Abstract

The DNA and RNA repair protein AlkB removes alkyl groups from nucleic acids by a unique iron- and α-ketoglutarate-dependent oxidation strategy. When alkylated adenines are used as AlkB targets, earlier work suggests that the initial target of oxidation can be the alkyl carbon adjacent to N1. Such may be the case with ethano-adenine (EA), a DNA adduct formed by an important anticancer drug, BCNU, whereby an initial oxidation would occur at the carbon adjacent to N1. In a previous study, several intermediates were observed suggesting a pathway involving adduct restructuring to a form that would not hinder replication, which would match biological data showing that AlkB almost completely reverses EA toxicity in vivo. The present study uses more sensitive spectroscopic methodology to reveal the complete conversion of EA to adenine; the nature of observed additional putative intermediates indicates that AlkB conducts a second oxidation event in order to release the two-carbon unit completely. The second oxidation event occurs at the exocyclic carbon adjacent to the N6 atom of adenine. The observation of oxidation of a carbon at N6 in EA prompted us to evaluate N6-methyladenine (m6A), an important epigenetic signal for DNA replication and many other cellular processes, as an AlkB substrate in DNA. Here we show that m6A is indeed a substrate for AlkB and that it is converted to adenine via its 6-hydroxymethyl derivative. The observation that AlkB can demethylate m6A in vitro suggests a role for AlkB in regulation of important cellular functions in vivo.
Introduction
Nuclear and mitochondrial DNA is damaged by radiation, by organic and inorganic chemical agents and by the misdirected activity of enzymes. Exogenous and endogenous processes that contribute to genomic damage include oxidation, alkylation, and deamination of DNA.1,2 Adducts arising from nucleic acid damage may cause mutations, jeopardize epigenetic patterns, block DNA and RNA synthesis, inhibit and alter the coding of mRNA transcription and translation, and promote strand breaks.2 Many chemical anticancer agents have been designed to form toxic DNA adducts. One example, BCNU (1,3-bis(2-chloroethyl)-1-nitrosourea), has been used to treat lymphoma, multiple myeloma, and several types of brain cancer.3−5 Among the toxic DNA adducts formed by this chemotherapeutic agent is 1,N6-ethanoadenine (EA, Figure 1a), which blocks DNA replication.6−8 In the EA DNA adduct, the exocyclic N6-nitrogen of adenine is connected to the N1 ring nitrogen by a saturated two-carbon bridge, creating a five-membered ring involving the N1 and N6 atoms that otherwise would be involved in canonical Watson–Crick H-bonding (Figure 1a). EA is repaired to some extent by the Escherichia coli repair protein 3-methyladenine DNA glycosylase (AlkA)9 and the human alkyladenine DNA glycosylase MPG (also called AAG, ANPG, or APNG).7 However, for both of the enzymes, the excision of EA is far less efficient than that of the structurally related 1,N6-ethenoadenine (eA), in which the two-carbon bridge is unsaturated (Figure 1d).9 Because EA lacks the structural features needed to form a Watson–Crick base pair with thymine, it is likely to be both toxic and mutagenic. In the absence of repair, EA blocks polymerase bypass and miscodes during attempted replication by mammalian DNA polymerases in vitro.8 The analogous eA adduct, which is also missing the identical base-pairing capabilities, is both toxic and mutagenic in E. coli in the absence of protection by the AlkB repair enzyme.10 Previously, we reported that AlkB can effectively alleviate the toxicity of the EA lesion in vivo; in AlkB-proficient cells, EA is easily tolerated and not significantly mutagenic.6 However, EA is extremely toxic to AlkB-deficient cells, showing an 86% reduction in replication.6 Preliminary in vitro experiments done in parallel show that AlkB could only partially convert EA to generate an open-ring intermediate 2 (Figure 1a, box), trappable with PFBHA. In an attempt to reconcile the potent ability of the protein to suppress toxicity in vivo with only partial conversion of the lesion in vitro, we speculated that intermediate 2, via flexible bond rotation, can take on a structure to form a regular Watson–Crick base pair with thymine (Figure 1a, box). That restructured base pair would not block DNA replication and hence enables tolerance of the lesion.
Figure 1.
Chemical structures of EA, m6A, m1A, and eA and proposed mechanisms of AlkB-mediated enzymatic reactions on alkyl-DNA adducts. (a) EA, (b) m6A, (c) m1A, and (d) eA.
