Scale bars are 20 μm.
Differential effect of DISC1 variants 37W and 607F on ATF4-mediated transcription
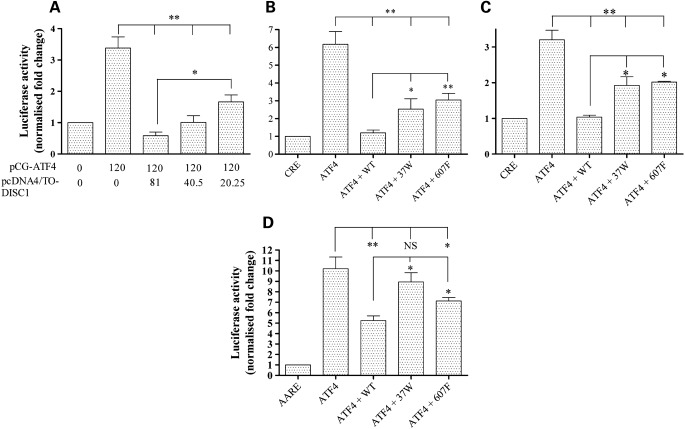
Sawamura et al. (3) showed that co-expression of DISC1 suppresses Gal4-ATF4-mediated transcription and that it enhances the ATF4-mediated inhibition of CRE-dependent transcription in response to increased intracellular cAMP levels. Since ATF4 has also been reported to activate CRE-mediated transcription under basal (low cAMP) conditions (23–25), we asked whether DISC1 regulates ATF4-mediated activation of the CRE at basal cAMP levels. As expected, in luciferase reporter assays carried out in HEK293 cells, we detected activation of CRE-driven transcription upon overexpression of ATF4, but not its dominant-negative mutant ATF4ΔRK, which lacks the DNA-binding domain (26,27) (Supplementary Material, Fig. S7). At basal cAMP levels, co-expression of DISC1 inhibits the ATF4-mediated transactivation of CRE-driven transcription in a dose-dependent manner (P < 0.01, Fig. 3A). Importantly, overexpression of DISC1 alone has no effect on the basal activity of the Som-CRE-luc reporter (Supplementary Material, Fig. S8), indicating that DISC1 acts via ATF4.
Figure 3.
Differential effect of DISC1 variants on ATF4-mediated transcription. (A) Relative CRE-dependent luciferase activity in HEK293 cells transfected with a fixed amount of pCG-ATF4 and decreasing amounts of wild-type DISC1. The numbers indicate the amount (ng/well) of plasmid DNA used for transfection. (B and C) Relative CRE-driven luciferase activity in HEK293 (B) or human oligodendrocyte MO3.13 cells (C) transfected with ATF4 only or in combination with the indicated DISC1 variants. (D) Relative CHOP AARE-driven luciferase activity in HEK293 cells transfected with ATF4 only or in combination with the indicated DISC1 variants. All data are normalized to the relative luciferase activity in cells transfected with the reporters only (CRE or AARE). The bars represent the average of at least three independent experiments. *P < 0.05, **P < 0.01.
Next, we tested the effect of DISC1 variants 37W and 607F. Both retain the ability to inhibit ATF4 transcriptional activity, but their inhibitory effect is significantly weaker compared with wild-type DISC1 (P < 0.05 for 37W, P < 0.01 for 607F, Fig. 3B). This is not caused by differences in expression levels (Supplementary Material, Fig. S9) nor it is limited to HEK293 cells, as we observed this same effect in MO3.13 human oligodendrocytes (P < 0.05 for both variants, Fig. 3C).
The transactivation activity of ATF4 is not limited to CRE-containing promoters. In fact, several ATF4 target genes involved in the response to cellular stresses such as amino acid limitation, oxidative stress or endoplasmic reticulum stress are activated through C/EBP-ATF Response Elements (CARE) in their promoters (28). For example, ATF4 induces expression of its target gene C/EBP homology protein (CHOP) by binding to a particular type of CARE in its promoter, the Amino Acid Response Element (AARE) (9,29,30). Thus, we sought to test whether DISC1 modulates the activity of ATF4 at the CHOP AARE. As expected, ATF4 strongly activates transcription from a CHOP AARE-luciferase reporter, but not from its mutant, non-responsive counterpart (31) (Supplementary Material, Fig. S10). As with the CRE, wild-type DISC1 significantly represses the ATF4-dependent transactivation of the CHOP AARE (P < 0.01, Fig. 3D). This inhibitory effect is reduced by DISC1-607F (P < 0.05, Fig. 3D) and DISC1-37W (P < 0.05, Fig. 3D).
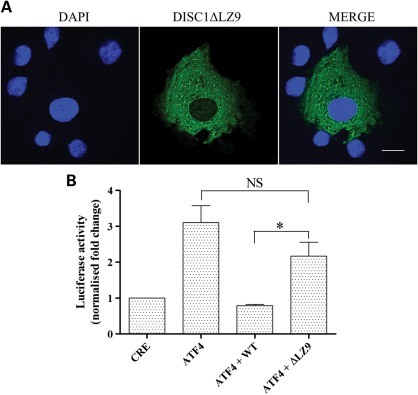
To further investigate the likely relationship between the decreased ability of DISC1 variants 37W and 607F to inhibit the transcriptional activity of ATF4 and their defective nuclear targeting, we tested a mutant form of DISC1 lacking the predicted leucine zipper in exon 9 (DISC1ΔLZ9), located between amino acids 607 and 628. Alanine substitutions at positions 614 and 621 prevent translocation of DISC1 to the nucleus (3), consistent with LZ9 being required for nuclear targeting. As expected, when expressed in COS7 cells, DISC1ΔLZ9 fails to accumulate in the nucleus, and assumes a diffuse distribution in the cytoplasm (Fig. 4A), closely resembling that of DISC1-607F (Fig. 1B, Supplementary Material, Figs S3 and S4). The remarkably similar effects of 607F and DISC1ΔLZ9 upon DISC1 nuclear localization are likely related to the predicted structural disruption of LZ9 by 607F (16). Consistent with its exclusion from the nucleus, DISC1ΔLZ9 does not significantly inhibit ATF4-mediated activation of CRE-dependent transcription (P < 0.05, Fig. 4B).
