Abstract
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is an inherited arrhythmogenic disease so far related to mutations in the cardiac ryanodine receptor (RYR2) or the cardiac calsequestrin (CASQ2) genes. Because mutations in RYR2 or in CASQ2 are not retrieved in all CPVT cases, we searched for mutations in the physiological protein partners of RyR2 and CSQ2 in a large cohort of CPVT patients with no detected mutation in these two genes. Based on a candidate gene approach, we focused our investigations on triadin and junctin, two proteins that link RyR2 and CSQ2. Mutations in the triadin (TRDN) and in the junctin (ASPH) genes were searched in a cohort of 97 CPVT patients. We identified three mutations in triadin which cosegregated with the disease on a recessive mode of transmission in two families, but no mutation was found in junctin. Two TRDN mutations, a 4 bp deletion and a nonsense mutation, resulted in premature stop codons; the third mutation, a p.T59R missense mutation, was further studied. Expression of the p.T59R mutant in COS-7 cells resulted in intracellular retention and degradation of the mutant protein. This was confirmed after in vivo expression of the mutant triadin in triadin knock-out mice by viral transduction. In this work, we identified TRDN as a new gene responsible for an autosomal recessive form of CPVT. The mutations identified in the two families lead to the absence of the protein, thereby demonstrating the importance of triadin for the normal function of the cardiac calcium release complex in humans.
INTRODUCTION
Catecholaminergic polymorphic ventricular tachycardia (CPVT; MIM #604772), a potentially fatal cardiac arrhythmia, is a genetic disorder characterized by stress-induced syncope and/or sudden death in juveniles and young adults in the absence of structural alteration of the heart (1,2). The two genes currently associated with CPVT, ryanodine receptor (RYR2) and calsequestrin (CASQ2), encode proteins responsible for sarcoplamic reticulum (SR) calcium release during cardiac muscle contraction. They belong to the macromolecular calcium release complex (CRC), composed mainly of following proteins: the SR calcium release channel ryanodine receptor (RyR2); the CSQ2, which acts as a calcium buffer inside the SR; triadin and junctin, two proteins that have been implicated in anchoring CSQ2 to RyR2, at the terminal cisternae of SR (3,4). Mutations have been identified in RYR2 and CASQ2 in only 70% of the patients affected by CPVT, indicating that other genes are involved in this disease. Therefore, the genes encoding triadin or junctin, the two linkers between RyR2 and CSQ2, naturally appeared as interesting candidate genes for CPVT.
Triadin is a multiprotein family arising from the alternative splicing of a single TRDN gene (MIM #603283) localized on Chr6 and containing 41 exons (5). Two isoforms, Trisk 95 and Trisk 51, are exclusively expressed in skeletal muscle. A third one, Trisk 32 (also called CT1), is expressed mainly in cardiac muscle (6). Two triadin knock-out (KO) mouse models have been developed, presenting with cardiac arrhythmia and skeletal muscle weakness (7–9). Therefore, the presence of triadin seems essential for normal cardiac and skeletal muscle function.
Junctin, a 26 kDa protein expressed both in skeletal and cardiac muscle (10) (MIM #600582), is an alternative splice product of the aspartyl-β-hydroxylase gene on Chr8 [ASPH, (11)]. Two junctin isoforms are expressed in human cardiac muscle (12). Junctin is involved in regulation of the calcium homeostasis, and deletion of junctin in a mouse model results in fatal arrhythmia (13).
This study presents the molecular investigations performed in a large panel of well-characterized CPVT patients in which mutations in RYR2 and CASQ2 genes had been excluded. Pathophysiological and clinical consequences of the mutations identified in the TRDN gene in two families have been analyzed and will be discussed.
RESULTS
Clinical phenotype
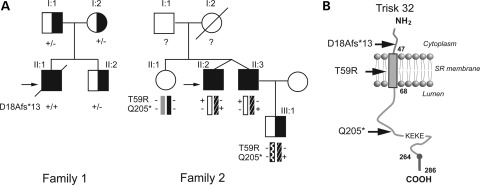
The proband of family 1 (ind II:1, Fig. 1) originated from French West Indies. He was a 2-year-old boy who experienced syncope followed by cardiac arrest after a shock while playing with his 7-year-old brother. Resting electrocardiogram following cardiac resuscitation showed numerous polymorphic or bidirectional ventricular extra beats and runs of polymorphic ventricular tachycardia (Supplementary Material, Fig. S1a). He died in the hospital 3 weeks following the initial cardiac arrest after a severe post-anoxic coma. There was no positive family history. The parents and the brother of the proband were considered healthy as their resting ECG, Holter recordings and exercise stress tests did not show any abnormality.
Figure 1.
Triadin mutations in two CPVT families. (A) Pedigrees of the two CPVT families. Filled squares indicated affected individuals and half filled symbols individuals heterozygous for a mutation. The proband in each Family is indicated by the black arrow. The genotype for each individual concerning the identified mutation is indicated as follow: ‘+’ in the presence of the mutation and ‘−’ in the absence of the mutation. (B) Membrane topology of cardiac isoform of triadin, Trisk 32, in the sarcoplasmic reticulum membrane with the localization of the three identified mutations. The ‘KEKE’ region is the interaction domain with RyR2 and CSQ2. Trisk 32 has a C-terminal specific region from amino acid 264–286.
The proband of Family 2 (ind II:2, Fig. 1) originated from Western France. He was a 26-year-old man who presented with recurrent syncopes at exercise since the infancy. Resting ECG was normal with no prolongation of the QT interval. Exercise testing showed numerous bidirectional ventricular extra beats (Supplementary Material, Fig. S1b). The patient also presented with proximal muscle weakness. CPVT was also diagnosed in his dizygotic twin-brother (ind II:3) after clinical examination. They were both treated by nadolol 40 mg/day, a non-selective β-blocker. Clinical evaluation, Holter recording and exercise testing did not show any abnormalities among other family members investigated (ind II:1 and III:1).