The AlkB protein is an α-ketoglutarate- and Fe(II)-dependent dioxygenase that can efficiently repair various alkyl lesions in both DNA and RNA, in single- and double-stranded contexts.11,12 It has at least nine human homologues: ABH1 through ABH8 and FTO, some of which have also been proposed or proven to act upon alkylated DNA and RNA.2,13−15 AlkB, together with Ada, AlkA, and AidB, is one of the four proteins in the E. coli adaptive response to alkylating agents.14 The reported substrate scope for the AlkB enzyme includes 1-methyladenine (m1A), 3-methylcytosine (m3C), 3-ethylcytosine (e3C), 1-methylguanine (m1G), 3-methylthymine (m3T), 3,N4-ethenocytosine (eC), eA, and EA.2,13−15 In the current work, we used high-resolution ESI-TOF mass spectrometry16 to investigate the ability of AlkB to repair EA. To our surprise we found that the oxidizing enzyme AlkB is capable of complete repair of the EA base (Figure 1a). The observation of AlkB’s complete conversion of EA to adenine along a defined reaction path makes plausible the strategy of designing inhibitors to block the repair activity, thus potentially increasing the anticancer efficacy of BCNU.
The EA dealkylation process requires two oxidation reactions, one at N1 and a second at N6, to achieve complete restoration of the undamaged adenine base. The observation of N6 oxidation in the reaction of AlkB with EA suggested that the enzyme might act on other N6-modifications of adenine. One such modification is N6-methyladenine (m6A) (Figure 1b). The m6A base is functionally similar to N4- and 5-methylcytosine (m4C and m5C), and is an important epigenetic signal for DNA replication and repair, protein–DNA interactions, host-pathogen interactions and other cellular processes;17−20 there is approximately one m6A for every 200 bases in the E. coli genome.21 As controls against which to compare the chemistry of AlkB on m6A (and EA), we used m1A and eA (Figure 1c and 1d), which are good substrates for AlkB both in vivo and in vitro.10,22 As we were preparing this manuscript, Jia et al. reported that the obesity-associated FTO protein (one of AlkB’s mammalian homologues) can remove the methyl group from m6A in both RNA and DNA; they indicated that the demethylation of FTO on m6A in mRNA correlates with epigenetic regulation in mammalian cells.23 In our present study, we observe that AlkB can also demethylate m6A. The implications of this discovery include the possibility that AlkB might be not only a DNA repair enzyme in E. coli but also a participant in cellular epigenetic control, which may help cells defend against alkylation damage to nucleic acids.
Results
This study utilized chemical synthesis to incorporate structurally defined lesions into specific sites of oligonucleotides. The modified oligonucleotides were allowed to react with purified AlkB and snapshots were taken at various time points to identify the products and intermediates of reactions. This method enabled us to efficiently monitor the ability of AlkB to repair alkylation damage using a high-resolution mass spectrometry tool employed by us earlier with a number of DNA lesions.6,10,16
Four 16mer oligonucleotides were chemically synthesized containing the alkyl-DNA adducts shown in Figure 1. The sequence used was 5′-GAAGACCTXGGCGTCC-3′; the flanking sequence of each lesion (X) was identical, fixing the local sequence context for the repair reaction. The 16mers with individual lesions were incubated with the cofactors for the AlkB reaction: Fe(II) and α-ketoglutarate (see Experimental Section). In the presence of all cofactors, two sets of experiments were conducted for each lesion, one with the AlkB protein and one with the purification buffer of AlkB but without the protein. Following the repair reaction, HPLC-ESI-TOF mass spectrometry was utilized to identify the products and intermediates. The 16mer oligonucleotides demonstrated a good signal in the −4 charge envelope of the mass spectra. To give one example to illustrate the method of analysis, the molecular weight (MW) of m1A in the 16mer is calculated as 4902.88 Da for the neutral species, and the MW of its monoisotopic peak (all 12C, 14N, etc.) when ionized with four negative charges (the −4 charge envelope) in the electric field of the spectrometer is calculated as having an m/z of 1224.71 (all MWs are shown in Table 1). The multiple peaks in each −4 charge envelope reflect the number of 13C or other heavier isotopes (Figure 2a). Keeping with m1A as the example, its monoisotopic peak is observed as 1224.67, consistent with the theoretical calculation. The next peak in that envelope has an m/z of 1224.92, 0.25 amu larger than the 1224.67 peak, which indicates a species containing 13C, 15N, or another isotope that adds a nominal mass of 1.0 to the total weight of the 16mer (e.g., 0.25 = m/z = one 13C/charge state of 4). Additional peaks in the spectrum represent additional heavy isotopes within the parental 16mer.
