Abstract
The goal of the present study was to design a vaccine that would provide universal protection against infection of humans with diverse influenza A viruses. Accordingly, protein sequences from influenza A virus strains currently in circulation (H1N1, H3N2), agents of past pandemics (H1N1, H2N2, H3N2) and zoonotic infections of man (H1N1, H5N1, H7N2, H7N3, H7N7, H9N2) were evaluated for the presence of amino acid sequences, motifs, that are predicted to mediate peptide epitope binding with high affinity to the most frequent HLA-DR allelic products. Peptides conserved among diverse influenza strains were then synthesized, evaluated for binding to purified HLA-DR molecules and for their capacity to induce influenza-specific immune recall responses using human donor peripheral blood mononuclear cells (PBMC). Accordingly, 20 epitopes were selected for further investigation based on their conservancy among diverse influenza strains, predicted population coverage in diverse ethnic groups and capacity to recall influenza-specific responses. A DNA plasmid encoding the epitopes was constructed using amino acid spacers between epitopes to promote optimum processing and presentation. Immunogenicity of the DNA vaccine was measured using HLA-DR4 transgenic mice and the TriGrid™ in vivo electroporation device. Vaccination resulted in peptide-specific immune responses, augmented HA-specific antibody responses and protection of HLA-DR4 transgenic mice from lethal PR8 influenza virus challenge. These studies demonstrate the utility of this vaccine format and the contribution of CD4+ T cell responses to protection against influenza infection.
Keywords: Influenza, CD4+ T cell epitopes, HA-specific antibodies
1. Introduction
Influenza viruses are single-stranded RNA viruses of the family Orthomyxoviridae. Of the 3 types (A, B, C) infectious in humans, only influenza A and B viruses occur in highly pathogenic forms and only the A virus is known to cause pandemics. Seasonal epidemics affect up to 15% of the world’s population resulting in 3–5 million cases of severe illness and approximately 500,000 deaths per year [1]. The very young, immunologically naïve, and very old, undergoing immune senescence, are most susceptible to infections. During epidemics, approximately 90% of all influenza-related deaths occur among people 65 years of age or older [2].
Pandemics of influenza A viruses continue to occur at sporadic intervals in human populations. Beveridge reported that at least 20 major influenza pandemics occurred between 1729 and 1968 at irregular intervals of between 3 and 28 years [3]. Three have occurred in the twentieth century alone in 1918, 1957 and 1968. These worldwide pandemics are noted for their high mortality and it is estimated that approximately 50 million people died in the 1918 pandemic and at least 1.5 million people in the 1957 and 1968 outbreaks combined [4]. The critical issue with development of a pandemic vaccine is the uncertainty of which subtype will cause the next event. Since 1996, several novel avian subtypes, H5N1, H7N1, H7N2, H7N3, H7N7 and H9N2 have crossed the species barrier from domestic poultry to humans and have caused a spectrum of mild to severe and even fatal human disease[5–7]. At present, a novel swine-associated H1N1 virus is being transmitted among humans. The severity of disease has been comparable to illness associated to seasonal influenza. However, it remains unknown whether this variant will further adapt to humans and become more virulent. Thus, in spite of the success of currently licensed influenza vaccines, development of new vaccines with increased effectiveness in specific age groups and development of pandemic influenza vaccines appear to be of significant interest.
Hemagglutinin (HA)-specific antibody response is the surrogate most commonly used to evaluate vaccine protection. Additional surrogate markers currently under study include matrix2-specific antibodies and CD4+ and CD8+ T cell responses [8,9]. Several lines of evidence support a role of cellular immune responses in protection against influenza. Epstein and colleagues demonstrated that either CD8+ or CD4+ T-lymphocytes promoted survival in mice immunized with an experimental DNA vaccine encoding internal viral proteins [10]. Crowe and colleagues have demonstrated that vaccination of a single HA-specific helper T-lymphocyte (HTL) epitope in mice increased epitope-specific T cell numbers and a significant reduction in viral titers following challenge [11].
