Abstract
RNase P derived from S. cerevisiae nuclei was tested for its ability to cleave a variety of naturally occurring and selectively altered precursor-tRNA molecules to yield matured 5' termini. Precursors were synthesized in vitro in order to test which aspects of substrate structure are crucial to recognition and cleavage by RNase P. Base modifications in the precursor substrates are not required for cleavage by the enzyme, but deletion and substitution mutations affecting any portion of the precursor tertiary structure reduce cleavage. In particular, a number of alterations in the intervening sequence (IVS) reduce the susceptibility of the substrate to cleavage by RNase P. The significance of these results is discussed in reference to the contribution of the IVS to the structure of the precursor-tRNA.
Full text
PDF


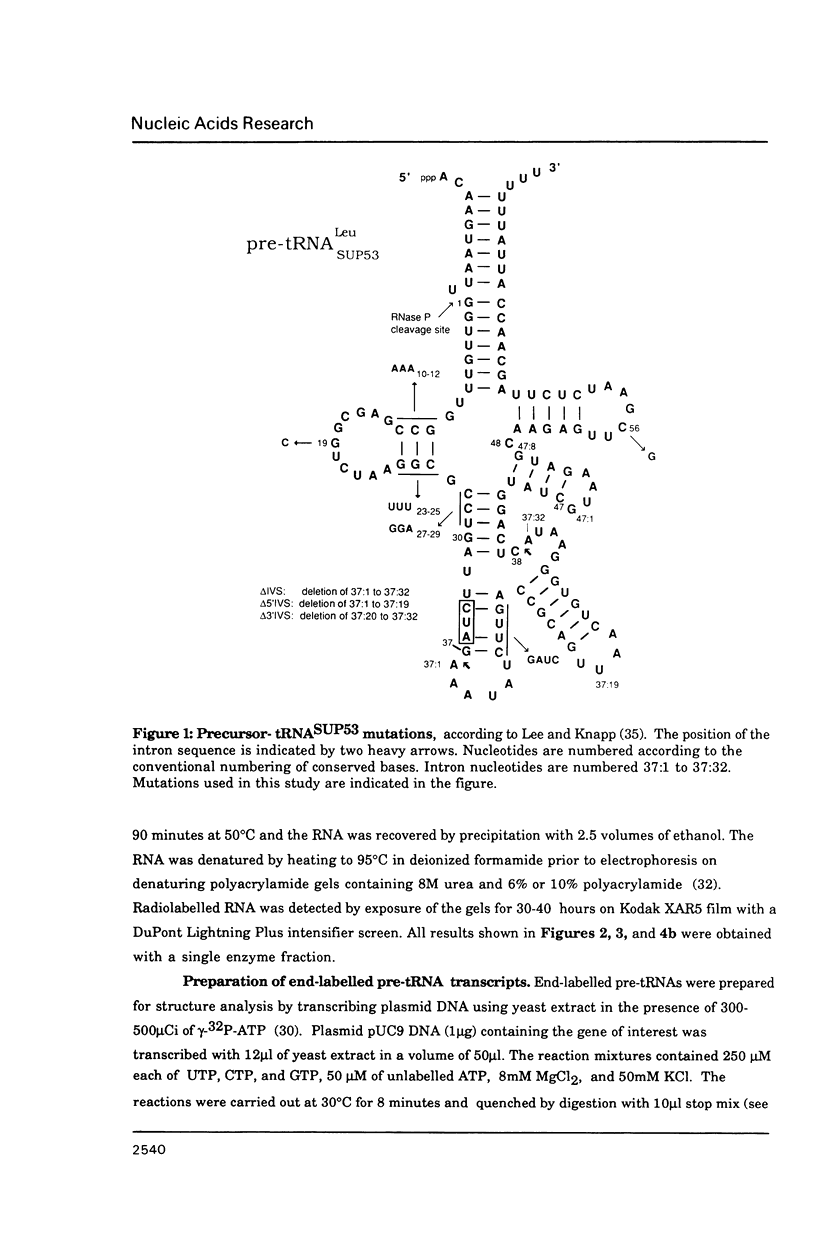





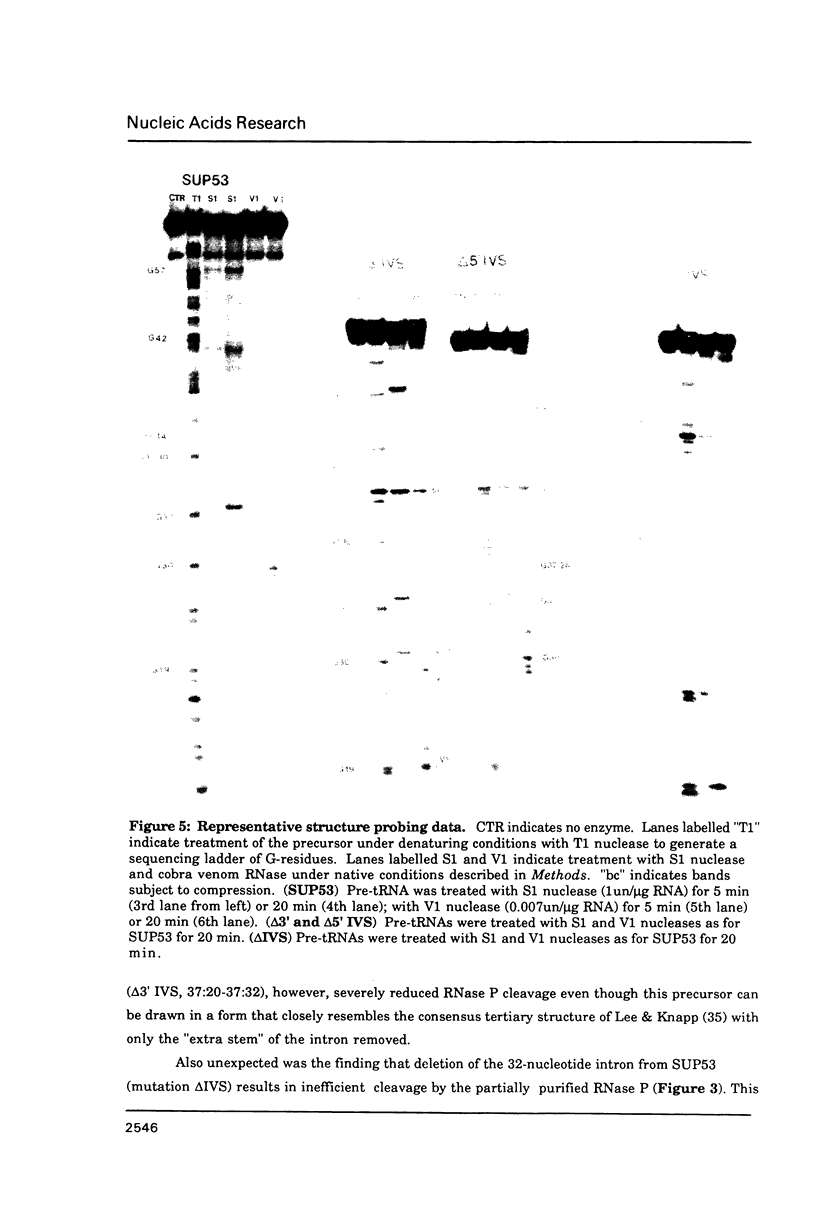

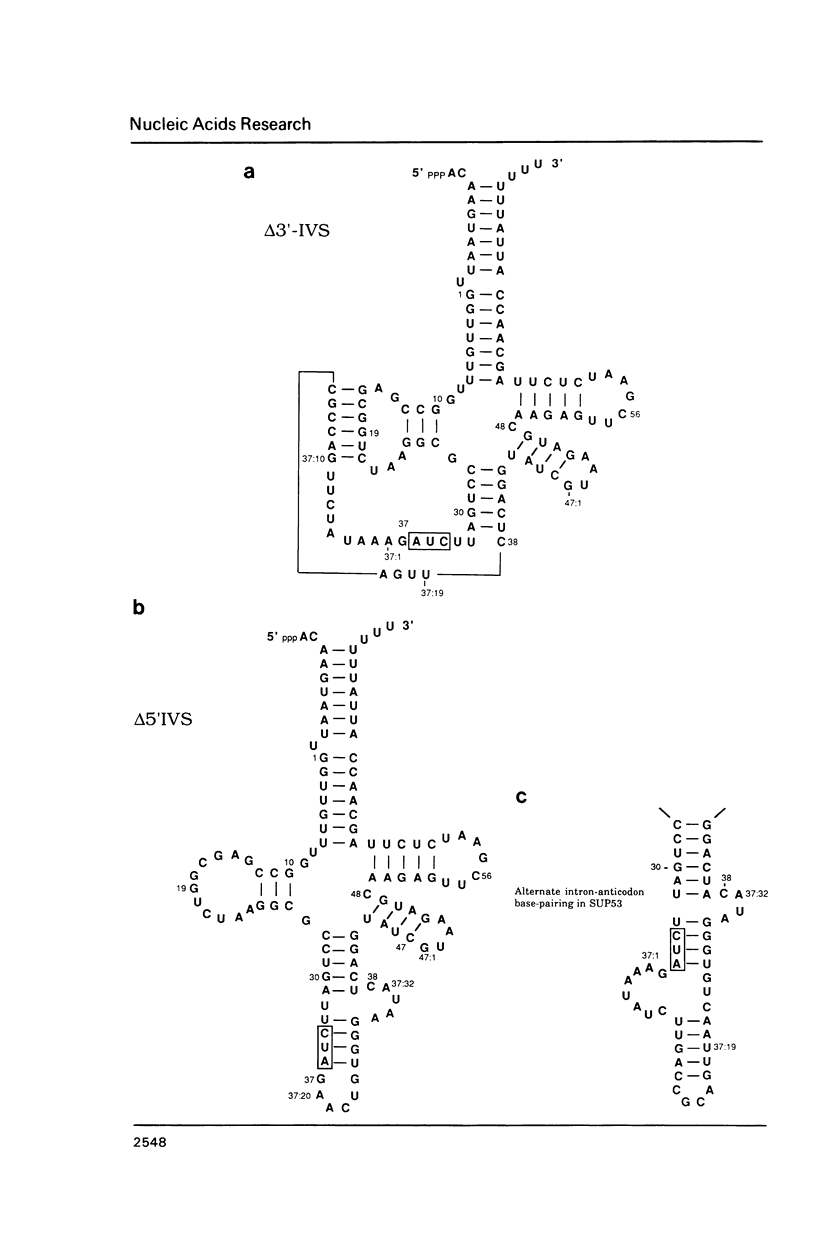




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akaboshi E., Guerrier-Takada C., Altman S. Veal heart ribonuclease P has an essential RNA component. Biochem Biophys Res Commun. 1980 Sep 30;96(2):831–837. doi: 10.1016/0006-291x(80)91430-8. [DOI] [PubMed] [Google Scholar]
- Bowman E. J., Altman S. Identification of ribonuclease P activity from chick embryos. Biochim Biophys Acta. 1980 Jun 13;613(2):439–447. doi: 10.1016/0005-2744(80)90098-4. [DOI] [PubMed] [Google Scholar]
- Butler E. T., Chamberlin M. J. Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. J Biol Chem. 1982 May 25;257(10):5772–5778. [PubMed] [Google Scholar]
