Abstract
Background
Diagnostic criteria of Alzheimer's disease (AD) emphasize the integration of clinical data and biomarkers. In practice, collection and analysis of patient data vary greatly across different countries and clinics. Objective: The goal was to develop a versatile and objective clinical decision support system that could reduce diagnostic errors and highlight early predictors of AD.
Methods
Novel data analysis methods were developed to derive composite disease indicators from heterogeneous patient data. Visualizations that communicate these findings were designed to help the interpretation. The methods were implemented with a software tool that is aimed for daily clinical practice.
Results
With the tool, clinicians can analyze available patients as a whole, study them statistically against previously diagnosed cases, and characterize the patients with respect to having AD. The tool is able to work with virtually any patient measurement data, as long as they are stored in electronic format or manually entered into the system. For a subset of patients from the test cohort, the tool was able to predict conversion to AD at an accuracy of 93.6%.
Conclusion
The software tool developed in this study provides objective information for early detection and prediction of AD based on interpretable visualizations of patient data.
Key Words: Alzheimer's disease, Biomarker, Decision support, Mild cognitive impairment, Memory
Introduction
Knowledge of Alzheimer's disease (AD) is increasing at a rate never seen before, with medical data acquisition methods and diagnostic procedures both driving and being evolved by the change. This is a blessing and a curse on clinicians, who now have an ever-increasing body of knowledge available on which to base diagnostic decisions. People can cognitively handle only few pieces of evidence at any single time [1], with information shown last influencing the decisions more [2]. Making an AD diagnosis is also a complex task, where interruptions can affect the quality of decisions [3].
Diagnostic decision support systems (DDSSs) have the potential to help manage large volumes of data. Traditionally, DDSSs have employed natural language processing, case-based reasoning, and rule-based expert systems to suggest potential diagnoses [4]. They have rarely used measurement data for the analyses, and the ones that do usually require a predefined set of measurements to be available.
The main contributions of this paper are the visualizations of patient data that have been generated with a novel DDSS, whose goal is to alleviate data-related issues in complex diagnostic decisions.
Materials and Methods
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). ADNI recruited approximately 400 people with mild cognitive impairment (MCI) to be followed for 4 years, with the primary goal to test multiple biological markers concerning progression of MCI and early AD.
A statistical decision support system, developed for early prediction of AD and described in detail in Mattila et al. [5], was used to evaluate patients' disease progression at baseline. The system compares each patient measure to the corresponding measure acquired from healthy and AD cases available in large databases, and computes an index reflecting the patient's match to these two populations. The index is computed for each measure, for each modality (e.g. MRI, cerebrospinal fluid, Alzheimer's Disease Assessment Scale) and finally for all data measured. The indices are visualized both numerically and graphically making fast interpretation of all heterogeneous patient data possible based on the principles of evidence-based medicine. The system is currently implemented as proprietary PC software running on a regular laptop computer. It is not restricted to ADNI and can be extended to work with any electronic datasets, consisting of textual, categorical, and scalar patient measurement values. A web-based version of the system is also in consideration.
Results
Using baseline data from all MCI cases in ADNI, the DDSS tool was able to discriminate stable MCI patients from those who converted to AD (average conversion time 19 months) at an accuracy of 68.6% representing the level reported typically for ADNI. However, if a reliability estimate is available, it becomes possible to identify a subset of patients for whom the diagnosis is possible with a much higher accuracy. By thresholding the index values given to patients by the DDSS tool, patients were automatically assigned to subgroups which attained considerably improved prediction of conversion to AD and also of non-conversion (table 1).
Table 1.