Genetic analysis
Molecular analysis of the TRDN gene in Family 1 identified a c.del53_56ACAG homozygous deletion in exon 2. This deletion of four nucleotides resulted in a frameshift in the amino acid sequence from amino acid 18 leading to a premature termination codon (PTC) at position 31, designed p.D18Afs*13 according to the nomenclature proposed by the Human Genome Variation Society (14) (http://www.hgvs.org/mutnomen/recs-prot.html). This mutation was identified at a heterozygous state in the three unaffected relatives tested (Fig. 1A). Haplotype analyses were performed in the family using polymorphic markers flanking the TRDN gene (cen-D6S1712-D6S979-TRDN-D6S1639-D6S408-tel) and were suggestive of a remote common ancestor.
Molecular analysis in Family 2 revealed that the two affected individuals II:2 and II:3 were compound heterozygous for a c.176C>G missense mutation in exon 2 that led to the substitution of a threonine by an arginyl residue at position 59 (p.T59R) and a c.613C>T nonsense mutation in exon 8 that introduced a PTC at position 205 in the amino acid sequence (p.Q205*). Unaffected individuals tested carried one (ind III:1) or no variant (ind II:1) (Fig. 1A).
In both the families, the phenotype–genotype analysis was in favor of a recessive form of CVPT. The frameshift deletion c.del53_56delACAG; p.D18Afs*13 inducing a stop codon in position 31 would result in deletion of the major part of the protein, and the production of a short cytosolic peptide as the stop was located before the transmembrane helix (Fig. 1B). The nonsense variation c.613C>T; p.Q205* generated a stop codon in position 205, just before the KEKE motif (domain composed of repetition of Lys and Glu) (15) located in position 210–224 responsible for interaction of Trisk 32 with RyR2 and CSQ2 (16,17). These two mutations resulting into a PTC were clearly pathogenic because they would lead either to an absence of protein synthesis via nonsense-mediated mRNA decay (18), or to the production of a non-functional protein. The p.T59R variation affected a well-conserved position in Trisk 32 and resulted in the introduction of a positively charged amino acid in the transmembrane domain of the protein. The deleterious effect of the substitution within the transmembrane domain of this conserved threonine at position 59 by an arginine was less predictable and was thus further studied.
Localization of the mutant p.T59R in a model cell line
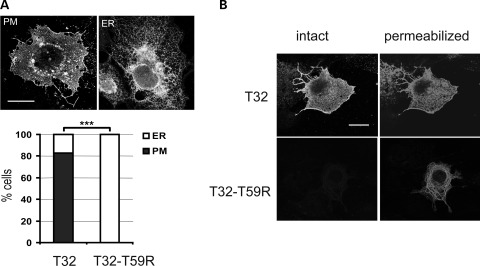
The effect of the p.T59R mutation on the behavior of Trisk 32 was first studied by expression in the non-muscle COS-7 cell line, and analysis of the localization of the wild-type (WT) (T32) or mutant protein (T32-T59R) by confocal microscopy. When expressed in COS-7 cells, the WT Trisk 32 was localized in the plasma membrane of 83% of the cells (Fig. 2A, panel PM) and in the endoplasmic reticulum for the remaining 17% of the cells (Fig. 2A, panel ER) (n = 500 cells). The p.T59R mutation induced the exclusive localization of Trisk 32 in an intracellular compartment (100%, n = 500 cells), identified as endoplasmic reticulum by co-localization with the endoplasmic reticulum (ER) resident protein calnexin (Supplementary Material, Fig. S2a). This observation was confirmed by differential immunofluorescent labeling before and after permeabilization of the cells (Fig. 2B). Whereas Trisk 32 was labeled both on intact and permeabilized cells, confirming its plasma membrane localization, the mutant Trisk 32-T59R was accessible only after permeabilization thus indicating an intracellular retention. This intracellular retention was also confirmed by biotinylation of cell surface proteins (Supplementary Material, Fig. S2b).
Figure 2.
Trisk 32 and Trisk 32-T59R cell localizations are different. (A) Trisk 32-T59R is located only at the endoplasmic reticulum, whereas Trisk 32 is mainly at the plasma membrane. WT Trisk 32 (T32) and mutant Trisk 32-T59R (T32–T59R) were transfected in COS-7 cells, and their localization analyzed by immunofluorescent labeling. Typical Trisk 32 labeling at the plasma membrane (PM) or in the ER are shown. Bar: 20 µm. The histogram shows the percentage of transfected cells exhibiting PM (closed square) or ER (open square) labeling, for a total of 500 transfected cells from three different experiments. ***P < 0.001, Fisher's test comparison of T32- vs. T32–T59R-transfected cells. (B) Immunofluorescent labeling on intact or permeabilized cells shows intracellular retention of Trisk 32-T59R. Transfected cells were fixed without permeabilization and stained with an antibody directed against the C-terminal end of Trisk 32, which is extracellular when the protein is in the plasma membrane (left panels, ‘intact’). Afterwards, cells were permeabilized and stained with an antibody directed against the N-terminal end of Trisk 32, which is cytosolic when the protein is in the plasma membrane or in the reticulum membrane (right panels, ‘permeabilized’). Bar: 20 µm.