Table 1. Calculated and Observed Monoisotopic Molecular Weights of Oligonucleotides and Intermediates Present in the AlkB Reactions.
| lesion or base | MW (calculated) of neutral species | m/z (calculated) –4 charge monoisotopic peak | m/z (observed) –4 charge monoisotopic peak |
|---|---|---|---|
| A | 4888.86 | 1221.21 | 1221.13/16/24 |
| m1A | 4902.88 | 1224.71 | 1224.67 |
| eA | 4912.86 | 1227.21 | 1227.22 |
| eA epoxide | 4928.86 | 1231.21 | 1231.22 |
| eA glycol | 4946.87 | 1235.71 | 1235.72 |
| EA | 4914.88 | 1227.71 | 1227.67 |
| EA+2 | 4916.86 | 1228.21 | 1228.16 |
| EA+16 | 4930.87 | 1231.71 | 1231.66 |
| EA+32 | 4946.87 | 1235.71 | 1235.67 |
| m6A | 4902.88 | 1224.71 | 1224.73 |
| HO-m6A | 4918.87 | 1228.71 | 1228.72 |
Figure 2.

ESI-TOF mass spectra from reactions of alkyl adducts and AlkB protein. Data represent the −4 charge envelopes and the monoisotopic peak (all 12C, 14N, etc.) value is labeled above each peak envelope. (a) m1A, (b) m1A + AlkB, (c) eA, and (d) eA + AlkB.
Model System: AlkB Repairs m1A and eA
Previously, we investigated m1A and eA lesions as substrates for AlkB both in vitro and in vivo;10,16,22 we used those two adducts as controls for the present study on the structurally similar compounds, m6A and EA.
In our in vitro study of m1A repair, the lesion was completely repaired to the undamaged base adenine (Figure 2b, m/z = 1221.13), while no change occurred in the absence of AlkB (Figure 2a, m/z 1224.67). The in vitro results confirm that m1A is a good substrate for AlkB, and reproduce our prior findings.16 Moreover, the current observations correlate very well with the strong reparability of m1A suggested by earlier in vivo lesion bypass studies.22 The second control compound in the present study was eA. eA was observed at m/z of 1227.22 for its −4 charge monoisotopic peak in the absence of AlkB (Figure 2c). In the presence of AlkB, the eA lesion was mostly converted to the undamaged product, adenine (m/z = 1221.24, Figure 2d). We also observed peak clusters consistent in MW with epoxide (1231.22) and glycol (1235.72) intermediates (Figure 1d and Figure 2d), which are consistent with previous observations.10
AlkB Repairs EA Completely
In our in vitro experiment with EA, the oligonucleotide with EA in the absence of AlkB was observed at m/z of 1227.67 for its −4 charge monoisotopic peak, which agrees well with the calculated m/z 1227.71 (Figure 3a and Table 1). In the presence of AlkB, the peak envelope of EA disappeared completely after the one hour incubation. Indeed, EA was converted to four new species in the mass spectrum (Figure 3b). The peak at m/z of 1221.16 corresponds to the 16mer with an undamaged adenine base, which we did not find in our previous study.6 One possible reason could be that the MALDI mass spectrometry tool we used earlier has poorer resolution than the current HPLC-ESI-TOF method. Another reason is that MALDI analyzes the samples immediately after the reactions, giving less time for conversion of intermediates to adenine. However, the HPLC-ESI-TOF method detects mass signals after the HPLC separation at room temperature, which might provide product and intermediate profiles different than those detected in the MALDI analyses. We also controlled the HPLC autosampler temperature at 5 °C and found the formation of undamaged adenine more limited under this condition (Figure 4). Detailed analyses of the product and intermediate kinetics will be presented in a later section. We also found peaks at m/z of 1231.66 and 1235.67, which match up with intermediates with one and two extra oxygen atoms added to the EA motif, respectively. By carefully comparing the high-resolution MWs between the new species with m/z of 1228.16 and the 16mer with adenine in the lesion position, we determined that this new species has two extra atoms, consistent with the addition of one oxygen and one carbon (intermediate 6 or 9 in Figure 1a).