- Castaño J. G., Ornberg R., Koster J. G., Tobian J. A., Zasloff M. Eukaryotic pre-tRNA 5' processing nuclease: copurification with a complex cylindrical particle. Cell. 1986 Aug 1;46(3):377–385. doi: 10.1016/0092-8674(86)90658-6. [DOI] [PubMed] [Google Scholar]
- Doersen C. J., Guerrier-Takada C., Altman S., Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985 May 25;260(10):5942–5949. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Furdon P. J., Guerrier-Takada C., Altman S. A G43 to U43 mutation in E. coli tRNAtyrsu3+ which affects processing by RNase P. Nucleic Acids Res. 1983 Mar 11;11(5):1491–1505. doi: 10.1093/nar/11.5.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K., Pace N. R. RNase P of Bacillus subtilis has a RNA component. J Biol Chem. 1980 Aug 25;255(16):7507–7509. [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Banks F. A yeast mutant which accumulates precursor tRNAs. Cell. 1978 Jun;14(2):211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Direct identification of small sequence changes in chromosomal DNA. Gene. 1986;44(1):151–158. doi: 10.1016/0378-1119(86)90056-9. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kline L., Nishikawa S., Söll D. Partial purification of RNase P from Schizosaccharomyces pombe. J Biol Chem. 1981 May 25;256(10):5058–5063. [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Kole R., Altman S. Properties of purified ribonuclease P from Escherichia coli. Biochemistry. 1981 Mar 31;20(7):1902–1906. doi: 10.1021/bi00510a028. [DOI] [PubMed] [Google Scholar]
- Kole R., Baer M. F., Stark B. C., Altman S. E. coli RNAase P has a required RNA component. Cell. 1980 Apr;19(4):881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Bothwell A. L., Altman S. Identification of a ribonuclease P-like activity from human KB cells. Cell. 