Early human studies also indicated cellular immune responses play a role in controlling influenza infection. McMichael and colleagues inoculated 63 volunteers intranasally with live unattenuated influenza A/Munich/1/79 virus and found that all subjects with demonstrable T cell responses cleared virus effectively [12]. Sonoguchi and colleagues found that students previously infected with H3N2 virus were partially protected against subsequent infection with H1N1 subtype virus suggesting cross-subtype protection in humans during sequential epidemics [13]. The protective effect of cellular immune responses may be particularly relevant in the elderly where studies suggest that cellular immune responses may be an important predictor of protection [14]. Recent studies have also demonstrated a correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children [15]. In part, the mechanisms employed by CD4+ T lymphocytes may be a result of ‘help’ for enhancing protective antibody responses and generation on optimal CD8+ T cell responses [16–20].
Taken together, these findings indicate cellular immunity contributes to the control and clearance of infection and reduces pathogenesis. Our working hypothesis is that vaccination with conserved CD4+ influenza-specific epitopes can establish a pool of memory T cells, resulting in more rapid and vigorous induction of immune responses upon vaccination with seasonal vaccines or infection with live virus. While vaccine induction of cellular immunity alone would not be expected to provide complete protection; upon infection, helper-T cells assisted, humoral and cytotoxic responses would mediate control of infection and prevent death. Because of T cell epitope conservation in different strains, such strategy can be tested with seasonal strains but success would be expected to extend to new pandemic strains. Accordingly, this report describes the design and evaluation of an HLA-DR epitope-based universal influenza plasmid DNA (pDNA) vaccine.
2. Materials and methods
2.1 Identification of peptides predicted to be conserved CD4+ helper T cell epitopes
Algorithm-based T cell epitope scans of amino acid sequences of the influenza proteins for A/HK/156/97 (H5N1) and A/Singapore/1/57 (H2N2) were performed to identify those peptides predicted to bind with high affinity to HLA-DR molecules [21,22]. The analysis included sequences for nucleoprotein (NP), matrix 1 (M1), matrix 2 (M2), nonstructural protein 1 (NS1), nonstructural protein 2 (NS2), polymerase acid (PA), polymerase basic 1 (PB1) and polymerase basic 2 (PB2). Influenza hemagglutinin (HA) and neuraminidase (NA) were not analyzed due to their high variability in different influenza isolates. These predicted epitope peptides, 14 – 19 amino acids in length, were then evaluated for amino acid sequence conservation amongst 52 diverse influenza virus strains (NCBI Taxonomy Browser); including: (1) strains that are currently in circulation (H1N1, H3N2); (2) strains with potential for becoming components of an influenza virus pandemic including agents of past pandemics (H1N1, H2N2, H3N2); and (3) strains associated with zoonotic influenza infections of man (H1N1, H5N1, H7N2, H7N3, H7N7, H9N2) (Table 1). Of note, due to the recent outbreak of swine-associated H1N1 virus, the A/California/7/09 strain was included in the analysis. The conservancy of each epitope was defined as the number of the strains containing the epitope at a 100% identity level divided by the total number of strains used in the analysis. Generally, sequences that were conserved in greater than 30% of the strains were selected for peptide synthesis. Peptides were produced using standard F-moc chemistry, purified by reversed-phase HPLC and characterized using mass spectrometry or sequencing. Peptide binding to HLA-DR molecules representing 13 of the most common HLA-DR types was then measured using standardized processes [21,22].
Table 1.