Figure 3.
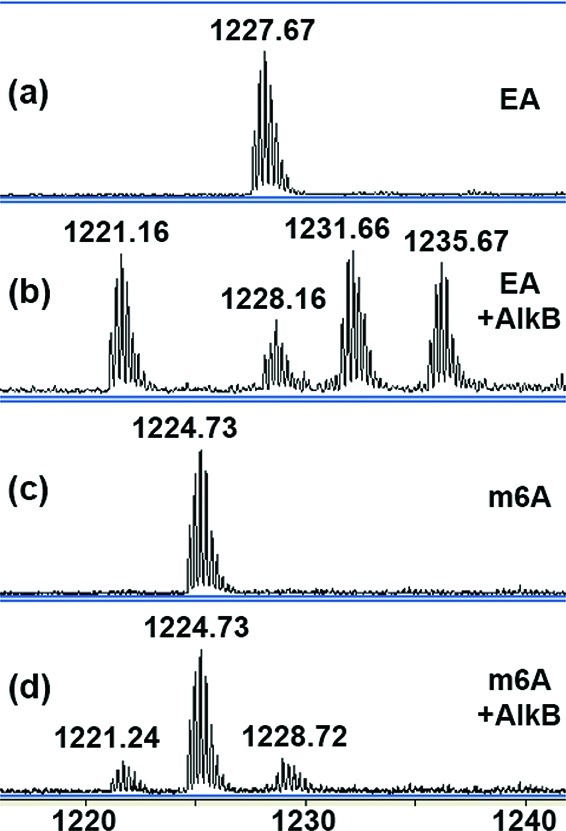
ESI-TOF mass spectra from reactions of alkyl adducts and AlkB protein. Data represent the −4 charge envelopes and the monoisotopic peak (all 12C, 14N, etc.) value is labeled above each peak envelope. (a) EA, (b) EA + AlkB, (c) m6A, and (d) m6A + AlkB.
Figure 4.
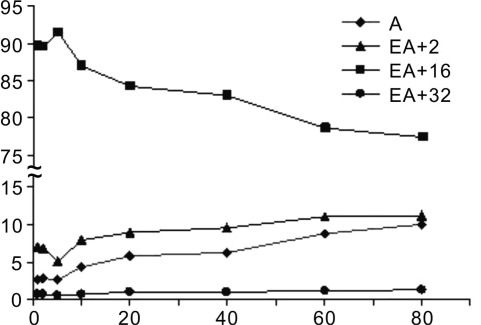
Percentages (y-axis) of the four species in the AlkB reactions on EA at 1, 2, 5, 10, 20, 40, 60, and 80 min incubation time points (x-axis) (see data in Table S1, SI ). The combined percentages of the four species are 100%.
In our previously proposed mechanism (Figure 1a, boxed), AlkB can oxidize the carbon atom attached to the N1 position of the adenine base. The mono-oxidized intermediates 1 and 2 (Figure 1a) equilibrate between the ring-closed and ring-opened forms, both bearing an observed m/z of 1231.66. The opened-ring intermediate 2 can take on a conformation in which it can form a Watson–Crick base pair and thereby not block DNA replication. This hypothesis is strongly supported by in vivo experiments showing EA is neither toxic nor mutagenic in AlkB-proficient E. coli cells.6 In the current study, we observed EA to be completely repaired to the adenine base, which requires oxidation reactions at both the N1 and N6-attached carbon atoms (Figures 1a and 3b). A clue as to the reaction path of EA was the observation of an AlkB-induced glycol (m/z of 1235.67) identical to the glycol formed during removal of the etheno bridge from eA (Figure 1d). In order for EA to form a glycol (intermediate 4, Figure 1a) such as that of eA, it would need to experience two oxidation reactions (Figure 1a). The presence of the glycol and the observation of other intermediates indicated by their masses in Figure 1a allowed us to assemble a reaction network that would explain the removal of the ethano-bridge from EA. As shown in Figure 1a, AlkB can oxidize each carbon of the saturated two carbon bridge and completely restore the undamaged adenine base. The first oxidation can occur either at the N1- or N6-attached carbon of the EA moiety and generate intermediate 1 or 3, respectively. Each ring-closed intermediate will equilibrate with its ring-opened form 2 or 7. Further oxidation of 1 or 3 will generate a common intermediate 4, which can also exist in the respective ring-opened form of 5 or 8. Further oxidation of the ring-opened intermediate 2 or 7 will also generate 5 or 8. The two-carbon unit attached to EA in intermediate 5 or 8 can then be released as glyoxal to form the undamaged adenine (Figure 1a), identical to the final step of eA repair after epoxide hydrolysis (Figure 1d).10 If the glycol 4 or the α-hydroxy aldehyde intermediate 5 or 8 is further oxidized, two aldehyde products will form: the terminal carbon will be released as formaldehyde, and the other carbon attached to either the N6- or N1-position of the adenine base will generate intermediates 6 or 9. The formation of intermediate 6 or 9 should adopt a mechanism similar to typical oxidative cleavage of a glycol to two aldehydes by sodium periodate.24,25 It is possible that intermediate 6 or 9 can be spontaneously converted to the adenine base by amide hydrolysis, or they may remain side products in the reaction rather than formal intermediates.