Stability of the mutant Trisk 32-T59R in a model cell line
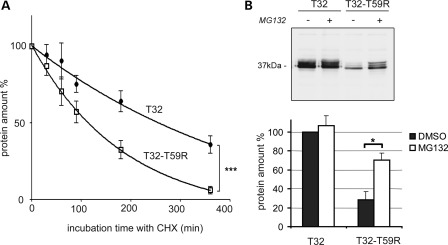
As a lower level of expression of Trisk 32-T59R was observed in COS-7 cells compared with the WT Trisk 32, the stability of the mutant protein was evaluated. After transfection in COS-7 cells, Trisk 32 and Trisk 32-T59R appeared as a triple band of centered on 37 kDa, due to post-translational modification (glycosylation), as observed previously (19,20). Transfected cells were incubated with cycloheximide (CHX), a protein synthesis inhibitor, and the amounts of Trisk 32 and Trisk 32-T59R were analyzed by quantitative western blotting after various incubation times. The relative amount of Trisk 32-T59R decreased faster than the relative amount of Trisk 32 (Fig. 3A), indicating a faster turnover of the mutant Trisk 32-T59R compared with the WT Trisk 32. This suggested that the abnormal localization observed for Trisk 32-T59R could result in protein instability and degradation. This degradation was further analyzed by proteasome inhibition by MG132. Proteasome inhibition did not increase the total amount of the WT Trisk 32, but it raised the amount of the mutant Trisk 32-T59R by 148% (Fig. 3B). This showed that Trisk 32-T59R was targeted for proteasome-mediated degradation. Altogether, results in COS-7 cells showed that mutant Trisk 32-T59R was unable to exit ER after synthesis and was prone to degradation most probably via the proteasomal pathway.
Figure 3.
The mutant protein Trisk 32-T59R is degraded faster, via the proteasome. (A) Trisk 32-T59R is degraded faster in the course of protein synthesis inhibition. WT Trisk 32 (T32) and mutant Trisk 32-T59R (T32-T59R) were first transfected in COS-7 cells. Transfected cells were then incubated with cycloheximine (CHX) to block protein synthesis for the indicated time. Cells were collected and analyzed by quantitative western blot with specific antibodies to evaluate the amount of protein. The curves show the quantification from four experiments of the amount of each protein (T32 -•- and T32-T59R -□-) compared with its initial amount. ***P < 0.001, F-test comparison of the two curves, based on decay and plateau parameters. (B) Inhibition of the proteasome raises the amount of the mutant protein Trisk 32-T59R. Twenty hours after transfection, the cells were incubated during 6 h with either dimethyl sulfoxide (DMSO) alone for controls (lanes ‘−’) or 50 µm MG132 to block proteasome (lanes ‘+’). The cells were then collected and analyzed by quantitative western blot with specific antibodies. The top panel shows a typical western blot, the bottom histogram (▪ DMSO; □ MG132) is a quantification (mean ± SEM) from three experiments. *P < 0.05, Mann and Whitney test comparison of DMSO- vs. MG132-treated cells.
Stability and localization of the mutant Trisk 32-T59R after in vivo expression
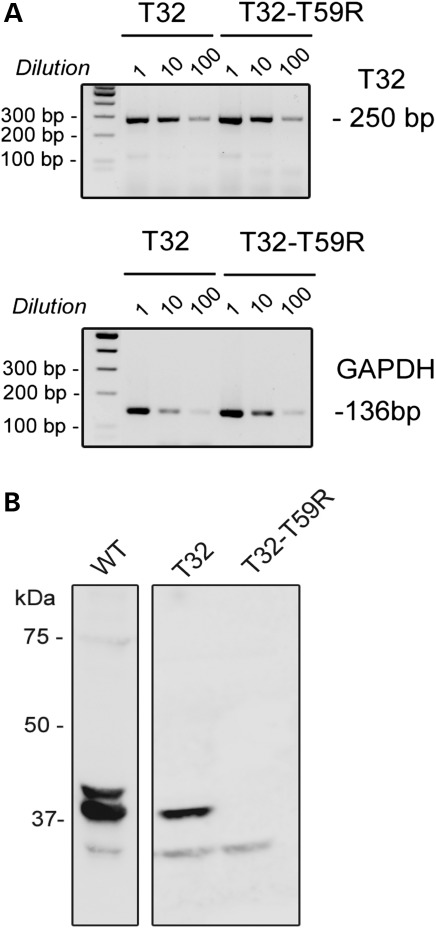
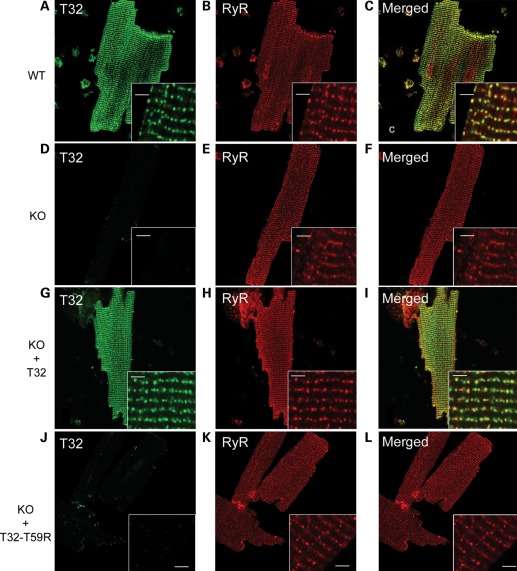
The behavior of the mutant Trisk 32-T59R was further studied after re-expression in the triadin KO mouse. The triadin KO mouse, a mouse line devoid of any triadin isoform, has been previously shown to present both cardiac arrhythmia (8) and muscle weakness (9). Viruses [AAV2/9, which preferentially target the heart in adult mice (21,22)] were developed to induce in vivo expression of the WT and mutant Trisk 32 proteins in the heart of triadin KO mice. One month after systemic injection of the viral vectors, the cardiomyocytes were isolated and characterized by different methodological approaches. The mRNA expression levels studied by semi-quantitative reverse transcription polymerase chain reaction (RT–PCR) of the WT Trisk 32 and the mutant Trisk 32-T59R were similar (Fig. 4A), showing that the mutation did not affect transcript stability. In contrast, the determination of the protein expression levels by quantitative western blot on cardiomyocytes homogenates (Fig. 4B) showed that no mutant protein could be detected in any of the six transduced mice, whereas the transduced WT Trisk 32 was detected in all the mice (Fig. 4B and Supplementary Material, Fig. S3). This difference in expression was confirmed by immunofluorescence labeling on fixed cardiomyocytes (Fig. 5). In WT cardiomyocytes (Fig. 5A–C), Trisk 32 presented a typical dyad labeling, a row of dots co-localized with RyR2. In the triadin KO cardiomyocytes (Fig. 5D–F), Trisk 32 labeling was absent and RyR2 labeling was not modified. After transduction of Trisk 32 or Trisk 32-T59R in the triadin KO mice, only the transduced WT Trisk 32 was detected, in the dyads of cardiomyocytes, where it perfectly colocalized with RyR2 (Fig. 5G–I) while the transduced mutant Trisk 32-T59R was not detectable (Fig. 5J–L). This suggests although the mRNA of Trisk 32-T59R is expressed in the cardiomyocytes of the mice, the protein is absent.