Fifty-two diverse influenza strains used for epitope conservancy analysis
| H1N1 | H2N2 | H3N2 |
|---|---|---|
| A/Brevig Mission/1/1918 | A/Leningrad/134/57 | A/Hong Kong/1/68 |
| A/PR/8/34 | A/Singapore/1/57 | A/Albany/1/76 |
| A/Fort Monmouth/1/47 | A/Ann Arbor/6/60 | A/Moscow/10/99 |
| A/USSR/90/1977 | A/Berlin/3/64 | A/Hong Kong/1774/99 |
| A/New Caledonia/20/1999 | A/Tokyo/3/67 | A/Wisconsin/67/2005 |
| A/California/UR06-0125/2007 | A/Brisbane/10/2007 | |
| A/California/07/09 | ||
| H5N1 | H7N1 & H7N2 & H7N3 & H7N7 | H9N2 |
| A/goose/Guangdong/1/1996 | A/chicken/Italy/2335/2000 | A/chicken/Beijing/1/94 |
| A/Hong Kong/483/97 | A/turkey/Italy/3675/99 | A/Hong Kong/1073/1999 |
| A/Hong Kong/156/97 | A/turkey/Virginia/55/02 | A/chicken/Hong Kong/G9/1997 |
| A/Hong Kong/213/03 | A/chicken/New York/21211-2/05 | A/quail/Hong Kong/G1/1997 |
| A/Vietnam/1203/04 | A/New York/107/03 | A/Korea/KBNP-0028/00 |
| A/Indonesia/5/2005 | A/chicken/Brit. Col./GSC human B/04 | A/ck/HK/CSW161/2003 |
| A/turkey/Turkey/1/2005 | A/Canada/rv504/04 | A/chicken/Shantou/6781/2005 |
| A/Egypt/902782/2006 | A/equine/San Paulo/4/76 | A/chicken/Jiangsu/L1/2004 |
| A/China/GD01/2006 | A/seal/Mass/1/1980 | |
| A/Bar-headed Goose/Qinghai/12/05 | A/chicken/Victoria/1/1985 | |
| A/Netherlands/219/203 | ||
| H4N6 | H6N1 & H6N2 | |
| A/swine/Ontario/01911-1/99 | A/teal/Hong Kong/W312/97 | |
| A/chicken/Taiwan/0204/05 | ||
| A/chicken/California/139/01 | ||
| A/chicken/Shantou/20741/05 | ||
2.2 Measurement of human memory CD4+ T cell responses
PBMC were obtained from HIV and HCV negative donors using apheresis (HemaCare Corporation, Van Nuys, CA), and purified using gradient separation prior to cryopreservation. The HLA-DR types of the donors were not known. Selected HLA-DR binding peptides were tested in two ways: (1) directly ex vivo where the PBMC were thawed, rested 5 days in media and responses measured by IFN-γ ELISPOT assay and (2) following a culture step with peptides to increase sensitivity. Specifically, the cryopreserved PBMC were thawed and rested overnight in media containing RPMI + 5% AB human serum/complete media followed by a 7 day expansion by culture with pools of 10 peptides, 1 µg/ml final concentration of each peptide. On days 1 and 3, cultures were fed with fresh media and IL2 (100U/ml). On day 7, the cultures were depleted of CD8+ lymphocytes using the MACS system (Miltenyi Biotec, Auburn, CA) prior to their use in an IFN-γ ELISPOT assay [22]. Spots were counted using an AID ELISpot Reader System (Straβberg, Germany). HLA-DR-restricted peptides from HIV, HCV and Plasmodium falciparum were used to determine background responses.
2.3 pDNA vaccine design and construction
A pDNA construct encoding 20 influenza-derived epitopes in addition to the universal Pan DR epitope (PADRE, seq; AKFVAAWTLKAAA) [23] was constructed. PADRE peptides were engineered by introducing anchor residues for different DR motifs within a poly-alanine background and introduction of bulky and charged residues at positions accessible to T cell recognition. PADRE elicits powerful responses in vitro from human PBMCs and in addition, cross-reacts on certain mouse Class II alleles [23]. CD4+ T cell epitopes were designed with GPGPG amino acid spacers to promote optimum epitope processing [24]. The gene was codon optimized for expression in mammalian cells (GeneArt, Regensburg, Germany) and was assembled using overlapping oligonucleotides in a PCR-based synthesis followed by subcloning into the pMB75.6 DNA plasmid vector.
The pDNA was produced by growth in Escherichia coli strain Stbl2 (Invitrogen, Carlsbad, CA) in terrific broth (TB, Becton Dickinson, Sparks, MD) with kanamycin (25 µg/ml) and was purified using EndoFree Plasmid Mega kit columns according to the manufacturer’s directions (Qiagen, Valencia, CA). The purified pDNA construct was dissolved in water and stored at −70°C.