To elucidate the kinetic distributions of the four new species in the AlkB reaction, we carried out a series of enzymatic reactions at 37 °C with varied time durations at a fixed enzyme–substrate ratio. We chose the time points at 1, 2, 5, 10, 20, 40, 60, and 80 min to quench the reactions and subsequently carried out HPLC-ESI-TOF analysis using an autosampler set to 5 °C in an effort to keep the products and intermediates intact. As presented in Figure 4, we observed that the EA starting material was completely consumed even after the 1 min incubation. This result indicates that the AlkB reaction on EA happened very fast under the current reaction conditions. We also observed that a major species in all mass spectra was the peak at m/z of 1231.66 (from 77.4% to 91.5%, Figure 4 and Table S1, SI), which matches up with intermediate with one extra oxygen atom added to EA. The percentage of this mono-oxidation intermediate decreased as incubation at 37 °C went to longer times. The intermediate with two extra oxygen atoms added to EA displayed a relative low percentage for all time periods. The ratios of the fully repaired product (undamaged adenine) and the EA+2 species (intermediate 6 or 9 in Figure 1a) gradually increased as the incubation time increased (Figure 4). The ratio of adenine increased from 2.6% to 10.0% during the 1–80 min interval. By performing the HPLC-ESI-TOF analysis of the incubation of EA with AlkB using an autosampler at room temperature, we observed a significantly increased amount of adenine (Figure 3b), which might be generated from the spontaneous decomposition of the oxidized intermediates.
AlkB Demethylates m6A
The ability of AlkB to demethylate an m6A-containing oligonucleotide was addressed. The observed m/z of 1224.73 for the −4 charge monoisotopic peak from the m6A starting material was seen in the absence of AlkB (Figure 3c). In the presence of the AlkB protein, three peak envelopes are seen in the mass spectrum (Figure 3d). The peak at 1224.73 corresponds to the unreacted 16mer containing m6A (about 68% for a 60 min incubation). The 1221.24 peak envelope is assigned to the unmodified base adenine (15%). The peak envelope at 1228.72 (17%) matches the theoretical MW of the HO-m6A very well (1228.71 at −4 charge state). It decomposes to A as the reaction time progresses (Figure S3, SI). We believe this to be the first instance of observing the hydroxymethyl intermediate in the AlkB reaction for simple methyl adducts, which strongly supports AlkB’s oxidative demethylation mechanism. With respect to the 100% conversion of m1A under similar reaction conditions, the incomplete repair and buildup of hydroxy intermediate seen for the AlkB reaction with m6A suggest that m6A is more sluggishly processed than m1A. Unlike the AlkB reaction on m6A, we did not observe a hydroxy intermediate (i.e., no HO-m1A) in the m1A reaction. The different behaviors of m1A and m6A could originate from the charge state of the lesions: at physiological pH, m1A is positively charged, whereas m6A is neutral. The positively charged HO-m1A intermediate should be less stable than the neutral HO-m6A, leading to faster accumulation of adenine and inability to observe the hydroxy intermediate.
Discussion
Biological Implications of AlkB Repair of EA
The alleviation of EA toxicity by AlkB in E. coli provides an opportunity to investigate further the repair efficiency of such lesions by human homologues, such as hABH2. If hABH2 or its homologues can efficiently repair EA in vivo, the anticancer efficacy of BCNU (and possibly other DNA damaging anticancer drugs) would be greatly reduced. The design of a chemical inhibitor to block the repair activity of AlkB or hABH2 on EA should enhance the anticancer efficacy of BCNU. Knowledge of intermediates along the repair pathway would enable those studies.
