Abstract
Friedreich's ataxia (FRDA) is the most common inherited human ataxia and is caused by a deficiency in the mitochondrial protein frataxin. Clinically, patients suffer from progressive spinocerebellar degeneration, diabetes and a fatal cardiomyopathy, associated with mitochondrial respiratory chain defects. Recent findings have shown that lysine acetylation regulates mitochondrial function and intermediary metabolism. However, little is known about lysine acetylation in the setting of pathologic energy stress and mitochondrial dysfunction. We tested the hypothesis that the respiratory chain defects in frataxin deficiency alter mitochondrial protein acetylation. Using two conditional mouse models of FRDA, we demonstrate marked hyperacetylation of numerous cardiac mitochondrial proteins. Importantly, this biochemical phenotype develops concurrently with cardiac hypertrophy and is caused by inhibition of the NAD+-dependent SIRT3 deacetylase. This inhibition is caused by an 85-fold decrease in mitochondrial NAD+/NADH and direct carbonyl group modification of SIRT3, and is reversed with excess SIRT3 and NAD+ in vitro. We further demonstrate that protein hyperacetylation may be a common feature of mitochondrial disorders caused by respiratory chain defects, notably, cytochrome oxidase I (COI) deficiency. These findings suggest that SIRT3 inhibition and consequent protein hyperacetylation represents a negative feedback mechanism limiting mitochondrial oxidative pathways when respiratory metabolism is compromised, and thus, may contribute to the lethal cardiomyopathy in FRDA.
INTRODUCTION
Friedreich's ataxia (FRDA) is an autosomal recessive mitochondrial disorder caused by a homozygous triplet nucleotide repeat (GAA·TTC) expansion in intron 1 of the FXN gene located on chromosome 9q21.11 (1). This intronic expansion causes impaired transcription of the FXN gene and, consequently, a pathologic deficiency of the FXN gene product, frataxin (2). Frataxin is targeted to the mitochondrial matrix, where it is known to act as an iron-binding protein and participate in the proper assembly and function of iron–sulfur cluster (ISC)-dependent proteins, including complexes I, II and III of the respiratory chain and aconitase of the tricarboxylic acid (TCA) cycle (3–5). Thus, frataxin deficiency severely compromises both cellular respiration and overall mitochondrial function, leading to energetic stress and ATP deficiency (6). Although patients develop multisystem disease, including early spinocerebellar degeneration, ataxia and diabetes, the primary cause of death is heart failure for nearly 85% of those afflicted (7). Similarly, although the phenotypes of the neuron-specific enolase (NSE) and muscle creatine kinase (MCK)-Cre conditional mouse models of FRDA differ, both models develop a fatal cardiomyopathy and impaired activity of ISC-dependent respiratory complexes consistent with the human disease (8).
Recent work has demonstrated that lysine acetylation is a highly conserved and abundant post-translational modification within mitochondria that is responsive to nutrient availability and may contribute to the physiologic adaptations of reduced caloric intake (9–14). Multiple investigations have demonstrated a role for reversible mitochondrial enzyme acetylation and, specifically, the NAD+-dependent deacetylase SIRT3, in the regulation of fatty acid oxidation, the TCA cycle, electron transport via respiratory complexes I and II and overall oxidative metabolism (15–20). These studies suggest that mitochondrial protein acetylation could mediate metabolic changes in pathologic states characterized by profound energetic stress and impaired cellular respiration, such as cardiomyopathies and inherited mitochondrial disorders, but this hypothesis remains unexplored (21). Using the above-mentioned mouse models, we investigated whether acetylation states are altered in the setting of FRDA, and if so, sought to determine the mechanism.
RESULTS
NSE- and MCK-Cre mouse heart homogenates display hyperacetylation
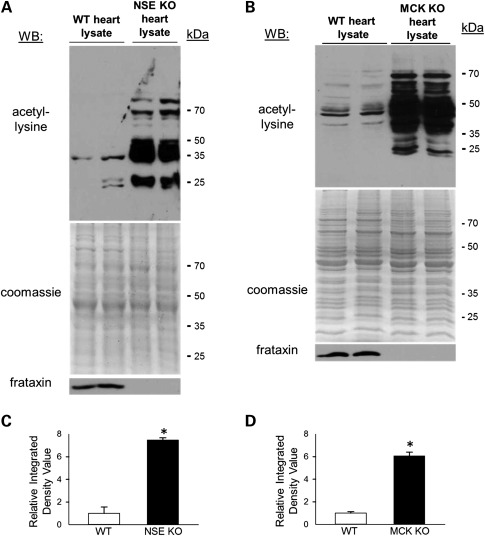
The heart contains the highest density of mitochondria of any organ in the mammalian body, and heart failure represents the primary cause of death for nearly 85% of FRDA patients. Hence, we prepared whole-heart lysates from WT, NSE and MCK conditional mouse models of FRDA and performed western blot analysis to assay protein acetyl-lysine modifications. Figure 1A and B shows that heart lysates from both the NSE and MCK mouse models of FRDA exhibited marked increases in acetyl-lysine modifications as compared with age-matched control hearts, and these are significant (Fig. 1C and D). The differences were most dramatic in proteins with an estimated molecular weight between ∼30 and 75 kDa.