2.4 Mouse immunogenicity and virus challenge studies
To study the efficacy of pDNA vaccine, HLA-DR4 transgenic mice (Taconic Farm, Hudson, NY) and the mouse adapted H1N1 PR8 virus (Charles River Laboratories, Wilmington, MA) were used. Studies were completed in accordance with the Guide for the Care and Use of Laboratory Animals with appropriate Institutional Animal Care and Use Committee review.
The pDNA vaccine was delivered intramuscularly (i.m.) using an in vivo electroporation device (TriGrid Delivery System®, Ichor Medical Systems, Inc., San Diego, CA) [25]. Briefly, the area proximal to Tibialis anterior muscles of anesthetized (Halocarbon, River Edge, NJ) mice weighing 25 to 30g were shaved and sterilized with isopropanol. The pDNA was delivered bilaterally, 25 µg DNA/20 µl in each muscle, 50 µg total per mouse using 0.3 ml syringes with attached ½” 30G needle (BD Ultra-Fine, 328431) and the TriGrid electode array, 2.5mm electrode length. Four seconds post pDNA injection, the electrical stimulation was applied at 62.5 Volts for a total duration of 40 mS over a 400 mS window. Mice in the positive control group were immunized once at the base of the tail with 15 µg of inactivated PR8 virus (Charles River Laboratories) in 100 µl of PBS. Cellular immune responses were measured using an IFN-γ ELISPOT assay following selection of mouse CD4+ MACS system as described [26].
HA antibody titers were measured using ELISA and hemagglutination inhibition (HAI). High-binding 96-well ELISA plates (Costar, Corning, NY) were coated with 10 µg purified inactivated PR8 virus (Charles River Laboratories) and incubated overnight at 4°C. The use of inactivated PR8 virus as a coating antigen will not only detect HA-specific but also presumably other antigen-specific antibody responses such as NA. Plates were washed and blocked with PBS + 3% BSA prior to use. Serial dilutions of immune sera were incubated for 2 hours followed by plate washing. Antigen-specific serum antibodies were detected using HRP-coupled secondary antibody (Invitrogen Life Science, Carlsbad, CA). The plates were developed with an HRP substrate reagent, stopped with acid, and absorbance measured at 450nm. End-point titers are expressed as the reciprocal of the highest dilution that gives a reading three standard deviations above the mean background.
HAI was performed using chicken RBCs (Charles River Laboratories) incubated at 22–25°C with 4 HA units of the virus in the presence of serially diluted sera, previously treated to destroy heat stable non-specific inhibitor in a 96 well plate for 1 hour. Plates were then visually inspected for agglutination. The highest dilution of antibody that causes inhibition of hemagglutination is considered the HI titer.
Protection of mice against influenza virus challenge was assessed using the PR8 virus strain; completed using biosafety level-2 containment. Virus was administered for challenge 2 weeks post-immunization. HLA-DR4 transgenic mice were lightly anesthetized using Methoxyflurane (Springvale, Australia) and infected intranasally (i.n.) with 100 HAU (4MLD50) of virus in a 20 µl volume. This dose was used to assure that a majority of the unvaccinated animals would not survive the challenge. Additionally, we assumed that the protection afforded by cellular immunity would not be as potent as HA-specific humoral immunity and a high dose of 10–100 MLD50 would be too high to measure any protective effects induced by the HLA-DR-restricted epitopes. Each test group contained 14 mice with experiments organized as follows: (1) 2 mice/group were used to complete IFN-γ ELISPOT assays on day 14 post immunization; (2) 2 mice/group were used to complete IFN-γ ELISPOT assays 6 days post virus challenge and (3) the remaining 10 mice were evaluated for survival for a duration of 14 days post-challenge.
2.5 Statistical analyses
Triplicate well testing was used in the ELISA and ELISPOT and Student t test was used to document significant levels of immunogenicity obtained using immunized vs. naïve (non-immunized) mice. A p value of < 0.05 indicated a positive response. Statistically significant differences in survival times were established using the Fisher’s exact test analysis. Differences were considered statistically significant if the p value was < 0.05.