Figure 4.
Effect of DISC1 LZ9 on nuclear distribution of DISC1 and transcriptional regulation of ATF4. (A) Confocal image of a representative COS7 cell expressing DISC1ΔLZ9 (green). Nuclei are stained with DAPI (blue). (B) Relative luciferase activity in HEK293 cells transfected with the reporters only (CRE) or in combination with ATF4 with or without wild-type DISC1 or DISC1ΔLZ9. The bars represent the average of four independent experiments. The data are normalized to the relative luciferase activity in cells expressing the reporters only (CRE). *P < 0.05, two-tailed paired Student's t-test. The scale bar is 20 μm.
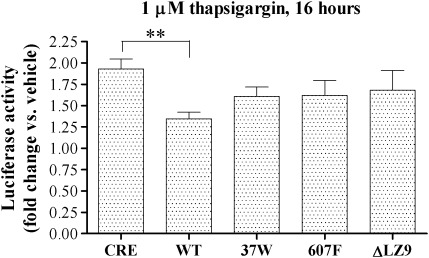
ATF4 plays a central role in mediating the cellular response to a range of damaging stimuli, including endoplasmic reticulum stress (7). Following on from our finding that DISC1 represses the transcriptional activity of exogenous ATF4, we examined the effect of DISC1 on the transcriptional activity of endogenously induced ATF4 using the endoplasmic reticulum stress inducer thapsigargin. In line with previously reported observations (32), thapsigargin treatment induces expression of endogenous ATF4 (Supplementary Material, Fig. S11) and determines a ∼2-fold activation of CRE-dependent transcription (Fig. 5). Overexpression of wild-type DISC1, but not variants 37W and 607F or the mutant DISC1ΔLZ9, significantly inhibits the thapsigargin-induced activation of CRE-dependent transcription (Fig. 5).
Figure 5.
Effect of DISC1 on the CRE-mediated transcriptional response to thapsigargin. HEK293 cells were transfected with the reporters only (CRE) or in combination with the indicated expression constructs. Starting from 32h post-transfection, the cells were exposed to thapsigargin or vehicle (DMSO) for the indicated time before being assayed for luciferase activity. The bars represent the average of at least three independent experiments. **P < 0.01.
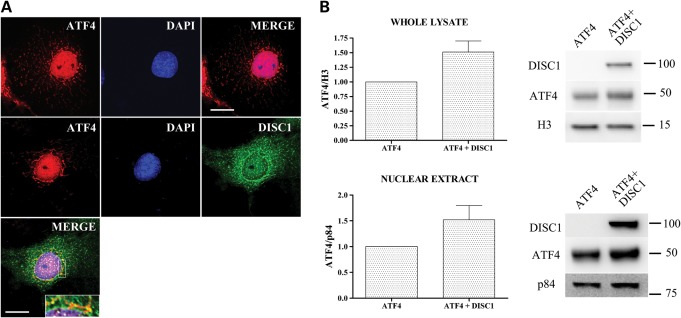
Morris et al. (4) showed that co-expression of full-length DISC1 inhibits accumulation of ATF5 in the cell nucleus. Since ATF4 and ATF5 are structurally closely related (33), we asked whether DISC1 impacts on the nuclear distribution and/or protein levels of ATF4, which may contribute to the observed transcriptional inhibition. Surprisingly, in cells transfected using the same ATF4/DISC1 DNA ratio used in the luciferase reporter assays, co-expression of DISC1 does not decrease the overall protein expression or nuclear targeting of exogenous ATF4, but instead seems to have the opposite effect, although this does not reach statistical significance (Fig. 6A and B). DISC1 therefore apparently does not inhibit ATF4-mediated transcription by reducing nuclear ATF4 expression.
Figure 6.
DISC1 does not affect nuclear targeting and protein levels of ATF4. (A) Representative COS7 cells transfected with ATF4 alone (top panels) or in combination with DISC1 (lower panels). The area delimited by a white rectangle is magnified in the inset. (B) ATF4 protein levels in whole-cell lysates or nuclear extracts prepared from HEK293 cells transfected with ATF4 alone or in combination with DISC1. The ATF4/DISC1 DNA ratio used in these experiments was the same as that used for the luciferase reporter assays. Loading controls are histone 3 (H3) for whole-cell lysates and nuclear matrix protein p84 for nuclear extracts. The bars represent the average of three independent experiments. Scale bars are 20 μm. The position and size (kDa) of the protein markers is indicated.
Although exogenous ATF4 is highly enriched in the nucleus, we noticed that it is also detectable in the perinuclear region in a pattern closely resembling the typical morphology of mitochondria (Fig. 6A), an organelle to which DISC1 is known to localize (34–36). Cytoplasmic ATF4 partially co-localizes with exogenous DISC1 in this location (Fig. 6A). To better test for a potential mitochondrial localization of exogenous ATF4, we used four different antibodies to detect the protein (both tagged and untagged) in transfected COS7 cells. In each case, we observed partial co-localization of ATF4 with mitochondria, particularly in the perinuclear region (Supplementary Material, Fig. S12).