Hypothesis on the Biological Function of m6A Demethylation by AlkB
Unlike m1A, eA, or EA, the m6A moiety is normally not considered a lesion. Rather, it is an important postreplicative DNA modification17−20 and is considered the “sixth element of DNA” (m5C being the fifth element in addition to the regular four DNA bases).19 The results presented here imply that AlkB might not only act as a repair enzyme to defend against exogenous or endogenous attacks by alkylating agents but may also help the cell to control its replication cycle. Besides its direct reversal repair of alkyl lesions, such as m1A and eA, AlkB, which is strongly induced by DNA damage, might also demethylate m6A to stop the cell replication cycle temporarily and provide extra time for the cell to repair alkyl adducts. After the majority of alkyl lesions are removed, AlkB and other repair enzymes can be degraded or down-regulated, followed by progression of the normal cell cycle. The epigenetic control of oxidative demethylation of m6A and its influence on cellular function regulation is also supported by the recent discovery of FTO’s demethylation of m6A.23
Conclusions
In this study, we demonstrated that AlkB can demethylate m6A and completely convert EA to undamaged adenine and other intermediates. These results indicate that AlkB can act not only as a DNA repair enzyme but also as a component that may modulate the cellular replication cycle, thus aiding E. coli in surviving alkylation attacks. Further investigation of the cellular functions of AlkB will provide insights into AlkB’s defense mechanisms for the cellular response to environmental attacks as well as the design and development of inhibitors for AlkB’s human homologues to improve treatments for cancer therapy.
Experimental Section
Oligonucleotide Synthesis
Oligonucleotides containing the adducts in Figure 1 were prepared using the solid-phase phosphoramidite method, and were deprotected, purified, and identified as described previously.10,22 For all four adducts, the oligonucleotide sequence of the 16mers was 5′-GAAGACCTXGGCGTCC-3′, (X = adduct). The calculated MWs of the oligonucleotides and intermediates are shown in Table 1. DNA concentration was measured by UV absorbance using the extinction coefficients (ε) at 260 nm. For any alkyl-modified base, we substituted its extinction coefficient with the extinction coefficient of its unmodified counterpart due to the tiny difference between the values in the context of a 16mer oligonucleotide.
In vitro Incubation Reactions of DNA Adducts with AlkB
All assays were carried out with AlkBΔN11 protein, a truncated version of AlkB in which the first eleven residues were removed. AlkBΔN11 protein was purified as described and shown previously to have similar activity to the wild type protein.6 All AlkB incubation reactions utilized similar conditions as previously described.10,16 Reactions were performed at 37 °C in 45 mM HEPES (pH 8.0), 0.9 mM α-ketoglutarate, 67 μM Fe(NH4)2(SO4)2·6H2O, and 1.8 mM ascorbate, followed by dry ice storage until HPLC-ESI-TOF MS analysis. A typical reaction was performed with 5 μM DNA, all cofactors, and with or without 2.5 μM AlkB in a 10-μL volume for 1 h. In the case of a reaction with no AlkB protein added, the elution buffer for AlkB purification was added instead. For the time and volumetric dependency reactions (Figures 4 in text and S1–S4 in SI), the reaction time and volumetric ratio varied correspondingly.
HPLC-ESI-TOF MS analysis
Oligonucleotide analyses were carried out on an Agilent ESI-TOF mass spectrometer (Palo Alto, CA). ESI was performed using a needle voltage of 3.5 kV. Nitrogen gas was set with drying at 10 L/min, the heated capillary at 325 °C and the nebulizer set at 15 psig. HPLC separations were performed by using a Zorbax SB-Aq column (2.1 mm × 150 mm; 3.5 μm; Agilent Technologies, Palo Alto, CA) with a flow rate of 0.2 mL/min. Solvent A used 10 mM ammonium acetate in water; solvent B used 100% acetonitrile. A linear gradient was carried out in the following steps: 2 to 30% B over 30 min, 30 to 2% B over 5 min, and 2 to 2% B over 10 min. LC–MS analyses were normally carried out at room temperature except for the data presented in Figure 4, which were collected with the temperature of the autosampler set at 5 °C. Data analyses used the Agilent MassHunter Workstation software7.