In the assays using human PBMC we knew the donors were not previously exposed to HIV, HCV and assume lack of infection with P. falciparium. Therefore responses to peptides derived from these pathogens were used to determine assay background. A mean influenza-specific response was considered significant if the net-SFC number was greater than the mean response of the background plus 2 standard deviations. In addition, a minimum positive response greater than 100 IFN-γ SFC/106 cells in the CD8+ cell depleted PBMC was considered the minimum for a positive response.
3. Results
3.1 Identification of influenza-derived CD4+ T cell epitopes
The HLA-DR peptide binding motif search of the selected influenza virus gene products resulted in the identification of 943 peptides. A total of 45 peptides were identified which bound with moderate to high affinity to at least 6 of the 13 most common HLA-DR molecules used in our study (Table 2). The 45 peptides were also highly conserved among the 52 diverse subtype strains with an indicated average conservation level of 74% (Table 2).
Table 2.
CD4+ epitope summary
| Human Recall | IC50 nM to purified HLAa) |
No. of HLA-DR |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Source | Cons. (%)b) | Positive Donors (19 Total) |
DRB1 *0101 |
DRB1 *0301 |
DRB1 *0401 |
DRB1 *0404 |
DRB1 *0405 |
DRB1 *0701 |
DRB1 *0802 |
DRB1 *0901 |
DRB1 *1101 |
DRB1 *1302 |
DRB1 *1501 |
DRB4 *0101 |
DRB5 *0101 |
alleles IC50≤1000nM |
| KGILGFVFTLTVPSEd,n) | M1.57 | 94 | 6 | 15 | 2610 | 5 | 1 | 6 | 11 | 33 | 33 | 37 | 10 | 127 | 356 | 2206 | 11 |
| YRKLKREITFHGAKEe) | M1.100 | 60 | 10 | 20 | 4633 | 13 | 20 | 951 | 976 | 47 | 432 | 28 | --c) | 327 | 58 | 459 | 11 |
| MGTVTTEVALGLVCA | M1.135 | 21 | 5 | 58 | 348 | 321 | 355 | -- | 924 | -- | 4234 | -- | 769 | 142 | 1589 | 7609 | 7 |
| NPLIRHENRMVLASTf) | M1.170 | 98 | 5 | 17 | 40 | 779 | 31 | 1588 | 298 | 108 | 95 | 32 | 6 | 3 | 690 | 3691 | 11 |
| AMEVASQARQMVQAMg) | M1.202 | 73 | 3 | 641 | 833 | 4252 | 1561 | 263 | 424 | 1142 | 1437 | 1406 | 185 | 310 | 759 | 786 | 8 |
| DPLVVAASIIGILHL | M2.24 | 46 | 0 | 78 | -- | 92 | 23 | 630 | 1304 | 3591 | 709 | 1804 | 1859 | 2196 | 252 | -- | 6 |
| IGRFYIQMCTELKLSDYEG | NP.36 | 65 | 4 | 31 | 291 | 53 | 116 | 29 | 443 | 486 | 193 | 236 | 2704 | 23 | 210 | 28 | 12 |
| QNSITIERMVLSAFD | NP.58 | 69 | 5 | 890 | 298 | 430 | 188 | 4137 | 806 | 534 | 5615 | 4231 | 1421 | 891 | 39 | -- | 8 |