Figure 4.
Stability analyses of transcripts and proteins after in vivo expression in triadin KO mice. (A) The mutant transcript is present at similar levels as the WT one. Messenger RNAs were purified from isolated cardiomyocytes of transduced mice. After reverse transcription, the cDNAs were either directly used or diluted 10 or 100 times before the PCR amplification of Trisk 32 (T32) and GAPDH. PCR products for Trisk 32 and GAPDH amplification were expected, respectively, at 250 and 136 bp. The PCR primers for GAPDH were designed to co-amplify possible contaminating genomic DNA as a 220 bp fragment, which was not observed. (B) The mutant protein Trisk 32-T59R was not detectable. Western blot analysis of 100 µg of cardiomyocytes homogenates from control WT mouse, with mouse-specific anti-Trisk 32 antibody (first lane), or 100 µg of cardiomyocytes from a Trisk 32-transduced mouse (second lane), or Trisk 32-T59R-transduced mouse (third lane) with rat-specific anti-Trisk 32 antibody. Trisk 32 appears as multiple bands (2/3) centered on 37 kDa, the higher bands, the glycosylated forms of Trisk 32, being in lower amount and visible in correlation with the intensity of the signal, as described before (6,20).
Figure 5.
Immunofluorescent analysis of Trisk 32 in isolated cardiomyocytes. Cardiomyocytes were isolated from a WT mouse, a triadin KO mouse (KO), a triadin KO mouse transduced with rat Trisk 32 (KO + T32) and a triadin KO mouse transduced with rat Trisk 32-T59R (KO + T32-T59R). They were labeled with antibodies against mouse Trisk 32 and RyR (A–F) or against rat Trisk 32 and RyR (G–L). In all cardiomyocytes, RyR labeling is typical of a dyad labeling showing aligned rows of dots, as observed in the inserts. Bar: 2 µm.
DISCUSSION
CPVT is a life-threatening inheritable disease that predisposes young individuals with normal cardiac structure to cardiac arrest. Pre-symptomatic genetic screening is of major importance for CPVT families, since life-saving preventive treatments are available. Therefore, the identification of causative mutations constitutes a major issue for CPVT families. Up to now, two genes have been involved in this disease. They encode proteins which constitute the core of the cardiac CRC: RyR2 in the dominant forms of CPVT and CSQ2 in the recessive forms. However, almost one-third of the patients remained without an identified molecular defect. In the present work, we searched for mutations in other genes in a panel of 97 CVPT patients without mutations in the RYR2 and CASQ2 genes. We focused our screening on triadin and junctin which are the physiological partners of RyR2 and CSQ2 in the CRC. No mutation was identified in junctin, whereas three mutations were identified in triadin. The characterization of the physiopathological mechanisms is of uppermost importance for a better understanding of the disease and for the development of new therapies, as the current therapies are not fully satisfying (1). Along this line, we searched for the functional consequences of the TRDN mutations. Two of these mutations resulted in a premature stop codon, and the corresponding proteins are most probably not produced as a consequence of NMD (18). Thus, we focused our investigation on the third mutation, the missense p.T59R.