Acknowledgments
This work was supported by National Institutes of Health Grants CA080024, CA26731, and ES02109. C.L.D. is a Howard Hughes Medical Institute Investigator. We thank Dr. John. S. Wishnok for helpful discussions. We also thank the Wishnok/Tannenbaum Mass Lab and the MIT Center for Environmental Health Sciences for providing the ESI-TOF mass spectrometry facility and Agilent Technologies for providing the UHPLC system.
Supporting Information Available
Table S1 and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# Visterra Inc., Cambridge, MA 02139.
Author Present Address
¶ Chemistry Department, Yale University, New Haven, CT, 06520.
Author Present Address
○ School of Pharmaceutical Science and Technology, Tianjin University, Tianjin, Peoples Republic of China 300072.
Author Present Address
□ Harvard Medical School, 25 Shattuck Street, Boston, MA 02115.
Author Present Address
△ Johnson & Johnson Pharmaceutical Research & Development, 930 Route 202 South, Raritan, NJ 08869.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Loeb L. A.; Harris C. C. Cancer Res. 2008, 68, 6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav N.; Li D.; Essigmann J. M. Carcinogenesis 2010, 31, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlum D. B. IARC Sci. Publ. 1986, 70, 137. [PubMed] [Google Scholar]
- Ludlum D. B. IARC Sci. Publ. 1986, 78, 71. [PubMed] [Google Scholar]
- Ludlum D. B. Mutat. Res. 1990, 233, 117. [DOI] [PubMed] [Google Scholar]
- Frick L. E.; Delaney J. C.; Wong C.; Drennan C. L.; Essigmann J. M. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guliaev A. B.; Hang B.; Singer B. Nucleic Acids Res. 2002, 30, 3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang B.; Chenna A.; Guliaev A. B.; Singer B. Mutat. Res. 2003, 531, 191. [DOI] [PubMed] [Google Scholar]
- Guliaev A. B.; Singer B.; Hang B. DNA Repair 2004, 3, 1311. [DOI] [PubMed] [Google Scholar]
- Delaney J. C.; Smeester L.; Wong C.; Frick L. E.; Taghizadeh K.; Wishnok J. S.; Drennan C. L.; Samson L. D.; Essigmann J. M. Nat. Struct. Mol. Biol. 2005, 12, 855. [DOI] [PubMed] [Google Scholar]
- Trewick S. C.; Henshaw T. F.; Hausinger R. P.; Lindahl T.; Sedgwick B. Nature 2002, 419, 174. [DOI] [PubMed] [Google Scholar]
- Falnes P. O.; Bjoras M.; Aas P. A.; Sundheim O.; Seeberg E. Nucleic Acids Res. 2004, 32, 3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y.; Duguid E. M.; He C. Chem. Rev. 2006, 106, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B.; Bates P. A.; Paik J.; Jacobs S. C.; Lindahl T. DNA Repair 2007, 6, 429. [DOI] [PubMed] [Google Scholar]
- Yi C.; Yang C. G.; He C. Acc. Chem. Res. 2009, 42, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Delaney J. C.; Page C. M.; Chen A. S.; Wong C.; Drennan C. L.; Essigmann J. M. J. Nucleic Acids 2010, 2010, 369434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G.; Casadesus J. FEMS Microbiol. Rev. 2009, 33, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion D.; Casadesus J. Nat. Rev. Microbiol. 2006, 4, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratel D.; Ravanat J. L.; Berger F.; Wion D. Bioessays 2006, 28, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F. Biochemistry (Moscow) 2005, 70, 488. [DOI] [PubMed] [Google Scholar]
- Nikolskaya I. I.; Tkatcheva Z. G.; Vanyushin B. F.; Tikchonenko T. I. Biochim. Biophys. Acta 1968, 155, 626. [DOI] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; Zhao X.; Dai Q.; Zheng G.; Yang Y.; Yi C.; Lindahl T.; Pan T.; Yang Y. G.; He C. Nat. Chem. Biol. 2011, 7, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost B. M., Fleming I., Eds. Comprehensive Organic Synthesis, 2nd ed.; Pergamon Press: Elmsford, NY, 1991. [Google Scholar]
- Carey F. A.; Sundberg R. J.. Advanced Organic Chemistry, 5th ed.; Springer: New York, 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.