Figure 1.
Frataxin-deficient hearts exhibit marked protein hyperacetylation. (A) Western blot (WB) probing for internal acetyl-lysine residues, using total heart homogenates derived from 24-day-old wild-type (WT, n = 2, lanes 1 and 2) and 24-day-old NSE-Cre mouse models of FRDA (n = 2, lanes 3 and 4). Below this, the corresponding SDS–PAGE gel stained with Coomassie Blue and a western blot probing for frataxin. (B) Similar to (A), only using total heart homogenates prepared from 9-week-old WT (n = 2, lanes 1 and 2) and 9- and 11-week-old MCK-Cre mouse models of FRDA (lanes 3 and 4, respectively). Below, the corresponding gel stained with Coomassie Blue and a western blot probing for frataxin. See also Supplementary Material, Figure S1. kDa, molecular weight in kilodaltons. (C) The average relative integrated densitometry values from the acetyl-lysine western blot in (A) are shown (mean ± SD; *P< 0.05). (D) The average relative integrated densitometry values from the acetyl-lysine western blot in (B) are shown (mean ± SD; *P< 0.05).
Hyperacetylation in frataxin-deficient hearts is localized to mitochondria and develops progressively with cardiac hypertrophy
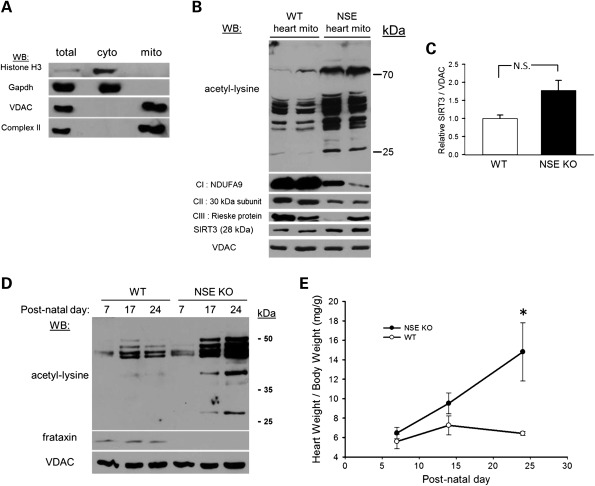
We performed sub-fractionation of heart samples to determine the sub-cellular distribution of hyperacetylated proteins. Analysis of the purity of these mitochondrial preparations showed that nuclear and cytosolic proteins were excluded and they were highly enriched for markers of both the outer and inner mitochondrial membranes (Fig. 2A). Using day-24 wild-type (WT, n = 2) control and NSE-Cre frataxin-deficient cardiac mitochondrial preparations (n = 2), we performed western blot analysis for acetyl-lysine modifications. Control cardiac mitochondria exhibited several acetylated proteins detectable by western blot, which is consistent with prior findings (10). However, frataxin-deficient cardiac mitochondria displayed marked hyperacetylation of numerous proteins (Fig. 2B and Supplementary Material, Fig. S1). This was accompanied by a characteristic downregulation of respiratory complexes I and II (succinate dehydrogenase) that was present as early as 7 days post-natal (Supplementary Material, Fig. S2). Levels of the dominant mitochondria-localized NAD+ -dependent deacetylase SIRT3 displayed a mild, though insignificant, increase in frataxin-deficient mitochondrial preparations (Fig. 2B and C), which is consistent with prior work demonstrating induction of SIRT3 in cultured cardiomyocytes subjected to various forms of stress (22). These results indicated that the hyperacetylation observed at the level of whole cardiac lysate is predominantly localized to mitochondria.
Figure 2.
Hyperacetylation in frataxin-deficient hearts is localized to mitochondria and develops progressively with cardiac hypertrophy. (A) Western blot (WB) demonstrating the purity of the cardiac mitochondrial preparations. Histone H3 was used as a nuclear marker, GAPDH as a cytoplasmic marker, VDAC as a mitochondrial outer membrane marker and complex II as a mitochondrial inner membrane marker. (B) Hyperacetylation is localized to cardiac mitochondria. Western blot probing for internal acetyl-lysine residues using 25 µg of isolated mitochondrial protein derived from two to three pooled hearts from 24-day-old wild-type (WT, n = 2, lanes 1 and 2) hearts and 24-day-old NSE frataxin KO hearts (n = 2, lanes 3 and 4). Below, a series of western blots probing for the respiratory complex I subunit NDUFA9, the complex II 30 kDa iron–sulfur subunit, the complex III Rieske protein and the NAD+-dependent deacetylase SIRT3. The mitochondrial outer membrane protein voltage-dependent anion channel (VDAC) was used as a loading control. See also Supplementary Material, Figures S1 and S2. kDa, molecular weight in kilodaltons. (C) The calculated densitometry for SIRT3 relative to the loading control VDAC shown in (B) (mean ± SD; N.S., not significant). (D) Hyperacetylation develops progressively. Western blot probing for internal acetyl-lysine residues in 25 μg of mitochondrial protein preparations isolated from two to three WT or NSE-Cre frataxin knockout hearts at day 7, day 17 and day 24 in their post-natal development. Below, a western blot probing for frataxin and VDAC was used as a loading control. Note: Owing to very small amounts of heart material and inefficient mitochondrial isolation at post-natal day 7, mitochondrial protein from day 7 WT and NSE animals was overloaded by 18.2 and 62%, respectively, with respect to the other samples to achieve normalization of mitochondrial protein. See also Supplementary Material, Figure S2. kDa, molecular weight in kilodaltons. (E) Cardiac hypertrophy develops concurrently with protein hyperacetylation. A graph assessing the post-natal development of cardiac hypertrophy in the NSE KO mice as measured by heart weight divided by body weight (mean ± SD; n = 3 measurements at each time point; *P< 0.05).