1976 Sep;9(1):101–116. doi: 10.1016/0092-8674(76)90056-8. [DOI] [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Söll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Jul;5(7):1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Mattoccia E., Baldi M. I., Pande G., Ogden R., Tocchini-Valentini G. P. Mutation in the a block of the yeast tRNAleu3 gene that allows transcription but abolishes splicing and 5'-end maturation. Cell. 1983 Jan;32(1):67–76. doi: 10.1016/0092-8674(83)90497-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983 Oct;34(3):911–917. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Nobrega F. G., Tzagoloff A. Assembly of the mitochondrial membrane system. Structure and location of the mitochondrial glutamic tRNA gene in Saccharomyces cerevisiae. FEBS Lett. 1980 Apr 21;113(1):52–54. doi: 10.1016/0014-5793(80)80492-3. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schaefer K. P., Altman S., Söll D. Nucleotide modification in vitro of the precursor of transfer RNA of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3626–3630. doi: 10.1073/pnas.70.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Meyhack B., Pace N. R. Recognition of local nucleotide conformation in contrast to sequence by a rRNA processing endonuclease. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5644–5648. doi: 10.1073/pnas.77.10.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark B. C., Kole R., Bowman E. J., Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Intron mutations affect splicing of Saccharomyces cerevisiae SUP53 precursor tRNA. Mol Cell Biol. 1986 Jul;6(7):2674–2683. doi: 10.1128/mcb.6.7.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]