| VGTMVMELIRMIKRGh) | NP.186 | 71 | 8 | 22 | 16 | 1092 | 13 | 1997 | 495 | 47 | 8886 | 32 | 256 | 738 | 10 | 25 | 10 |
| DLIFLARSALILRGSi) | NP.255 | 92 | 4 | 7 | 857 | 315 | 9 | 348 | 13 | 15 | 365 | 321 | 6 | 8 | 2243 | 35 | 12 |
| RSALILRGSVAHKSCj) | NP.261 | 100 | 2 | 13 | 1482 | 269 | 17 | 599 | 14 | 16 | 63 | 265 | 268 | 119 | 480 | 61 | 12 |
| KSQLVWMACHSAAFEk) | NP.325 | 71 | 2 | 35 | 1118 | 194 | 34 | 127 | 172 | 324 | 201 | 472 | -- | 259 | 149 | 462 | 11 |
| AGQISVQPTFSVQRNl) | NP.403 | 62 | 5 | 12 | 1632 | 41 | 11 | 335 | 290 | -- | 589 | 3744 | 55 | 9 | 34 | 163 | 10 |
| EGAIVGEISPLPSLPGHTD | NS1.153 | 27 | 5 | 64 | 8334 | 14 | 6 | 98 | -- | 227 | 6365 | 88 | 1773 | 7 | 131 | 1142 | 8 |
| SLKLYRDSLGEAVMR | NS2.37 | 44 | 3 | 11 | 46 | 903 | 996 | 5993 | 790 | -- | 707 | -- | 800 | 19 | -- | -- | 8 |
| IRWLIEEVRHRLRIT | NS2.76 | 8 | 4 | 137 | 73 | 1045 | 3841 | 3511 | 877 | 1672 | 6597 | 703 | 932 | 189 | 2944 | 39 | 7 |
| FEQITFMQALQLLLE | NS2.94 | 60 | 1 | 5 | 4551 | 345 | 160 | 335 | 38 | 2696 | 47 | 765 | 1083 | 10 | 387 | 314 | 10 |
| ITFMQALQLLLEVEQ | NS2.97 | 60 | 1 | 13 | 2511 | 161 | 124 | 224 | 26 | 2426 | 20 | 1375 | 242 | 104 | 149 | 46 | 10 |
| RREVHIYYLEKANKI | PA.124 | 77 | 4 | 34 | -- | 63 | 181 | 1490 | 23 | 95 | 851 | 96 | 150 | 4 | 5 | 25 | 11 |
| LFTIRQEMASRGLWD | PA.175 | 69 | 3 | 25 | 523 | 53 | 4 | 768 | 3317 | 88 | 2428 | 890 | 304 | 237 | 15 | 43 | 11 |
| EPFLKTTPRPLRLPD | PA.258 | 35 | 2 | 17 | 373 | 961 | 2021 | -- | 51 | 117 | 109 | 131 | 412 | 570 | 7660 | 62 | 10 |
| RSKFLLMDALKLSIED | PA.279 | 90 | 2 | 1 | 125 | 53 | 14 | 35 | 25 | 96 | 15 | 49 | 110 | 1 | 639 | 16 | 13 |
| VAPIEHIASMRRNYF | PA.432 | 75 | 4 | 375 | 114 | 503 | 41 | 7658 | 543 | 643 | 7437 | 70 | 221 | 147 | 529 | 4 | 11 |
| EYIMKGVYINTALLN | PA.457 | 98 | 4 | 22 | 3758 | 1064 | 67 | 425 | 1041 | 133 | 1200 | 2732 | 7 | 70 | 1337 | 585 | 7 |
| RPMFLYVRTNGTSKI | PA.559 | 90 | 3 | 5 | -- | 7 | 7 | 400 | 15 | 12 | 82 | 8 | 1 | 83 | 694 | 6 | 12 |
| PTLLFLKVPAQNAIST | PB1.5 | 69 | 2 | 8 | -- | 17 | 9 | 152 | 34 | 15 | 69 | 49 | 1380 | 285 | 64 | 50 | 11 |
| SYLIRALTLNTMTKD | PB1.216 | 87 | 2 | 7 | 4900 | 42 | 14 | 429 | 76 | 56 | 121 | 280 | 87 | 7 | 3 | 386 | 12 |
| FLAMITYITRNQPEW | PB1.318 | 77 | 3 | 8 | -- | 126 | 64 | 200 | 590 | 65 | 769 | 59 | 174 | 74 | 1140 | 60 | 11 |