Because the NSE-Cre mouse models of FRDA begin to develop cardiac hypertrophy in their second week of life (8), we next sought to examine the developmental profile of mitochondrial protein acetylation. We prepared cardiac mitochondria for western analysis from WT control and NSE-Cre mice at post-natal days 7, 17 and 24. At post-natal day 7, NSE-Cre cardiac mitochondrial proteins exhibit only a mildly increased acetylation state when compared with their wild-type counterparts (Fig. 2D and Supplementary Material, Fig. S2). However, at post-natal day 17, NSE-Cre cardiac mitochondria display increased acetylation of cardiac mitochondrial proteins when compared with their WT counterparts, and this difference becomes more dramatic by post-natal day 24. Importantly, the progressive increase in cardiac mitochondrial protein acetylation over this time frame corresponded with the development of cardiac hypertrophy in the NSE-Cre models (Fig. 2E).
Hyperacetylation is caused by SIRT3 inhibition
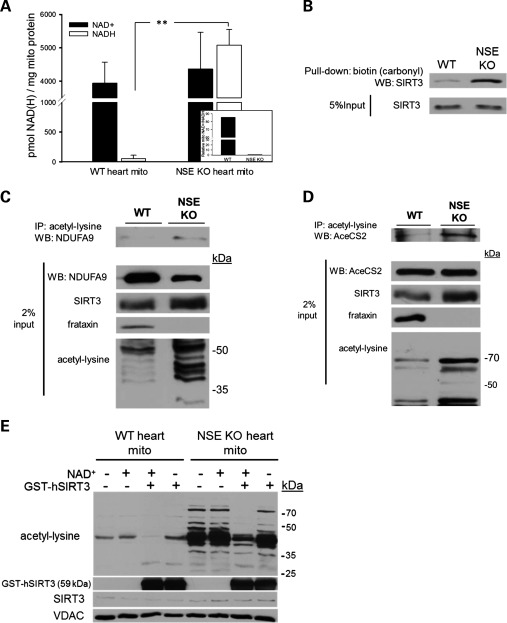
Prior work had demonstrated that genetic ablation of the deacetylase SIRT3, but not SIRT4 or SIRT5, caused hyperacetylation of mitochondrial proteins in liver and brown adipose tissue (23). These findings immediately suggested that hyperacetylation in frataxin deficiency could be caused by impairment or inhibition of SIRT3. Respiratory chain dysfunction is known to alter the cellular and mitochondrial redox state (NAD+/NADH) via impaired oxidation and consequent accumulation of NADH generated by the TCA cycle (24–26). Importantly, sirtuin activity is responsive to redox state (27), although this finding is somewhat controversial (28). Thus, we determined the redox state of WT and frataxin-deficient cardiac mitochondria preparations. WT heart mitochondria displayed robust NAD+ levels and >100-fold less NADH by comparison, which is consistent with highly oxidative cardiac catabolism and a continuous demand for carbon fuels (Fig. 3A). Frataxin-deficient mitochondria displayed a mild, though insignificant, increase in NAD+ levels when compared with WT. In striking contrast, frataxin-deficient mitochondrial NADH levels were, on average, over 95-fold greater than those in WT mitochondria (P <0.005), resulting in a corresponding NAD+/NADH ratio that was 85-fold less than those in WT animals (Fig. 3A inset). The observed accumulation of NADH and consequent shift in mitochondrial redox state are consistent with the impairments of mitochondrial respiration found in both human FRDA patients and the NSE and MCK animal models (3,8).
Figure 3.