Prediction accuracy for several subgroups of patients
| Group of patients based on DDSS evaluation | Share of patients assigned into the group, % | Prediction accuracy for the group of patients, % |
|---|---|---|
| Very strong evidence of having AD | 2.9 | 79.8 |
| Very strong evidence of not having AD | 6.3 | 100.0 |
| Very strong evidence (AD/not AD, total) | 9.2 | 93.7 |
| Strong evidence of having AD | 11.7 | 79.7 |
| Strong evidence of not having AD | 18.3 | 87.5 |
| Strong evidence (AD/not AD, total) | 30.1 | 84.4 |
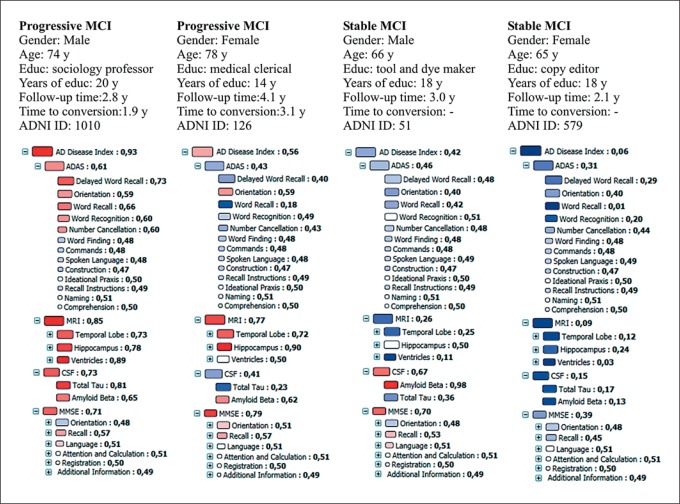
Figure 1 shows the disease indices and graphical fingerprint visualization computed from baseline measurements for 4 MCI cases of whom 2 converted to AD later (progressive MCI) and 2 remained stable.
Fig. 1.
Disease state fingerprint and background information for 4 MCI cases at baseline: 2 cases converted later to AD and 2 remained stable. The size of the box indicates the relevance/importance of the parameter in separating healthy controls and AD cases, and the color indicates the patient's match to healthy (more blue) and AD (more red) populations.
Discussion
Most publications reporting on the performance of certain patient measures in diagnostics do not estimate the reliability of classification for each individual but report only classification accuracy, sensitivity, specificity or ROC curves for the whole population. This may be one reason for the low adaptation of these modern and objective techniques into clinical practice; the classification accuracy is just too low. We have developed a DDSS which allows clinicians to make observations from all available data simultaneously, and provides evidence-based and objective information about the status of the patient for improving the diagnostic accuracy and confidence. We reported prediction accuracies of 93.7% (for 9.2% of cases) and 84.4% (for 30.1% of cases). Whether 93.6 or 84.0% are acceptable accuracies for making the diagnosis earlier is difficult to define, especially when the ground truth diagnoses from ADNI are clinical, not pathologically confirmed diagnoses. Nevertheless, the diagnosis is not based on data alone but requires interviewing the patient and relatives. Knowing that the prediction accuracy, based only on data, is 93.6 or 84.0% rather than 68.6% can give more confidence for making the diagnosis earlier.
Acknowledgements
This work was partially funded under the 7th Framework Program by the European Commission (http.//cordis.europa.eu/ist; EU-Grant-224328-PredictAD; Name: From Patient Data to Personalized Healthcare in Alzheimer's Disease). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the private sector as well as non-profit partners. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
References
- 1.Cowan N. The magical number 4 in short-term memory: a reconsideration of mental storage capacity. Behav Brain Sci. 2001;24:87–114. doi: 10.1017/s0140525x01003922. [DOI] [PubMed] [Google Scholar]
- 2.Bergus GR, Chapman GB, Gjerde C, Elstein AS. Clinical reasoning about new symptoms in the face of pre-existing disease: sources of error and order effects. Fam Med. 1995;27:314–320. [PubMed] [Google Scholar]
- 3.Speier C, Valacich JS, Vessey I. The influence of task interruption on individual decision making: an information overload perspective. Decision Sci. 1999;30:337–360. [Google Scholar]
- 4.Musen MA, Sharar Y, Shortliffe EH. Biomedical Informatics. New York: Springer; 2006. Clinical Decision-Support Systems; pp. 698–736. [Google Scholar]
- 5.Mattila J, Koikkalainen J, Virkki A, Simonsen A, van Gils M, Waldemar G, Soininen H, Lötjönen J. A disease state fingerprint for evaluation of Alzheimer's diseases. J Alzheimers Dis. [DOI] [PubMed]