DISC1 variants affect its interaction with ATF4
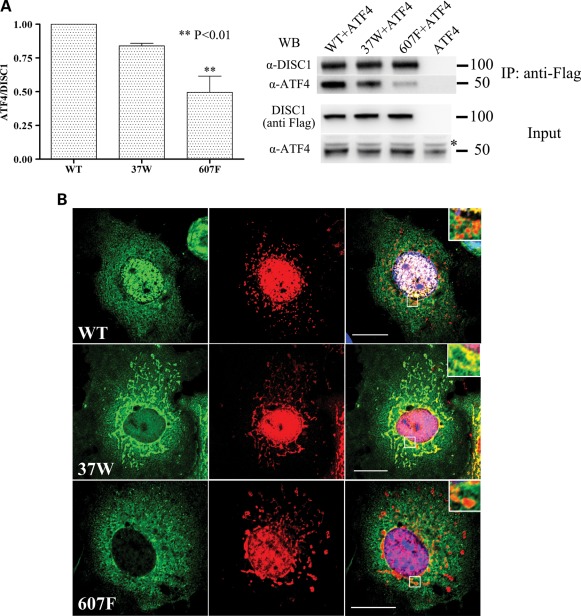
The L607F substitution is predicted to disrupt the Leucine packing in DISC1 LZ9, a region identified as essential to mediate binding to ATF4 (3), and could therefore directly impair the DISC1–ATF4 interaction. In addition, by introducing a physical segregation between the two proteins, the defective nuclear localization of DISC1 variants 37W and 607F might in itself impair their interaction with ATF4, the majority of which is found in the nucleus. This mechanism could contribute to their blunted inhibitory effect on ATF4-mediated transcription. As expected, in co-immunoprecipitation assays performed with exogenous proteins in HEK293 cells, DISC1-607F shows significantly impaired binding to ATF4 (Fig. 7A). However, the R37W substitution produces only a slight, non-significant decrease in DISC1 binding to ATF4 (Fig. 7A).
Figure 7.
Effect of DISC1 variants on the interaction between DISC1 and ATF4. (A) HEK293 cells were transfected with pCG-ATF4 alone or in combination with the indicated DISC1variants. Exogenous DISC1 was immunoprecipitated using an anti-Flag antibody and detected using an anti-DISC1 antibody (α-DISC1). Co-precipitating ATF4 was detected with an anti-ATF4 antibody (sc-200). The bars represent the average of three independent experiments. *Non-specific band. The position and size (kDa) of the protein markers is indicated. (B) COS7 cells transfected with the indicated variants of DISC1 (green) and ATF4 (red). The right-hand panels represent the corresponding merged images. The areas delimited by a white square are magnified in the insets. Scale bars are 20 μm.
The observed discrepancy between the defective nuclear targeting of DISC1-37W and its largely preserved interaction with ATF4 prompted us to analyse the sub-cellular distribution of the two proteins in co-transfected cells. Interestingly, unlike wild-type DISC1 and DISC1-607F, which exhibit limited co-localization with ATF4 outside the nucleus, DISC1-37W clearly co-distributes with ATF4 at mitochondria (Fig. 7B). We therefore conclude that the reduced capacity of DISC1-37W to inhibit ATF4-mediated transcription is due to its exclusion from the nucleus, rather than to reduced interaction with ATF4.
DISCUSSION
Growing evidence indicates that by establishing dynamic interactions with multiple binding partners, DISC1 functions as a hub protein whose principal role is to modulate various cellular processes in a space- and time-regulated manner. Sequence changes in DISC1 that disrupt its normal compartmentalization and protein interactions are therefore likely to have functional consequences, and may highlight biological processes involved in psychopathology.
In this study, we demonstrated that the putatively causal variant 37W and the common variant 607F both induce a ∼50% depletion of the nuclear pool of DISC1, and perturb the nuclear targeting of wild-type DISC1 in a dominant-negative fashion. In addition, both variants negatively impact on the ability of DISC1 to regulate transcription in response to exogenous ATF4 and endoplasmic reticulum stress. 37W was identified in a patient diagnosed with schizophrenia and not in 10 000 control alleles (18), and is thus a rare, putatively causal variant. In contrast, 607F is a common variant, present in ∼10% of the population. The single nucleotide polymorphism (SNP) determining variation at this position, or haplotypes including this SNP, is associated with schizophrenia, schizoaffective disorder, bipolar disorder and depression and correlate with symptom severity in schizophrenia, the P300 waveform mental illness endophenotype and influence brain structure and function (37–46). Singh et al. (17) recently assayed several DISC1 variants, including L607F and two other common variants R264Q and S704C for their effect on wnt signalling. Interestingly, they reported abnormal wnt signalling for 264Q and 607F, but not 704C, while we report abnormal nuclear localization and ATF4 binding for 607F, but not 264Q or 704C, consistent with a differential effect of non-synonymous amino acid substitutions on the varied and distinct functions of DISC1.
The 37W sequence change disrupts a highly conserved tetra-arginine nuclear localization signal in the otherwise poorly conserved head region of DISC1 (3), while the common variant 607F is located in a conserved predicted leucine zipper which was previously shown to contribute to DISC1 nuclear targeting and to be essential for interaction with ATF4 (3). The predicted disruption of this leucine zipper structural feature by L607F (16) therefore likely explains the reduced nuclear targeting and ATF4 binding of DISC1-607F, two effects that may potentially be interrelated. For both 37W and 607F sequence variants, the end result of reduced nuclear DISC1 expression most likely explains their decreased ability to repress ATF4 transcriptional activity.
Depending on the construct tested, DISC1 is capable of forming dimers, octamers and other oligomers and multimers (19,20,22,47,48). Thus, in heterozygous cells expressing wild-type and variant DISC1, hetero/oligomerization will likely occur, accounting for the dominant-negative effects reported here. Indeed, we have recently observed that DISC1-37W recruits wild-type DISC1 to perinuclear mitochondrial aggregates (F. Ogawa, unpublished data). It is therefore possible that the dominant-negative effect exerted by DISC1-37W on the nuclear targeting of wild-type DISC1 results from redistribution of wild-type DISC1 to mitochondria. We observed a significant, but milder reduction in nuclear abundance of wild-type DISC1 upon co-expression of 607F DISC1, and a change in the cytoplasmic distribution of wild-type DISC1 from punctate to diffuse. 607F is located close to a region of DISC1 (668–747) identified as essential for oligomerization (20) and it resides within a predicted oligomerization-promoting leucine zipper (16). Thus, the dominant-negative effect of DISC1-607F on the nuclear and cytoplasmic distribution of wild-type DISC1 may be related to the potentially altered oligomerization propensity of this variant form of DISC1. These observations indicate that nuclear expression of wild-type and variant DISC1 will likely be reduced in 37W or 607F carriers, with consequent effects for the role of DISC1 in transcription.