Hyperacetylation in frataxin-deficient hearts is caused by impaired SIRT3 activity. (A) Determination of nicotinamide adenine dinucleotide (NAD+) and reduced NAD+ (NADH) levels in WT and frataxin-deficient cardiac mitochondrial preparations. Individual measurements were performed on a pooled sample of at least 300 μg of fresh cardiac mitochondria derived from two to three hearts and normalized to total mitochondrial protein input (n = 3–5 biologic replicates per condition, mean ± SD; **P< 0.005). Inset: The ratio of NAD+ to NADH for WT and NSE-KO heart mitochondria. (B) SIRT3 is differentially modified by a carbonyl group in frataxin-deficient mitochondria. This modification was previously shown to allosterically inhibit SIRT3 activity (32). The figure shown is representative of two independent experiments. (C) Targets of SIRT3-mediated deacetylation are hyperacetylated in frataxin-deficient hearts. Immunoprecipitation of 500 μg of WT and NSE-Cre cardiac mitochondrial proteins with an acetyl-lysine antibody and subsequent western blot probing for a known target of SIRT3-mediated deacetylation, the respiratory complex I subunit NDUFA9. Below, western blots of 2% of the total mitochondrial lysate input used for the immunoprecipitations showing NDUFA9, SIRT3, frataxin and the acetylation states of both samples. Cardiac mitochondrial preparations were derived from two to three pooled hearts per condition. The figure shown is representative of three independent experiments. (D) Similar to (C), only examining AceCS2 as a target of SIRT3-mediated deacetylation. (E) Recombinant SIRT3 in the presence of 10 mm NAD+ reverses mitochondrial protein hyperacetylation in frataxin-deficient hearts. Below are western blots showing recombinant SIRT3, endogenous SIRT3 and VDAC which was used as a loading control. kDa, molecular weight in kilodaltons.
An increased sensitivity to oxidative stress is believed to contribute to the pathogenesis of FRDA (29). A proteomic investigation of the MCK mouse heart revealed upregulation of the antioxidant enzymes glutathione S-transferase (GST) and DJ-1 (30). Furthermore, FRDA patients were shown to have increases in plasma levels of malondialdehyde, a product of lipid peroxidation (31). A recent report demonstrated that SIRT3 can be directly modified and allosterically inhibited by sub-physiologic levels of 4-hydroxy-2-nonenal (4-HNE), an endogenous product of lipid peroxidation (32). Therefore, we investigated whether SIRT3 is differentially modified by 4-HNE in frataxin-deficient cardiac mitochondria. We pulled down all reactive aldehyde-containing mitochondrial proteins, following their derivatization to biotin hydrazide, and then performed western blot analysis for SIRT3. SIRT3 in WT heart mitochondria exhibited a small amount of 4-HNE modification. In contrast, SIRT3 in frataxin-deficient mitochondria exhibited a marked increase in 4-HNE modification (Fig. 3B), suggesting that SIRT3 may be directly inhibited via carbonyl group adduction in the setting of frataxin deficiency.
We next analyzed the acetylation states of known targets of SIRT3-mediated deacetylation. The complex I subunit NDUFA9 was previously found to be a specific target of SIRT3-mediated deacetylation (16). To test the hypothesis that SIRT3 is inhibited in frataxin-deficient hearts, we immunoprecipitated mitochondrial acetyl-lysine proteins followed by western blot analysis for NDUFA9. Consistent with previous findings, we observed a minimal amount of NDUFA9 acetyl-lysine signal in the WT mitochondrial immunoprecipitate. However, the frataxin-deficient mitochondrial immunoprecipitate displayed a greater acetylated NDUFA9 signal despite downregulation of NDUFA9 as seen in the input (Fig. 3C). We additionally analyzed the acetylation state of a second specific target of SIRT3-mediated deacetylation, acetyl-CoA synthetase 2 (AceCS2) (11,13). Similarly, this analysis revealed a greater amount of acetylated AceCS2 in the frataxin-deficient condition when compared with the WT (Fig. 3D). The increased acetylation states of two known targets of SIRT3-mediated deacetylation indirectly demonstrated that SIRT3 is inhibited in frataxin-deficient cardiac mitochondria. Importantly, increases in the acetylation states of NDUFA9 and AceCS2 are linked to a decrease in the activity of respiratory complex I and the synthesis of activated acetate, respectively (11,16).
Prior studies have demonstrated that SIRT3 exhibits acetyl-lysine target specificity and does not deacetylate all acetyl-lysines present on a target protein (15,33). Based on these findings, we reasoned that if hyperacetylation of frataxin−/− mitochondrial protein lysine residues were caused by an inhibition of endogenous SIRT3, then incubating with excess SIRT3 and NAD+ in vitro would restore SIRT3 activity, resulting in deacetylation of its specific acetyl-lysine targets. Accordingly, we incubated solubilized WT and frataxin-deficient cardiac mitochondrial proteins with GST-tagged, processed human recombinant SIRT3 (GST-hSIRT3) in the presence or absence of NAD+ to assay for changes in acetylation states. Incubating solubilized WT or frataxin−/− cardiac mitochondrial homogenates with NAD+ alone caused no change in acetylation signal. In contrast, incubating frataxin-deficient mitochondrial protein with 3 μg of GST-hSIRT3 and NAD+ resulted in a marked reduction of acetyl-lysine signal from nearly every protein band and completely eliminated the acetyl-lysine signal from multiple bands (Fig. 3E). Furthermore, the observed reduction in acetylation signal was abolished upon withdrawal of NAD+ from the incubation buffer, demonstrating that the hyperacetylated protein lysine residues in frataxin-deficient cardiac mitochondria are, indeed, specifically sensitive to NAD+-dependent SIRT3-mediated deacetylation and that the observed hyperacetylation is not caused by a general increase in non-specific lysine acetylation. Taken together, these data strongly suggest that hyperacetylation in frataxin-deficient mitochondria is due to both redox state and lipid-peroxidation-mediated inhibition of endogenous SIRT3.