To study the consequences of this mutation, we analyzed the localization and the stability of the mutant protein in COS-7 cells and in cardiomyocytes. All the experiments we performed, either in transfected COS-7 cells or in cardiomyocytes from triadin KO mice transduced in vivo, showed that the p.T59R mutation resulted in instability of the protein, leading to its degradation. It would be interesting in this context to assay the ability of blockers of ER quality control or of molecular chaperones to restore the folding of Trisk 32-T59R and its correct targeting instead of its degradation, as done for α-sarcoglycan mutations in Limb Girdle Muscular Dystrophy type 2D (23,24). The likely absence of the protein as a result of the mutation in patient's heart could only be further confirmed by western blot on a cardiac biopsy, which was not available. As the two other mutations (p.D18Afs*13, homozygous mutation in Family 1 and p.Q205*, compound heterozygous mutation associated to p.T59R in Family 2) most likely resulted in the absence of the protein, all the affected patients in this study can be considered natural triadin KO cases. The severe CPVT phenotype observed in these patients thus highlights the role of triadin in the regulation of the calcium release during cardiac contraction. In addition, the identified mutations mapped to a region of the TRDN gene which is common to all triadin isoforms, including the skeletal muscle Trisk 95 and Trisk 51. Thus, these three mutations would also lead to an absence of triadin in the skeletal muscle. This would give a possible explanation for the associated muscle weakness that was clinically observed in individual II:2 in Family 2. The affected patient in Family 1 died so young (2 years) that muscle weakness could not have been detected, but it could be envisaged that CPVT related to triadin mutation systematically affect both cardiac and skeletal muscle. Therefore, when a CPVT patient presents also with a moderate muscle weakness, screening of the TRDN gene should be performed at first. In this context, the triadin KO mouse is a good model to study the consequences of triadin deletion in human, as it presents with the same disease hallmarks: muscle weakness (9) and cardiac arrhythmia induced by β-adrenergic stimulation (8). Different mechanisms have been proposed to explain the cardiac arrhythmia resulting of RYR2 or CASQ2 mutations (2,25). Among the possible mechanisms, one is related to the dissociation of FKBP12.6 from RyR2 in the case of RYR2 mutation, which would result in a leaky Ca2+ channel (26). This mechanism could be envisaged for the triadin deletion resulting from the mutations identified in the present study, as it has been proposed in the triadin KO mouse that triadin ablation could impair FKBP12/RyR1 interaction in skeletal muscle (27). Such a hypothesis should nevertheless be confirmed in the heart for FKBP12.6/RyR2 interaction. Another mechanism proposed as a result of CASQ2 mutations is a reduction in the amount of CSQ2 or in the calcium-binding capacity of the mutant CSQ2. Such a mechanism could also be proposed in the case of the identified triadin mutations, as a CSQ2 reduction has been observed in the triadin KO mouse heart (8). Therefore, the triadin mutations identified in the present study and the characterization of the triadin KO mice lines performed previously (8,9) confirmed the hypothesis that CPVT could be more generally the result of a functional defect in the CRC (2,28). Our study clearly showed that mutations in the TRDN gene are responsible for CPVT in the human and is therefore a gene to systematically investigate when screening familial CPVT. Nevertheless, the absence of mutation in RYR2, CASQ2, ASPH and TRDN in the 94 remaining patients of this cohort points to the limitation of the gene candidate approach. To search for other genes that might be involved, a large-scale screening should be performed in these patients. Finally, this paper described for the first time an involvement of the TRDN gene in human pathology, and interestingly, although this gene encodes for cardiac and muscle triadins, the pathology is mainly a cardiac one.
MATERIALS AND METHODS
Ethics statement
Investigations of patient population reported in the paper received a formal approval from the Regional Medical Ethical Committee (Comité de Protection des Personnes, CHU de Saint-Etienne; France). All procedures using animals were approved by the Institutional ethics committee and followed the guidelines of the National Research Council Guide for the care and use of laboratory animals.
Clinical evaluation of probands
The patient population consisted in 97 CPVT probands carefully characterized after clinical examination and electrocardiographic records. Referred to the clinical centers for syncope with or without seizures, or cardiac arrest, they have been diagnosed on the basis of typical polymorphic ventricular arrhythmia recorded on 12-lead ECG, and after 24 h Holter ECG and exercise stress test. Family members were evaluated at least by resting ECG, and when possible a 24 h ECG Holter recordings and exercise stress test were performed.
Genetic analysis
Blood samples used for genetic evaluation were obtained after a written informed consent was signed by patients according to the French regulation for genetic studies. Molecular screening of RYR2 and CASQ2 genes has been performed as described before (29). Screening of the two junctin isoforms was performed by sequencing the six exons (NM_02164.4) corresponding to the longest isoform. Molecular analysis of the TRDN gene was performed by sequencing all the exons that encode the cardiac human isoform Trisk 32. Because of the large number of triadin transcripts described in the cardiac tissue originating from different species (19,30,31) without the experimental evidences for the existence of the corresponding proteins, we first characterized the human Trisk 32 sequence both at the cDNA and at the protein level using human cardiac tissue (Supplementary Material, Fig. S4 and Supplementary Methods). The presence of the corresponding protein was confirmed using an antibody developed against the specific C-terminal end of the identified sequence (Supplementary Material, Fig. S4c). The sequence of human Trisk 32 identified in this study is accessible in GenBank database under accession number JN900469. Primers sequences used for molecular investigation of the TRDN and ASPH genes are listed in Supplementary Material, Table S1. Family members were screened for the mutation(s) identified in probands. A panel of 300 control chromosomes was screened for each novel variant reported in this study.
Plasmids, viruses and antibodies
Full-length cDNA coding for rat Trisk 32 (accession number: EMBL AJ812276), with or without c.176C>G mutation, corresponding to p.T59R change in the protein, was inserted into pcDNA3.1 (Invitrogen, Cergy Pontoise, France) for cell transfection, or in pZac2.1 for AAV development. The rat Trisk 32 nucleotide sequence used in this study has 84% identity with the human sequence and 92% with the mouse sequence, and was chosen because of the availability of efficient antibodies (20) developed in different species and necessary for the double immunofluorescent labeling. All viruses were engineered and produced by the Penn Vector Core (Philadelphia, PA, USA). Two recombinant adeno-associated viruses of serotype 2/9, encoding respectively the WT and the mutant T59R–Trisk 32 were used in this study: AAV-Trisk 32 and AAV-Trisk 32-T59R. All the transgenes were under the control of a CMV promoter.
Rabbit anti-RyR antibodies have been described previously (32), as well as rabbit anti-peptide antibodies against the N-terminal end of triadins (33), and anti-peptide antibodies against the rat Trisk 32 C-terminal end (20), developed in rabbit or guinea pig. We also used anti-peptide antibodies against the mouse Trisk 32 (9), and an anti-RyR developed in guinea pig by immunization with the C-terminal peptide of RyR1, as described before (34). The rabbit polyclonal antibody to calnexin (SPA-860) is from Stressgen (Victoria, Canada).