ATF4 transcriptional activity is modulated at multiple levels, including transcription and translation, post-translational modification and repression of its transcriptional activity through protein interactions (7,8,23,49–51). Mammalian cells respond to different types of environmental stressors by activating distinct stress-responsive kinases, all converging on phosphorylation of the α subunit of eukaryotic initiation factor 2 α (eIF2-α) (52). Phosphorylation of eIF2-α inhibits general protein synthesis while favouring the preferential translation of ATF4 (11,53,54). The regulation of ATF4 expression in response to stress also occurs at the transcriptional level, with different environmental stressors either activating or suppressing ATF4 mRNA synthesis (8). At the post-translational level, ATF4 is regulated by phosphorylation at multiple sites, which controls both the protein stability by regulating its ubiquitination (55–57), and its transcriptional activity (49,51). One further level of control of ATF4 resides in its interaction with binding partners that can directly inhibit its transcriptional activity, such as CHOP (58), neuronal cell death inducible putative kinase (NIPK) (23) and, as we and others (3) have reported, DISC1. The existence of multiple mechanisms controlling the production, persistence and activity of ATF4 indicates the critical importance of tight regulation of ATF4-mediated gene transcription. We and others (23–25,29,30) have demonstrated that ATF4 activates CRE- and AARE-mediated transcription at basal cAMP levels, while, in contrast, ATF4 represses CRE-mediated transcription in response to elevated cAMP (3,59). DISC1 inhibits both the transactivation and repression (3) activities of ATF4, and is thus likely to be an important modulator of ATF4-mediated transcription in the brain. Our observation that the repressive activity of DISC1 is weakened by sequence variants that influence risk of mental illness suggests that altered ATF4-mediated transcription may be a contributing factor to the overall disease risk.
ATF4 antagonizes the transcriptional activity of CREB, a key mediator of LTP and LTM (13,15,60,61). Therefore, through its modulatory effect on ATF4, DISC1 might be involved in the regulation of the transcriptional events that mediate synaptic plasticity. This in turn implies that DISC1 variants 37W and 607F may deregulate synaptic plasticity and cognitive processes through their defective modulation of ATF4 transcriptional activity. Since cognitive impairment is a core feature of schizophrenia, this is a potential route by which these DISC1 variants influence risk of mental illness.
Both DISC1 and ATF4 are implicated in the regulation of emotional behaviour in rodent models (12,62–67). ATF4 expression in the nucleus accumbens (NA), a key reward region in the brain, is induced by amphetamine administration or restraint stress, and ATF4 overexpression in this region decreases the behavioural responsiveness to amphetamine (12). Like its related transcription factor CREB, ATF4 attenuates emotional reactivity and induces depression-like behaviours when overexpressed in the NA, clearly indicating that ATF4 functions as an activator of CRE-driven transcription in this brain area (12). Like ATF4, DISC1 modulates the behavioural responsiveness to amphetamine in rodent models (66,68,69), and DISC1 mutations or altered expression are associated with depression-like behaviours in several mouse models (62–67). In the light of these findings, it is tempting to speculate that the role of DISC1 in emotional behaviour may be at least partly mediated by its modulation of ATF4 transcriptional activity on the CRE. If true, this would imply that DISC1 variants that interfere with this particular function of DISC1, such as 37W and 607F, may directly impact on the regulation of emotional behaviour.
ATF4 is also a key mediator of the integrated stress response, and its transcription and translation are strongly upregulated in response to a range of stressors, including amino acid deprivation, oxidative stress, hypoxia, viral infections, endoplasmic reticulum stress and mitochondrial dysfunction (28,70–73). The contribution of damaging environmental exposures to the risk of developing schizophrenia is well established, but mechanistically unclear. Maternal starvation, viral infections and perinatal hypoxia are among the best supported risk factors for schizophrenia (74), and each of these exposures could potentially activate the stress responses mediated by ATF4. It is intriguing to speculate that, by modulating the transcriptional activity of ATF4, DISC1 might contribute to regulation of cellular responses to stress, tipping the balance towards adaptation or apoptosis. In this scenario, 37W and 607F substitutions in DISC1 could increase the risk of mental illness by rendering the brain more susceptible to stress.
In conclusion, we have identified novel cellular and molecular phenotypes associated with common and rare DISC1 variants, and highlighted routes by which these psychiatric illness-associated variants could influence emotional and cognitive processes that are characteristically dysfunctional in mental illness, and the effects of environmental stressors that increase the risk of mental illness. Formal testing of gene–environment interactions has rarely been possible (75), but is of fundamental importance. Identification of a molecular mechanism that links the common DISC1 L607F polymorphism to the ATF4-mediated stress response provides an exciting opportunity to do so in cohorts with appropriate epidemiological data.
MATERIALS AND METHODS
Reagents and antibodies
DMSO and thapsigargin were from Sigma. Primary antibodies were: anti-Flag (F7425 and F3165) (Sigma), c-Myc (sc-40), anti-ATF4 (sc-200) (Santa Cruz) and anti-ATF4 (WH0000468M1) (Sigma), α-DISC1 (76), anti-p84 (ab487), anti-H3 (ab1791), anti-Calreticulin (ab22683), anti-Vimentin (ab8978) (Abcam) and anti-GAPDH (MAB347) (Millipore). TO-PRO3 and Mito Tracker Red were from Invitrogen.
Cell culture and transfection
MO3.13 cells (77) were a gift from Adrian Walmsley (Novartis Institute for Biomedical Research, Basel). All cell lines were cultured in DMEM supplemented with 10% FBS (Gibco) and maintained in a humidified incubator at 37°C and 5% CO2. Unless otherwise stated, HEK293 and COS7 cells were transfected with Lipofectamine 2000 (Invitrogen) and SH-SY5Y cells were transfected with FugeneHD (Roche), according to the manufacturer's directions.