Altered protein acetylation may be a common feature of respiratory chain compromise
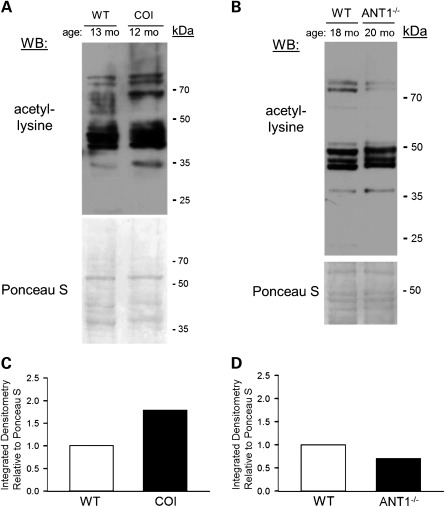
FRDA is a mitochondrial disorder that has a distinctive cause and unique pathogenesis. However, the observed accumulation of NADH in frataxin-deficient mitochondria and consequent drop in NAD+/NADH ratio is often a common biochemical feature of respiratory chain disorders, regardless of mutational origin (26,34,35). Taking this common feature into consideration with the findings that SIRT3 deacetylase activity is NAD+-dependent and likely to be responsive to this change in mitochondrial NAD+/NADH balance, we hypothesized that other mitochondrial disorders caused by respiratory chain defects would display alterations in cardiac acetylation profiles. To test this, we obtained whole cardiac lysates from the COI mouse model, which harbors an mtDNA mutation causing a defect of respiratory complex IV, and the adenine nucleotide translocase knockout (ANT1−/−) mouse model, which models a non-respiratory chain defect with a deficiency of ATP translocation (36,37). These mice model the human mitochondrial disorders—autosomal dominant progressive external ophthalmoplegia, and Leber's hereditary optic neuropathy, respectively, and both models develop cardiomyopathy (38,39). Analyzing cardiac acetylation via western blot revealed mild to moderate increases in acetylation in 12-month-old COI hearts when compared with 13-month-old control hearts (Fig. 4A and C). Interestingly, 20-month-old ANT1−/− hearts exhibited a small decrease in acetyl-lysine profiles when compared with 18-month-old control hearts (Fig. 4B and D). Taken together with our data from the frataxin-deficient mice, these results suggest that increases in protein acetylation may be a common feature of respiratory chain malfunction in the mammalian heart.
Figure 4.
Respiratory chain defects may cause protein hyperacetylation. (A) Western blot probing for internal acetyl-lysine residues, using 25 μg of whole-heart lysate derived from a 13-month-old wild-type (WT) animal and a 12-month-old animal harboring a missense mutation in the mitochondrial genome-encoded protein subunit of the respiratory chain enzyme cytochrome c oxidase (COI). Below, the same membrane used for western blotting stained with the non-specific protein marker Ponceau S. (B) Western blot probing for internal acetyl-lysine residues, using 25 μg of whole-heart lysate prepared from an 18-month-old WT animal and a 20-month-old animal lacking the non-respiratory chain mitochondrial inner membrane protein adenine nucleotide translocase 1 (ANT1−/−). Below, the same membrane used for western blotting stained with Ponceau S. (C) The calculated integrated densitometry values for acetyl-lysine signal relative to Ponceau S signal from (A). (D) Similar to (C), only calculated from (B). kDa, molecular weight in kilodaltons.
DISCUSSION
In this study, we have demonstrated that the respiratory chain defects accompanying frataxin deficiency cause progressive hyperacetylation of cardiac mitochondrial proteins due to the inhibition of a major positive regulator of oxidative metabolism, the SIRT3 deacetylase (20,40). These findings present a novel disease phenotype and reveal a negative feedback mechanism that may serve to coordinately suppress mitochondrial oxidative pathways, and that may be hyperactive in the setting of respiratory chain malfunction.