Cell culture, transfection and treatment
COS-7 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). Cells were transfected using Exgen 500 (Euromedex, Mundolsheim, France) for 28 h for immunofluorescent labeling experiments and for 20 h for western blot analysis after CHX or MG132 treatment.
For triadin stability experiment, transfected COS-7 cells were treated with 50 µg/ml of CHX for 0–6 h, or with 50 µm MG132 for 6 h, or with 0.2% dimethyl sulfoxide (DMSO) for 6 h (control). Cells were then lysed with radioimmuno precipitation assay buffer [25 mm Tris, pH 7.6, 150 mm NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)], centrifuged and proteins in the supernatant were precipitated by chloroform/methanol for further analysis by quantitative western blot.
Immunofluorescent labeling on COS-7 cells and on cardiomyocytes
Transfected COS-7 cells were fixed either 6 min in cold methanol at −20°C, or 10 min in 4% paraformaldehyde (PFA). For detection of Trisk 32 at the cell surface, cells fixed with PFA were incubated with a guinea pig antibody against the C-terminus of rat Trisk 32. After permeabilization with 0.05% saponin, cells were incubated with a rabbit antibody against the N-terminus of rat Trisk 32 and then with secondary antibodies against both species.
Primary cardiomyocytes were seeded 1 h on laminin-coated glass cover slip, fixed 15 min in 4% PFA, permeabilized for 30 min in phosphate buffered saline (PBS)—0.5% Triton X-100, then saturated 1 h in PBS—0.1% Triton X-100—0.5% bovine serum albumin—2% goat serum, and incubated with primary antibodies.
The cells were observed under a LEICA SPE confocal microscope.
Mice and in vivo transduction
Triadin KO mice developed on C57Bl6 background were described previously (9). Two-month-old triadin KO male mice (19–24 g) were anesthetized with xylazine (10 µg/g) and ketamine (100 µg/g), and injected with 2 × 1011 viral genome copy diluted in 100 µl of 150 mm NaCl, by a single injection in the tail vein.
Cardiomyocytes preparation
One month after virus injection, the hearts of transduced KO mice of non-transduced KO mice or of WT littermate mice were collected and cardiac ventricular myocytes were enzymatically dissociated as previously reported (35). Briefly, mice were euthanized by cervical dislocation, the heart rapidly excised and retrogradely perfused at 37°C for 6–8 min with a modified tyrode solution (in mm: NaCl 113, KCl 4.7, KH2PO4 0.6, Na2HPO4 0.6, MgSO4 1.2, NaHCO3 12, KHCO3 10, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid 10 Taurine 30, pH = 7.4) containing 0.1 mg/ml liberase (Roche, France). Isolated cardiomyocytes were then transferred to the same solution, free of enzyme, containing 1 mm CaCl2.
RT–PCR
mRNA expression levels in transduced cardiomyocytes were determined by semi-quantitative RT–PCR. Briefly, total RNA was extracted from isolated cardiomyocytes using TRIzol Reagent and PureLink RNA Mini Kit (Invitrogen). First-strand cDNA was synthesized from 500 ng of total RNA using specific primer mixes followed by cDNA amplification. Primer sequences were as follows: rat Trisk 32: forward, 5′-AGCCAAAGAAACTCCGAAAA-3′; reverse, 5′-TCCAGTGGC CGTATGTACTTC-3′; mouse GAPDH: forward, 5′-CGTGCCGCCTGGAGAAAC-3′; reverse, 5′-TGGGAGTTGCTGTTGAAGTCG-3′. Equal amounts of cDNA were used in each reaction sample.
Western blot
Cardiomyocytes or transfected cells were homogenized by 15 s sonication at 0°C in the presence of 1 mm DFP and 200 µm PMSF as proteases inhibitors. One hundred micrograms of cardiomyocyte homogenates were analyzed by quantitative western blot analysis as described previously (9), with antibodies against rat Trisk 32 (20) or mouse Trisk 32 (9). Secondary antibodies used for western blot were labeled with horseradish peroxydase (Jackson ImmunoResearch Laboratories, Immunotech S.A.S., Marseille, France). The quantification was performed with a Chemidoc and the Quantity One software (BioRad, Marnes la Coquette, France).