Plasmids
The ATF4 expression construct pCG-ATF4 (24), encoding human ATF4, was donated by Adrian Harris (University of Oxford). pCG-ATF4ΔRK was obtained by mutating the DNA-binding domain of human ATF4 in pCG-ATF4 (294RYRQKKR300 to 294GYLEAAA300) by site-directed mutagenesis. The reporter Som-CRE-luc (Stratagene) and the TK-Renilla control luciferase vector were gifted by Richard Killick (King's College London). The reporter pGL4.23-CRE was generated by inserting four copies of the Somatostatin CRE (underlined) and its flanking regions (5′-AGCCTGACGTCAGAG-3′) upstream of the minimal promoter of the vector pGL4.23[luc2/minP] (Promega). The reporter vector pGL4.23-CHOP AARE and its mutant, non-responsive version pGL4.23-mutCHOP AARE, were generated by site-directed mutagenesis from pGL4.23[luc2/minP]. Mutagenic primers contained two copies of the core CHOP AARE (underlined) and its flanking regions: CHOP AARE, 5′-AACATTGCATCATCCCCGC-3′ and mut CHOP AARE, 5′-AACAATGCATCATCCCCGC-3′ (31), which only differed for the base in bold. All DISC1 expression constructs were generated from the plasmid pcDNA4/TO-Flag DISC1, coding for N-terminus Flag-tagged full-length human DISC1 (isoform L), by site-directed mutagenesis. All the site-directed mutagenesis reactions were performed using the QuikChange II or QuikChange Lightning site-directed mutagenesis kit (Stratagene), according to the manufacturer's directions. All the reporters and expression constructs were verified by direct sequencing.
Luciferase reporter assays
Most luciferase reporter assays were performed in HEK293 cells because they are a well-characterized model for the study of the transcriptional effects of ATF4 (30,78) and they consistently achieve high transfection rates, generating a strong and replicable luminescence signal. Cells were seeded in black-walled 96-well plates at a density of 6 × 104/well (HEK293) or 2.5 × 104/well (MO3.13) and transfected with Fugene HD (Roche Applied Science) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were transfected with 90 ng/well of CRE-luciferase reporter plasmid and 9 ng/well of TK Renilla luciferase vector to control for variation in transfection efficiency. The CRE-luciferase reporters were Som-CRE-luc for HEK293 cells and pGL4.23-CRE for MO3.13 cells. The ATF4 expression plasmids were used at a concentration of 120 ng/well and 15 ng/well for CRE-reporter and CHOP AARE-reporter assays, respectively. Unless otherwise stated, DISC1 expression plasmids were used at 81 ng/well. Where necessary, the empty vector pcDNA4/TO was added to the transfection mix to bring the total amount of transfected DNA to 300 ng/well. Background luminescence was measured in cells transfected with the empty vector pcDNA4/TO, and subtracted from the mean readings of each sample. Each transfection was performed in triplicate. Where indicated, drug treatments were started 32 h after transfection. When DMSO was used as vehicle, its final concentration in the medium was 0.1% or lower, and it did not affect cell viability or morphology. Luciferase activity was measured with the Dual Glo Luciferase Assay System (Promega), following the manufacturer's protocol.
Immunocytochemistry
To stain mitochondria, cells were incubated in 50 nm Mito Tracker Red in DMEM 10% FBS for 30 min before fixation. Cells were then fixed in methanol for 5 min at −20°C, followed by four washes in cold PBS. Fixed cells were blocked for 30 min in PBS containing 3% bovine serum albumin (PBS/BSA), and then incubated for 2 h at room temperature with the primary antibodies diluted in PBS/BSA. Anti-Flag antibodies F7425 and F3165 were used at 1:2000 and 1:10.000, respectively. The anti-c-Myc antibody was used at 1:500. The anti-ATF4 antibodies were used at 1:4000 (sc-200) and 1:2.500 (WH0000468M1). Secondary antibodies were Alexa Fluor goat anti-rabbit IgG 488 and 594 and Alexa Fluor goat anti-mouse IgG 488 and 594, all used at 1:1000. TO-PRO3 was used at 1:500. Images were acquired with a Zeiss LSM510 or a Nikon A1R confocal microscope.
Analysis of DISC1 subcellular distribution by immunofluorescence
COS7 cells transfected with equal amounts of the indicated DISC1 expression constructs were stained using an anti-Flag antibody (F7425) to detect exogenous DISC1, and nuclei were counterstained with TO-PRO3. Single plane confocal images of individual transfected cells were acquired using a Zeiss LSM510 confocal microscope (Zeiss). In each experiment, all the images were acquired using the same confocal settings. The investigator who acquired the images and performed the analysis was blinded to which variant of DISC1 had been transfected in each cell sample analysed. Image analysis was performed with IPLab version 3.9.5 r5 (BD Biosciences). For each cell image analysed, the total pixel intensity of DISC1 staining was measured both in the whole cell and in the nucleus only. The proportion of DISC1 staining localized in the nucleus was then calculated as the nucleus/cell ratio of total pixel intensities in individual cells.
Subcellular fractionation
Subcellular fractions were prepared from SH-SY5Y or HEK293 cells transfected with the indicated constructs for 72 or 24 h, respectively. In each fractionation experiment, a small proportion of the cell sample was lysed in RIPA buffer (150 mm NaCl, 50 mm Tris–HCl, pH 7.5, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with a protease inhibitor cocktail (Roche) for subsequent analysis of the expression level of exogenous proteins by western blotting. Subcellular fractions were obtained using the Subcellular Protein Fractionation Kit or the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Thermo Scientific) according to the manufacturer's instructions.