Defects in cellular respiration can be inherited as mitochondrial disease, or acquired over a lifetime via somatic mutations, and are linked to many conditions, including neurodegenerative disease, diabetes, heart failure, cancer and indeed, the aging process in general (34). The involvement of cellular respiration in numerous common human pathologies emphasizes the need for greater understanding of the pathophysiologic processes that occur in response to respiratory chain compromise. SIRT3-mediated deacetylation has recently emerged as a major mechanism regulating the activity of mitochondrial oxidative and intermediary metabolism. SIRT3 is also uniquely poised to respond to the flux of mitochondrial NAD+ and NADH, which is determined, in large part, by the capacity of the respiratory chain to oxidize NADH. This capacity is severely decreased in FRDA, as well as in other mitochondrial defects such as cytochrome c oxidase (complex IV) deficiency (24), causing an accumulation of NADH and, consequently, a redox state of perceived nutrient excess. In this setting, it is logical to predict that mitochondria would have a mechanism to restrain oxidative metabolism and the further generation of NADH, as well as spare the cell from damaging ROS that would be produced by forcing electrons through a dysfunctional respiratory chain. Indeed, it has been known for several decades that a decrease in NAD+/NADH redox balance has a largely inhibitory effect on many enzymes of intermediary mitochondrial metabolism (41). Based on these observations and predictions, our data suggest that this mechanism involves inhibition of the NAD+-dependent SIRT3 deacetylase mediated through a dramatic decrease in mitochondrial redox state, as measured by the NAD+/NADH ratio, and direct modification by lipid peroxidation.
Recent studies have shown that SIRT3-mediated deacetylation activates the respiratory chain via complexes I and II, fatty acid oxidation via long-chain acyl-CoA dehydrogenase, the TCA cycle via isocitrate dehydrogenase and the production of acetyl-CoA via acetyl-CoA synthetase, among others (11,15–17). Additionally, SIRT3-deficient fibroblasts show a decrease of carbon intermediates in the TCA cycle (42). Together, these studies indicate a key role for SIRT3 in the maintenance of oxidative metabolism (20) and may be especially important in the heart, which derives as much as 90% of its ATP from oxidative phosphorylation (43). Indeed, SIRT3-deficient mice exhibit reduced cardiac ATP levels, are hypersensitive to cardiac stress and develop an early onset cardiac hypertrophy (16,44,45). These findings suggest that SIRT3 inhibition, in addition to impaired ISC biosynthesis, may contribute to FRDA cardiomyopathy by further limiting key components of oxidative and intermediary metabolism, such as the respiratory chain, the TCA cycle and the β-oxidation of fatty acids (Fig. 5). In addition to its metabolic functions, SIRT3 is known to directly activate antioxidant defense mechanisms via deacetylation of mitochondrial manganese superoxide dismutase and isocitrate dehydrogenase of the glutathione system (19,46,47). These important findings suggest that inhibition of SIRT3 in FRDA could underlie the increased sensitivity to oxidative stress (29). Thus, it is logical to predict that frataxin deficiency promotes a vicious cycle whereby improper assembly of ISC-dependent respiratory complexes and their consequently reduced activity cause SIRT3 inhibition, leading to the accumulation of protein lysine acetylation, which further inhibits the activity of the respiratory chain and oxidative metabolism (Fig. 5). This model would be consistent with the progressive increase in mitochondrial protein acetylation we observe during the post-natal development of the NSE-Cre Frataxin−/− mice (Fig. 2C). Interestingly, a recent report noted that frataxin-deficient yeast show decreased acetylation of mitochondrial proteins (48), which may reflect the yeast preference for glucose as a carbon source instead of fatty acids utilized by heart mitochondria, or that no sirtuins are known to function within yeast mitochondria. It is important to note that the present findings may also involve a putative mammalian mitochondrial acetyltransferase(s), a component of which was recently identified as GCN5L1 (49). The extent to which SIRT3 inhibition and mitochondrial protein hyperacetylation influence these diverse processes and the pathogenesis of FRDA cardiomyopathy is likely to be very important and remains to be explored.
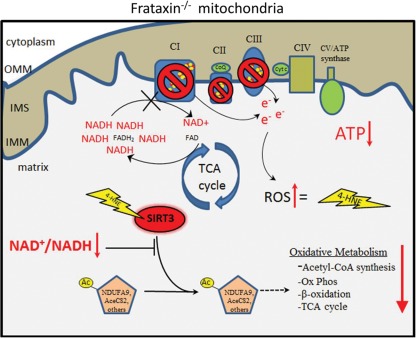
Figure 5.
Cartoon schematic summarizing the mechanism of SIRT3 inhibition in frataxin-deficient cardiac mitochondria. Impaired ISC biosynthesis in frataxin−/− mitochondria results in defects of respiratory complexes I, II and III, which cause impaired oxidation and accumulation of NADH generated by the TCA cycle. The resulting dramatic decrease in the mitochondrial NAD+/NADH ratio (redox state) creates a reducing environment mimicking nutrient excess. In addition, an increased sensitivity to oxidative stress consequently promotes lipid peroxidation and the formation of 4-HNE protein carbonyl adducts. The dramatic decrease in mitochondrial redox state and increase in 4-HNE modified SIRT3 work in concert to inhibit SIRT3 activity, causing protein hyperacetylation and most likely reduced activity of SIRT3 target proteins which have been shown to play key roles in the maintenance of oxidative and intermediary metabolism. OMM, outer mitochondrial membrane; IMS, intermembrane space; IMM, inner mitochondrial membrane; ROS, reactive oxygen species; 4-HNE, 4-hydroxy-2-nonenal; NDUFA9, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9; AceCS2, acetyl-CoA synthetase 2.