Statistics
Statistics were performed using either Fisher's test (Figs 2A and 3A) or Mann and Whitney's test (Fig. 3B) on Prism 4.0 software (GraphPad, San Diego, CA, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Association Française contre les Myopathies (AFM), the Centre National de la Recherche Scientifique (CNRS), the Fondation Daniel Ducoin, the Programme Hospitalier de Recherche Clinique (PHRC), the Direction de la Recherche Clinique du CHU Grenoble and the Société Française de Myologie (SFM). Funding to pay the Open Access publication charges for this article was provided by Foundation Daniel Ducoin.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all family members for their contribution to this study. We thank Mrs F. Berthoin, D. Martinez and Mr J. Trapani for their technical assistance. We thank Dr Jacques Brocard for his help in the statistical analysis of the data, the Penn Vector Core (Philadelphia, USA) for viruses production, and the Myocastor study group (MSG) for fruitful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hayashi M., Denjoy I., Extramiana F., Maltret A., Buisson N.R., Lupoglazoff J.M., Klug D., Hayashi M., Takatsuki S., Villain E., et al. Incidence and risk factors of arrhythmic events in catecholaminergic polymorphic ventricular tachycardia. Circulation. 2009;119:2426–2434. doi: 10.1161/CIRCULATIONAHA.108.829267. doi:10.1161/CIRCULATIONAHA.108.829267. [DOI] [PubMed] [Google Scholar]
- 2.Priori S.G., Chen S.R. Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ. Res. 2011;108:871–883. doi: 10.1161/CIRCRESAHA.110.226845. doi:10.1161/CIRCRESAHA.110.226845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo W., Campbell K.P. Association of triadin with the ryanodine receptor and calsequestrin in the lumen of the sarcoplasmic reticulum. J. Biol. Chem. 1995;270:9027–9030. doi: 10.1074/jbc.270.16.9027. doi:10.1074/jbc.270.16.9027. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Kelley J., Schmeisser G., Kobayashi Y.M., Jones L.R. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 1997;272:23389–23397. doi: 10.1074/jbc.272.37.23389. doi:10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 5.Thevenon D., Smida-Rezgui S., Chevessier F., Groh S., Henry-Berger J., Romero N.B., Villaz M., De Waard M., Marty I. Human skeletal muscle triadin: gene organization and cloning of the major isoform, Trisk 51. Biochem. Biophys. Res. Commun. 2003;303:669–675. doi: 10.1016/s0006-291x(03)00406-6. doi:10.1016/S0006-291X(03)00406-6. [DOI] [PubMed] [Google Scholar]
- 6.Marty I., Fauré J., Fourest-Lieuvin A., Vassilopoulos S., Oddoux S., Brocard J. Triadin: what possible function 20 years later? J. Physiol. 2009;587:3117–3121. doi: 10.1113/jphysiol.2009.171892. doi:10.1113/jphysiol.2009.171892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen X., Franzini-Armstrong C., Lopez J.R., Jones L.R., Kobayashi Y.M., Wang Y., Kerrick W.G., Caswell A.H., Potter J.D., Miller T., et al. Triadins modulate intracellular Ca(2+) homeostasis but are not essential for excitation-contraction coupling in skeletal muscle. J. Biol. Chem. 2007;282:37864–37874. doi: 10.1074/jbc.M705702200. doi:10.1074/jbc.M705702200. [DOI] [PubMed] [Google Scholar]
- 8.Chopra N., Yang T., Asghari P., Moore E.D., Huke S., Akin B., Cattolica R.A., Perez C.F., Hlaing T., Knollmann-Ritschel B.E., et al. Ablation of triadin causes loss of cardiac Ca2+ release units, impaired excitation-contraction coupling, and cardiac arrhythmias. Proc. Natl Acad. Sci. USA. 2009;106:7636–7641. doi: 10.1073/pnas.0902919106. doi:10.1073/pnas.0902919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oddoux S., Brocard J., Schweitzer A., Szentesi P., Giannesini B., Brocard J., Fauré J., Pernet-Gallay K., Bendahan D., Lunardi J., et al. Triadin deletion induces impaired skeletal muscle function. J. Biol. Chem. 2009;284:34918–34929. doi: 10.1074/jbc.M109.022442. doi:10.1074/jbc.M109.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones L.R., Zhang L., Sanborn K., Jorgensen A.O., Kelley J. Purification, primary structure, and immunological characterization of the 26-kDa calsequestrin binding protein (junctin) from cardiac junctional sarcoplasmic reticulum. J. Biol. Chem. 1995;270:30787–30796. doi: 10.1074/jbc.270.51.30787. doi:10.1074/jbc.270.51.30787. [DOI] [PubMed] [Google Scholar]
- 11.Dinchuk J.E., Henderson N.L., Burn T.C., Huber R., Ho S.P., Link J., O'Neil K.T., Focht R.J., Scully M.S., Hollis J.M., et al. Aspartyl beta-hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J. Biol. Chem. 2000;275:39543–39554. doi: 10.1074/jbc.M006753200. doi:10.1074/jbc.M006753200. [DOI] [PubMed] [Google Scholar]
- 12.Lim K.Y., Hong C.S., Kim D.H. cDNA cloning and characterization of human cardiac junctin. Gene. 2000;255:35–42. doi: 10.1016/s0378-1119(00)00299-7. doi:10.1016/S0378-1119(00)00299-7. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Q., Fan G.C., Dong M., Altschafl B., Diwan A., Ren X., Hahn H.H., Zhao W., Waggoner J.R., Jones L.R., et al. Sarcoplasmic reticulum calcium overloading in junctin deficiency enhances cardiac contractility but increases ventricular automaticity. Circulation. 2007;115:300–309. doi: 10.1161/CIRCULATIONAHA.106.654699. doi:10.1161/CIRCULATIONAHA.106.654699. [DOI] [PubMed] [Google Scholar]
- 14.den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. doi:10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 15.Realini C., Rogers S.W., Rechsteiner M. KEKE motifs. Proposed roles in protein-protein association and presentation of peptides by MHC class I receptors. FEBS Lett. 1994;348:109–113. doi: 10.1016/0014-5793(94)00569-9. doi:10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y.M., Alseikhan B.A., Jones L.R. Localization and characterization of the calsequestrin-binding domain of triadin 1. Evidence for a charged beta-strand in mediating the protein-protein interaction. J. Biol. Chem. 2000;275:17639–17646. doi: 10.1074/jbc.M002091200. doi:10.1074/jbc.M002091200. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.M., Rho S.H., Shin D.W., Cho C., Park W.J., Eom S.H., Ma J., Kim D.H. Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J. Biol. Chem. 2004;279:6994–7000. doi: 10.1074/jbc.M312446200. [DOI] [PubMed] [Google Scholar]