Immunoprecipitation and immunoblotting
For immunoprecipitation, HEK293 cells were lysed in PBS containing 1% Triton X-100 supplemented with a protease inhibitor cocktail (Roche) 24 h after transfection with the indicated plasmids. Immunoprecipitation was performed using an anti-Flag (F3165) antibody, following a standard protocol (79). Immunoblotting was performed as described (36). Chemiluminescent images were captured using the GeneGnome imaging system and densitometry was performed using the Gene Tools software, both from Syngene (Cambridge, UK). Densitometry was performed in parallel on duplicate western blots, in a minimum of three independent experiments. For the western blotting analysis of subcellular fractions, exogenous DISC1 band densities were first corrected using appropriate loading controls (nuclear matrix protein p84 for nuclear extracts, GAPDH for cytoplasmic extracts, calreticulin for membrane-bound extracts, vimentin for cytoskeletal extracts), and then divided for the respective GAPDH-corrected DISC1 band intensities in the corresponding whole-cell lysates.
Statistical analysis
Unless otherwise specified, data were analysed by one-way ANOVA followed by Dunnett's multiple comparison test.
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported by grants from The Wellcome Trust (083210/Z/07/Z) and the Medical Research Council (G0600214, G0902166). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust (083210/Z/07/Z) and the Medical Research Council (G0902166).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dinesh Soares for critical comments on the manuscript. We also thank Richard Killick for providing reagents and technical advice on luciferase assays, and Paul Perry for advice and technical support on image analysis.
Conflict of Interest statement. The authors declare no conflict of interest.
REFERENCES
- 1.Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol. Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. doi:10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw N.J., Porteous D.J. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;31:9043–9054. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawamura N., Ando T., Maruyama Y., Fujimuro M., Mochizuki H., Honjo K., Shimoda M., Toda H., Sawamura-Yamamoto T., Makuch L.A., et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol. Psychiatry. 2008;13:1138–1148. doi: 10.1038/mp.2008.101. 1069 doi:10.1038/mp.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris J.A., Kandpal G., Ma L., Austin C.P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. doi:10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 5.Ozeki Y., Tomoda T., Kleiderlein J., Kamiya A., Bord L., Fujii K., Okawa M., Yamada N., Hatten M.E., Snyder S.H., et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc. Natl Acad. Sci. USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. doi:10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millar J.K., Christie S., Porteous D.J. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem. Biophys. Res. Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. doi:10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- 7.Ameri K., Harris A.L. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. doi:10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Dey S., Baird T.D., Zhou D., Palam L.R., Spandau D.F., Wek R.C. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. doi:10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawcett T.W., Martindale J.L., Guyton K.Z., Hai T., Holbrook N.J. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 1999;339:135–141. doi:10.1042/0264-6021:3390135. [PMC free article] [PubMed] [Google Scholar]
- 10.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. doi:10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. doi:10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 12.Green T.A., Alibhai I.N., Unterberg S., Neve R.L., Ghose S., Tamminga C.A., Nestler E.J. Induction of activating transcription factors (ATFs) ATF2, ATF3, and ATF4 in the nucleus accumbens and their regulation of emotional behavior. J. Neurosci. 2008;28:2025–2032. doi: 10.1523/JNEUROSCI.5273-07.2008. doi:10.1523/JNEUROSCI.5273-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. doi:10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel T., Martin K.C., Bartsch D., Kandel E.R. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279:338–341. doi: 10.1126/science.279.5349.338. doi:10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- 15.Chen A., Muzzio I.A., Malleret G., Bartsch D., Verbitsky M., Pavlidis P., Yonan A.L., Vronskaya S., Grody M.B., Cepeda I., et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. doi:10.1016/S0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 16.Soares D.C., Carlyle B.C., Bradshaw N.J., Porteous D.J. DISC1: structure, function, and therapeutic potential for major mental illness. ACS Chem. Neurosci. 2011;2:609–632. doi: 10.1021/cn200062k. doi:10.1021/cn200062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh K.K., De Rienzo G., Drane L., Mao Y., Flood Z., Madison J., Ferreira M., Bergen S., King C., Sklar P., et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron. 2011;72:545–558. doi: 10.1016/j.neuron.2011.09.030. doi:10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song W., Li W., Feng J., Heston L.L., Scaringe W.A., Sommer S.S. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem. Biophys. Res. Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. doi:10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- 19.Leliveld S.R., Bader V., Hendriks P., Prikulis I., Sajnani G., Requena J.R., Korth C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 2008;28:3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. doi:10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leliveld S.R., Hendriks P., Michel M., Sajnani G., Bader V., Trossbach S., Prikulis I., Hartmann R., Jonas E., Willbold D., et al. Oligomer assembly of the C-terminal DISC1 domain (640–854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48:7746–7755. doi: 10.1021/bi900901e. doi:10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- 21.Ottis P., Bader V., Trossbach S.V., Kretzschmar H., Michel M., Leliveld S.R., Korth C. Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol. Psychiatry. 2011;70:604–610. doi: 10.1016/j.biopsych.2011.03.027. doi:10.1016/j.biopsych.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Narayanan S., Arthanari H., Wolfe M.S., Wagner G. Molecular characterization of disrupted in schizophrenia-1 risk variant S704C reveals the formation of altered oligomeric assembly. J. Biol. Chem. 2011;286:44266–44276. doi: 10.1074/jbc.M111.271593. doi:10.1074/jbc.M111.271593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ord D., Ord T. Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp. Cell Res. 2003;286:308–320. doi: 10.1016/s0014-4827(03)00070-3. doi:10.1016/S0014-4827(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 24.Liang G., Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J. Biol. Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. doi:10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo M., Ron D., Miller C.P., Habener J.F. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc. Natl Acad. Sci. USA. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. doi:10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu F., Bain P.J., LeBlanc-Chaffin R., Chen H., Kilberg M.S. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. doi:10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 27.He C.H., Gong P., Hu B., Stewart D., Choi M.E., Choi A.M., Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. doi:10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 28.Kilberg M.S., Shan J., Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. doi:10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Brewer J.W., Diehl J.A., Hendershot L.M. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. doi:10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 30.Shan J., Ord D., Ord T., Kilberg M.S. Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J. Biol. Chem. 2009;284:21241–21248. doi: 10.1074/jbc.M109.011338. doi:10.1074/jbc.M109.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruhat A., Averous J., Carraro V., Zhong C., Reimold A.M., Kilberg M.S., Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. doi:10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- 32.Luo S., Baumeister P., Yang S., Abcouwer S.F., Lee A.S. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J. Biol. Chem. 2003;278:37375–37385. doi: 10.1074/jbc.M303619200. doi:10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- 33.Vinson C., Myakishev M., Acharya A., Mir A.A., Moll J.R., Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. doi:10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park Y.U., Jeong J., Lee H., Mun J.Y., Kim J.H., Lee J.S., Nguyen M.D., Han S.S., Suh P.G., Park S.K. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc. Natl Acad. Sci. USA. 2010;107:17785–17790. doi: 10.1073/pnas.1004361107. doi:10.1073/pnas.1004361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millar J.K., James R., Christie S., Porteous D.J. Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol. Cell Neurosci. 2005;30:477–484. doi: 10.1016/j.mcn.2005.08.021. doi:10.1016/j.mcn.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 36.James R., Adams R.R., Christie S., Buchanan S.R., Porteous D.J., Millar J.K. Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell Neurosci. 2004;26:112–122. doi: 10.1016/j.mcn.2004.01.013. doi:10.1016/j.mcn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Hodgkinson C.A., Goldman D., Jaeger J., Persaud S., Kane J.M., Lipsky R.H., Malhotra A.K. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 2004;75:862–872. doi: 10.1086/425586. doi:10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lepagnol-Bestel A.M., Dubertret C., Benmessaoud D., Simonneau M., Ades J., Kacha F., Hamdani N., Gorwood P., Ramoz N. Association of DISC1 gene with schizophrenia in families from two distinct French and Algerian populations. Psychiatr. Genet. 2010;20:298–303. doi: 10.1097/YPG.0b013e32833aa5c4. doi:10.1097/YPG.0b013e32833aa5c4. [DOI] [PubMed] [Google Scholar]
- 39.Schosser A., Gaysina D., Cohen-Woods S., Chow P.C., Martucci L., Craddock N., Farmer A., Korszun A., Gunasinghe C., Gray J., et al. Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol. Psychiatry. 2009;15:844–849. doi: 10.1038/mp.2009.21. doi:10.1038/mp.2009.21. [DOI] [PubMed] [Google Scholar]
- 40.Szeszko P.R., Hodgkinson C.A., Robinson D.G., Derosse P., Bilder R.M., Lencz T., Burdick K.E., Napolitano B., Betensky J.D., Kane J.M., et al. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol. Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. doi:10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaikh M., Hall M.H., Schulze K., Dutt A., Li K., Williams I., Walshe M., Constante M., Broome M., Picchioni M., et al. Effect of DISC1 on the P300 waveform in psychosis. Schizophr. Bull. 2011 doi: 10.1093/schbul/sbr101. [Epub ahead of print]. doi: 10.1093/schbul/sbr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon T.D., Hennah W., van Erp T.G., Thompson P.M., Lonnqvist J., Huttunen M., Gasperoni T., Tuulio-Henriksson A., Pirkola T., Toga A.W., et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. doi:10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 43.Raznahan A., Lee Y., Long R., Greenstein D., Clasen L., Addington A., Rapoport J.L., Giedd J.N. Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents. Mol. Psychiatry. 2011;16:917–926. doi: 10.1038/mp.2010.72. doi:10.1038/mp.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brauns S., Gollub R.L., Roffman J.L., Yendiki A., Ho B.C., Wassink T.H., Heinz A., Ehrlich S. DISC1 is associated with cortical thickness and neural efficiency. Neuroimage. 2011;57:1591–1600. doi: 10.1016/j.neuroimage.2011.05.058. doi:10.1016/j.neuroimage.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whalley H.C., Sussmann J.E., Johnstone M., Romaniuk L., Redpath H., Chakirova G., Mukherjee P., Hall J., Johnstone E.C., Lawrie S.M., et al. Effects of a mis-sense DISC1 variant on brain activation in two cohorts at high risk of bipolar disorder or schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012 doi: 10.1002/ajmg.b.32035. [Epub ahead of print]. doi: 10.1002/ajmg.b.32035. [DOI] [PubMed] [Google Scholar]
- 46.Hennah W., Thomson P., McQuillin A., Bass N., Loukola A., Anjorin A., Blackwood D., Curtis D., Deary I.J., Harris S.E., et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol. Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. doi:10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- 47.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. doi:10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 48.Brandon N.J., Handford E.J., Schurov I., Rain J.C., Pelling M., Duran-Jimeniz B., Camargo L.M., Oliver K.R., Beher D., Shearman M.S., et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol. Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. doi:10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Elefteriou F., Ahn J.D., Takeda S., Starbuck M., Yang X., Liu X., Kondo H., Richards W.G., Bannon T.W., Noda M., et al. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514–520. doi: 10.1038/nature03398. doi:10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- 50.Lassot I., Estrabaud E., Emiliani S., Benkirane M., Benarous R., Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. doi:10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. doi:10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 52.Dever T.E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. doi:10.1016/S0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 53.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. doi:10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 54.