MATERIALS AND METHODS
Please see Supplementary Material for full Materials and Methods.
Animal use statement
The use of laboratory mice in this study was approved by the Indiana University School of Medicine's Institutional Animal Care and Use Committee (IACUC) and Laboratory Animal Resource Center (LARC). All conducted experiments conform to the American Veterinary Medical Association's Panel on Euthanasia.
Isolation of cardiac mitochondria
Briefly, heart tissue was homogenized with a motor-driven Potter–Elvehjem homogenizer in mitochondrial isolation buffer [220 mm mannitol, 70 mm sucrose, 30 mm Tris–Cl (pH 7.4), 0.5 mm EGTA and 0.1% BSA] supplemented with an EDTA-free protease inhibitor cocktail (Roche) and 10 mm nicotinamide (NAM), 200 nm trichostatin A and 5 mm sodium butyrate as deacetylase inhibitors. Mitochondria were isolated using a standard differential centrifugation method as described previously (50). Full methods are available in Supplementary Material, Methods.
Statistical analysis
Statistical significance was determined using a two-sample t-test, assuming unequal variances. Any P-value <0.05 was judged to be significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a Kyle Bryant Award from the Friedreich's Ataxia Research Alliance (R.M.P.); the National Institutes of Health (1P01HL085098 to R.M.P.); an American Heart Association Grant in Aid (0855646G to R.M.P.) and an American Heart Association Fellowship (11PRE7290079 to G.R.W.). The funding sources had no role in the conduct or presentation of this research.
Supplementary Material
ACKNOWLEDGEMENTS
We kindly acknowledge Dr Brittney-Shea Herbert and Dr X. Charlie Dong for thoughtful review of the manuscript. We would also like to thank Dr Eric Verdin and colleagues for their generous gift of the SIRT3 antibody, Dr Douglas C. Wallace for his generous donation of the ANT−/− and COI mouse model cardiac lysates, Dr Grazia Isaya for the frataxin antibody and Dr Helene Puccio for the conditional mouse models of FRDA.
Conflict of Interest statement. G.R.W., P.M.P., C.M.B. and R.M.P. declare no conflicts of interest.
REFERENCES
- 1.Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 2.Punga T., Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 4.Tsai C.L., Barondeau D.P. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 5.Bulteau A.-L., O'Neill H.A., Kennedy M.C., Ikeda-Saito M., Isaya G., Szweda L.I. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 6.Lodi R., Cooper J.M., Bradley J.L., Manners D., Styles P., Taylor D.J., Schapira A.H. Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc. Natl Acad. Sci. USA. 1999;96:11492–11495. doi: 10.1073/pnas.96.20.11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durr A., Cossee M., Agid Y., Campuzano V., Mignard C., Penet C., Mandel J.-L., Brice A., Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N. Engl. J. Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 8.Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 9.Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Schwer B., Eckersdorff M., Li Y., Silva J.C., Fermin D., Kurtev M.V., Giallourakis C., Comb M.J., Alt F.W., Lombard D.B. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B., Bunkenborg J., Verdin R.O., Andersen J.S., Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallows W.C., Lee S., Denu J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C.F., Grishin N.V., Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B., Grueter C.A., Harris C., Biddinger S., Ilkayeva O.R., et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn B.-H., Kim H.-S., Song S., Lee I.H., Liu J., Vassilopoulos A., Deng C.-X., Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl Acad. Sci. USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimen H., Han M.J., Yang Y., Tong Q., Koc H., Koc E.C. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.S., Patel K., Muldoon-Jacobs K., Bisht K.S., Aykin-Burns N., Pennington J.D., van der Meer R., Nguyen P., Savage J., Owens K.M., et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Someya S., Yu W., Hallows W.C., Xu J., Vann J.M., Leeuwenburgh C., Tanokura M., Denu J.M., Prolla T.A. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdin E., Hirschey M.D., Finley L.W., Haigis M.C. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner G.R., Payne R.M. Mitochondrial acetylation and diseases of aging. J. Aging Res. 2011;2011:234875. doi: 10.4061/2011/234875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundaresan N.R., Samant S.A., Pillai V.B., Rajamohan S.B., Gupta M.P. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombard D.B., Alt F.W., Cheng H.L., Bunkenborg J., Streeper R.S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung H.J., Ma W., Wang P.Y., Hynes J., O'Riordan T.C., Combs C.A., McCoy J.P., Jr., Bunz F., Kang J.G., Hwang P.M. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat. Commun. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayevsky A., Rogatsky G.G. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am. J. Physiol. Cell Physiol. 2007;292:C615–C640. doi: 10.1152/ajpcell.00249.2006. [DOI] [PubMed] [Google Scholar]