- 18.Bhuvanagiri M., Schlitter A.M., Hentze M.W., Kulozik A.E. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010;430:365–377. doi: 10.1042/BJ20100699. doi:10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi Y.M., Jones L.R. Identification of triadin 1 as the predominant triadin isoform expressed in mammalian myocardium. J. Biol. Chem. 1999;274:28660–28668. doi: 10.1074/jbc.274.40.28660. doi:10.1074/jbc.274.40.28660. [DOI] [PubMed] [Google Scholar]
- 20.Vassilopoulos S., Thevenon D., Smida Rezgui S., Urbani-Brocard J., Chapel A., Lacampagne A., Lunardi J., DeWaard M., Marty I. Triadins are not triad specific proteins: two new skeletal muscle triadins possibly involved in the architecture of sarcoplasmic reticulum. J. Biol. Chem. 2005;280:28601–28609. doi: 10.1074/jbc.M501484200. doi:10.1074/jbc.M501484200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacak C.A., Mah C.S., Thattaliyath B.D., Conlon T.J., Lewis M.A., Cloutier D.E., Zolotukhin I., Tarantal A.F., Byrne B.J. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 2006;99:e3–e9. doi: 10.1161/01.RES.0000237661.18885.f6. doi:10.1161/01.RES.0000237661.18885.f6. [DOI] [PubMed] [Google Scholar]
- 22.Inagaki K., Fuess S., Storm T.A., Gibson G.A., Mctiernan C.F., Kay M.A., Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. doi:10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartoli M., Gicquel E., Barrault L., Soheili T., Malissen M., Malissen B., Vincent-Lacaze N., Perez N., Udd B., Danos O., Richard I. Mannosidase I inhibition rescues the human alpha-sarcoglycan R77C recurrent mutation. Hum. Mol. Genet. 2008;17:1214–1221. doi: 10.1093/hmg/ddn029. doi:10.1093/hmg/ddn029. [DOI] [PubMed] [Google Scholar]
- 24.Soheili T., Gicquel E., Poupiot J., N'Guyen L., Le Roy F., Bartoli M., Richard I. Rescue of sarcoglycan mutations by inhibition of endoplasmic reticulum quality control is associated with minimal structural modifications. Hum. Mutat. 2012;33:429–439. doi: 10.1002/humu.21659. doi:10.1002/humu.21659. [DOI] [PubMed] [Google Scholar]
- 25.Liu N., Rizzi N., Boveri L., Priori S.G. Ryanodine receptor and calsequestrin in arrhythmogenesis: what we have learnt from genetic diseases and transgenic mice. J. Mol. Cell Cardiol. 2009;46:149–159. doi: 10.1016/j.yjmcc.2008.10.012. doi:10.1016/j.yjmcc.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Wehrens X.H., Lehnart S.E., Huang F., Vest J.A., Reiken S.R., Mohler P.J., Sun J., Guatimosim S., Song L.S., Rosemblit N., et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. doi:10.1016/S0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 27.Eltit J.M., Feng W., Lopez J.R., Padilla I.T., Pessah I.N., Molinski T.F., Fruen B.R., Allen P.D., Perez C.F. Ablation of skeletal muscle triadin impairs FKBP12/RyR1 channel interactions essential for maintaining resting cytoplasmic Ca2+ J. Biol. Chem. 2010;285:38453–38462. doi: 10.1074/jbc.M110.164525. doi:10.1074/jbc.M110.164525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maclennan D.H., Zvaritch E. Mechanistic models for muscle diseases and disorders originating in the sarcoplasmic reticulum. Biochim. Biophys. Acta. 2011;1813:948–964. doi: 10.1016/j.bbamcr.2010.11.009. doi:10.1016/j.bbamcr.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Roux-Buisson N., Rendu J., Denjoy I., Guicheney P., Goldenberg A., David N., Faivre L., Barthez O., Danieli G.A., Marty I., et al. Functional analysis reveals splicing mutations of the CASQ2 gene in patients with CPVT: implication for genetic counselling and clinical management. Hum. Mutat. 2011;32:995–999. doi: 10.1002/humu.21537. doi:10.1002/humu.21537. [DOI] [PubMed] [Google Scholar]
- 30.Guo W., Jorgensen A.O., Jones L.R., Campbell K.P. Biochemical characterization and molecular cloning of cardiac triadin. J. Biol. Chem. 1996;271:458–465. doi: 10.1074/jbc.271.1.458. doi:10.1074/jbc.271.1.458. [DOI] [PubMed] [Google Scholar]
- 31.Hong C.S., Ji J.H., Kim J.P., Jung D.H., Kim D.H. Molecular cloning and characterization of mouse cardiac triadin isoforms. Gene. 2001;278:193–199. doi: 10.1016/s0378-1119(01)00718-1. doi:10.1016/S0378-1119(01)00718-1. [DOI] [PubMed] [Google Scholar]
- 32.Marty I., Robert M., Villaz M., Lai Y., De Jongh K.S., Catterall W.A., Ronjat M. Biochemical evidence for a complex involving dihydropyridine receptor and ryanodine receptor in triad junctions of skeletal muscle. Proc. Natl Acad. Sci. USA. 1994;91:2270–2274. doi: 10.1073/pnas.91.6.2270. doi:10.1073/pnas.91.6.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marty I., Robert M., Ronjat M., Bally I., Arlaud G., Villaz M. Localization of the N-terminal and C-terminal ends of triadin with respect to the sarcoplasmic reticulum membrane of rabbit skeletal muscle. Biochem. J. 1995;307:769–774. doi: 10.1042/bj3070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marty I., Villaz M., Arlaud G., Bally I., Ronjat M. Transmembrane orientation of the N-terminal and C-terminal ends of the ryanodine receptor in the sarcoplasmic reticulum of rabbit skeletal muscle. Biochem. J. 1994;298:743–749. doi: 10.1042/bj2980743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fauconnier J., Thireau J., Reiken S., Cassan C., Richard S., Matecki S., Marks A.R., Lacampagne A. Leaky RyR2 trigger ventricular arrhythmias in Duchenne muscular dystrophy. Proc. Natl Acad. Sci. USA. 2010;107:1559–1564. doi: 10.1073/pnas.0908540107. doi:10.1073/pnas.0908540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.