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. doi:10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lassot I., Segeral E., Berlioz-Torrent C., Durand H., Groussin L., Hai T., Benarous R., Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol. Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. doi:10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pons J., Evrard-Todeschi N., Bertho G., Gharbi-Benarous J., Benarous R., Girault J.P. Phosphorylation-dependent structure of ATF4 peptides derived from a human ATF4 protein, a member of the family of transcription factors. Peptides. 2007;28:2253–2267. doi: 10.1016/j.peptides.2007.09.016. doi:10.1016/j.peptides.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 57.Frank C.L., Ge X., Xie Z., Zhou Y., Tsai L.H. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J. Biol. Chem. 2010;285:33324–33337. doi: 10.1074/jbc.M110.140699. doi:10.1074/jbc.M110.140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gachon F., Gaudray G., Thebault S., Basbous J., Koffi J.A., Devaux C., Mesnard J. The cAMP response element binding protein-2 (CREB-2) can interact with the C/EBP-homologous protein (CHOP) FEBS Lett. 2001;502:57–62. doi: 10.1016/s0014-5793(01)02646-1. doi:10.1016/S0014-5793(01)02646-1. [DOI] [PubMed] [Google Scholar]
- 59.Karpinski B.A., Morle G.D., Huggenvik J., Uhler M.D., Leiden J.M. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc. Natl Acad. Sci. USA. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. doi:10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa-Mattioli M., Sonenberg N. Translational control of long-term synaptic plasticity and memory storage by eIF2alpha. Crit. Rev. Neurobiol. 2006;18:187–195. doi: 10.1615/critrevneurobiol.v18.i1-2.190. [DOI] [PubMed] [Google Scholar]
- 61.Bartsch D., Ghirardi M., Skehel P.A., Karl K.A., Herder S.P., Chen M., Bailey C.H., Kandel E.R. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. doi:10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 62.Clapcote S.J., Lipina T.V., Millar J.K., Mackie S., Christie S., Ogawa F., Lerch J.P., Trimble K., Uchiyama M., Sakuraba Y., et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. doi:10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 63.Shen S., Lang B., Nakamoto C., Zhang F., Pu J., Kuan S.L., Chatzi C., He S., Mackie I., Brandon N.J., et al. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J. Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. doi:10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hikida T., Jaaro-Peled H., Seshadri S., Oishi K., Hookway C., Kong S., Wu D., Xue R., Andrade M., Tankou S., et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc. Natl Acad. Sci. USA. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. doi:10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W., Zhou Y., Jentsch J.D., Brown R.A., Tian X., Ehninger D., Hennah W., Peltonen L., Lonnqvist J., Huttunen M.O., et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc. Natl Acad. Sci. USA. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. doi:10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayhan Y., Abazyan B., Nomura J., Kim R., Ladenheim B., Krasnova I.N., Sawa A., Margolis R.L., Cadet J.L., Mori S., et al. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol. Psychiatry. 2011;16:293–306. doi: 10.1038/mp.2009.144. doi:10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao Y., Ge X., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K., et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. doi:10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niwa M., Kamiya A., Murai R., Kubo K., Gruber A.J., Tomita K., Lu L., Tomisato S., Jaaro-Peled H., Seshadri S., et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. doi:10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipina T.V., Niwa M., Jaaro-Peled H., Fletcher P.J., Seeman P., Sawa A., Roder J.C. Enhanced dopamine function in DISC1-L100P mutant mice: implications for schizophrenia. Genes Brain Behav. 2010;9:777–789. doi: 10.1111/j.1601-183X.2010.00615.x. doi:10.1111/j.1601-183X.2010.00615.x. [DOI] [PubMed] [Google Scholar]
- 70.Badiola N., Penas C., Minano-Molina A., Barneda-Zahonero B., Fado R., Sanchez-Opazo G., Comella J.X., Sabria J., Zhu C., Blomgren K., et al. Induction of ER stress in response to oxygen-glucose deprivation of cortical cultures involves the activation of the PERK and IRE-1 pathways and of caspase-12. Cell Death Dis. 2011;2:e149. doi: 10.1038/cddis.2011.31. doi:10.1038/cddis.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granberg F., Svensson C., Pettersson U., Zhao H. Adenovirus-induced alterations in host cell gene expression prior to the onset of viral gene expression. Virology. 2006;353:1–5. doi: 10.1016/j.virol.2006.06.019. doi:10.1016/j.virol.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 72.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M., Geschwind D.H., Ratan R.R. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. doi:10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva J.M., Wong A., Carelli V., Cortopassi G.A. Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol. Dis. 2009;34:357–365. doi: 10.1016/j.nbd.2009.02.005. doi:10.1016/j.nbd.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 74.Brown A.S. The environment and susceptibility to schizophrenia. Prog. Neurobiol. 2011;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. doi:10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caspi A., Moffitt T.E., Cannon M., McClay J., Murray R., Harrington H., Taylor A., Arseneault L., Williams B., Braithwaite A., et al. Moderation of the effect of adolescent-onset cannabis use on adult psychosis by a functional polymorphism in the catechol-O-methyltransferase gene: longitudinal evidence of a gene X environment interaction. Biol. Psychiatry. 2005;57:1117–1127. doi: 10.1016/j.biopsych.2005.01.026. doi:10.1016/j.biopsych.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 76.Ogawa F., Kasai M., Akiyama T. A functional link between Disrupted-In-Schizophrenia 1 and the eukaryotic translation initiation factor 3. Biochem. Biophys. Res. Commun. 2005;338:771–776. doi: 10.1016/j.bbrc.2005.10.013. doi:10.1016/j.bbrc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 77.McLaurin J., Trudel G.C., Shaw I.T., Antel J.P., Cashman N.R. A human glial hybrid cell line differentially expressing genes subserving oligodendrocyte and astrocyte phenotype. J. Neurobiol. 1995;26:283–293. doi: 10.1002/neu.480260212. doi:10.1002/neu.480260212. [DOI] [PubMed] [Google Scholar]
- 78.Ord D., Meerits K., Ord T. TRB3 protects cells against the growth inhibitory and cytotoxic effect of ATF4. Exp. Cell Res. 2007;313:3556–3567. doi: 10.1016/j.yexcr.2007.07.017. doi:10.1016/j.yexcr.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Bradshaw N.J., Ogawa F., Antolin-Fontes B., Chubb J.E., Carlyle B.C., Christie S., Claessens A., Porteous D.J., Millar J.K. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophys. Res. Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. doi:10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.