- 26.Munnich A., Rustin P., Rotig A., Chretien D., Bonnefont J.P., Nuttin C., Cormier V., Vassault A., Parvy P., Bardet J., et al. Clinical aspects of mitochondrial disorders. J. Inherit. Metab. Dis. 1992;15:448–455. doi: 10.1007/BF01799603. [DOI] [PubMed] [Google Scholar]
- 27.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt M.T., Smith B.C., Jackson M.D., Denu J.M. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 29.Schmucker S., Puccio H. Understanding the molecular mechanisms of Friedreich's ataxia to develop therapeutic approaches. Hum. Mol. Genet. 2010;19:R103–R110. doi: 10.1093/hmg/ddq165. [DOI] [PubMed] [Google Scholar]
- 30.Sutak R., Xu X., Whitnall M., Kashem M.A., Vyoral D., Richardson D.R. Proteomic analysis of hearts from frataxin knockout mice: marked rearrangement of energy metabolism, a response to cellular stress and altered expression of proteins involved in cell structure, motility and metabolism. Proteomics. 2008;8:1731–1741. doi: 10.1002/pmic.200701049. [DOI] [PubMed] [Google Scholar]
- 31.Emond M., Lepage G., Vanasse M., Pandolfo M. Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology. 2000;55:1752–1753. doi: 10.1212/wnl.55.11.1752. [DOI] [PubMed] [Google Scholar]
- 32.Fritz K.S., Galligan J.J., Smathers R.L., Roede J.R., Shearn C.T., Reigan P., Petersen D.R. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem. Res. Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimazu T., Hirschey M.D., Hua L., Dittenhafer-Reed K.E., Schwer B., Lombard D.B., Li Y., Bunkenborg J., Alt F.W., Denu J.M., et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watmough N.J., Bindoff L.A., Birch-Machin M.A., Jackson S., Bartlett K., Ragan C.I., Poulton J., Gardiner R.M., Sherratt H.S., Turnbull D.M. Impaired mitochondrial beta-oxidation in a patient with an abnormality of the respiratory chain. Studies in skeletal muscle mitochondria. J. Clin. Invest. 1990;85:177–184. doi: 10.1172/JCI114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan W., Waymire K.G., Narula N., Li P., Rocher C., Coskun P.E., Vannan M.A., Narula J., Macgregor G.R., Wallace D.C. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narula N., Zaragoza M.V., Sengupta P.P., Li P., Haider N., Verjans J., Waymire K., Vannan M., Wallace D.C. Adenine nucleotide translocase 1 deficiency results in dilated cardiomyopathy with defects in myocardial mechanics, histopathological alterations, and activation of apoptosis. JACC Cardiovasc. Imaging. 2011;4:1–10. doi: 10.1016/j.jcmg.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deschauer M., Hudson G., Muller T., Taylor R.W., Chinnery P.F., Zierz S. A novel ANT1 gene mutation with probable germline mosaicism in autosomal dominant progressive external ophthalmoplegia. Neuromuscul. Disord. 2005;15:311–315. doi: 10.1016/j.nmd.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Brown M.D., Voljavec A.S., Lott M.T., MacDonald I., Wallace D.C. Leber's hereditary optic neuropathy: a model for mitochondrial neurodegenerative diseases. FASEB J. 1992;6:2791–2799. doi: 10.1096/fasebj.6.10.1634041. [DOI] [PubMed] [Google Scholar]
- 40.Guarente L. The logic linking protein acetylation and metabolism. Cell Metab. 2011;14:151–153. doi: 10.1016/j.cmet.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Krebs H.A., Veech R.L. Equilibrium relations between pyridine nucleotides and adenine nucleotides and their roles in the regulation of metabolic processes. Adv. Enzyme Regul. 1969;7:397–413. doi: 10.1016/0065-2571(69)90030-2. [DOI] [PubMed] [Google Scholar]
- 42.Finley L.W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P.I., Cardoso S.M., Clish C.B., et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura-Clapier R., Garnier A., Veksler V. Energy metabolism in heart failure. J. Physiol. 2004;555:1–13. doi: 10.1113/jphysiol.2003.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hafner A.V., Dai J., Gomes A.P., Xiao C.Y., Palmeira C.M., Rosenzweig A., Sinclair D.A. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundaresan N.R., Gupta M., Kim G., Rajamohan S.B., Isbatan A., Gupta M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao R., Coleman M.C., Pennington J.D., Ozden O., Park S.H., Jiang H., Kim H.S., Flynn C.R., Hill S., Hayes McDonald W., et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu X., Brown K., Hirschey M.D., Verdin E., Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Sliwa D., Dairou J., Camadro J.M., Santos R. Inactivation of mitochondrial aspartate aminotransferase contributes to the respiratory deficit of yeast frataxin-deficient cells. Biochem. J. 2012;441:945–953. doi: 10.1042/BJ20111574. [DOI] [PubMed] [Google Scholar]
- 49.Scott I., Webster B.R., Li J.H., Sack M.N. Identification of a molecular component of the mitochondrial acetyl transferase program; a novel role for GCN5L1. Biochem. J. 2012 doi: 10.1042/BJ20120118. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crowley K.S., Payne R.M. Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J. Biol. Chem. 1998;273:17278–17285. doi: 10.1074/jbc.273.27.17278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.