| QPEWFRNVLSIAPIMF | PB1.329 | 75 | 2 | 37 | 8991 | 100 | 236 | 374 | 12 | 228 | 197 | 288 | 98 | 723 | 104 | 656 | 12 |
| FRNVLSIAPIMFSNKM | PB1.333 | 75 | 1 | 10 | 1450 | 25 | 8 | 409 | 10 | 71 | 88 | 68 | 77 | 62 | 43 | 33 | 12 |
| IAPIMFSNKMARLGK | PB1.339 | 83 | 1 | 58 | 51 | 404 | 180 | -- | 315 | 27 | 1836 | 140 | 77 | 25 | 3733 | 100 | 10 |
| KGYMFESKSMKLRTQI | PB1.352 | 75 | 2 | 7 | 547 | 173 | 63 | 806 | 6 | 60 | 80 | 33 | 9 | 2 | 34 | 8 | 13 |
| MMGMFNMLSTVLGVSm) | PB1.408 | 100 | 10 | 3 | 9191 | 6 | 1 | 72 | 85 | 15 | 20 | 13 | 40 | 48 | 37 | 42 | 12 |
| DFALIVNAPNHEGIQ | PB1.446 | 85 | 3 | 47 | -- | 37 | 10 | 52 | 8663 | 295 | 724 | 153 | 9 | 26 | 46 | 208 | 11 |
| YGFVANFSMELPSFG | PB1.499 | 90 | 3 | 5 | 5376 | 40 | 61 | 617 | 8 | 1177 | 13 | 700 | 1 | 17 | 2632 | 1320 | 9 |
| GVTVIKNNMINNDLGP | PB1.526 | 92 | 2 | 37 | -- | 956 | 180 | 457 | 153 | 239 | 1880 | -- | 8 | 8 | 3762 | -- | 8 |
| PNLYNIRNLHIPEVC | PB1.596 | 81 | 2 | 68 | -- | 1159 | 9 | 259 | 10 | 71 | 914 | 1535 | 148 | 9 | 409 | 2309 | 9 |
| ISSMVEAMVSRARID | PB1.711 | 79 | 1 | 112 | 4284 | 93 | 10 | -- | 2942 | 310 | 1076 | 294 | 1108 | 902 | 108 | 13 | 8 |
| KWMMAMKYPITADKR | PB2.48 | 81 | 1 | 415 | 5884 | -- | 182 | 8335 | 114 | 13 | 446 | 19 | 6773 | 198 | 1257 | 101 | 8 |
| GARILTSESQLTITK | PB2.173 | 81 | 2 | 46 | 967 | 103 | 65 | 5479 | 376 | -- | 300 | 4800 | 19 | 19 | 1181 | 7735 | 8 |
| KAAMGLRISSSFSFG | PB2.312 | 77 | 3 | 37 | 501 | 47 | 4 | 452 | 4 | 466 | 58 | 398 | 3 | 1 | 79 | 45 | 13 |
| IKAVRGDLNFVNRAN | PB2.411 | 90 | 4 | 729 | 123 | 75 | 17 | 510 | -- | 419 | -- | 1914 | 264 | 517 | 515 | -- | 9 |
| LRHFQKDAKVLFQNW | PB2.435 | 88 | 1 | 153 | 51 | 1431 | 1348 | 9917 | 578 | 536 | 5727 | 318 | 240 | 207 | 979 | 1412 | 8 |
| QWIIRNWETVKIQWS | PB2.551 | 75 | 3 | 47 | 4422 | 49 | 16 | 124 | 24 | 1573 | 243 | 729 | 9 | 2 | 49 | 156 | 11 |
| RMQFSSLTVNVRGSG | PB2.630 | 96 | 2 | 9 | 9323 | 33 | 71 | 988 | 310 | 833 | 306 | 437 | 149 | 102 | 3725 | 8 | 11 |
IC50 nM to purified HLA (In general, IC50 nM < 1,000 is immunogenic)
amino acid conservancy among H1N1, H2N2, H3N2, H5N1, H7N7, H9N2, etc. (52 strains total)
--indicates IC50 nM > 10,000
20 epitopes selected for inclusion in ‘prototype’ DNA construct are indicated by shading
The 45 peptides were next evaluated for CD4+ T cell recognition using PBMC from 19 human volunteers. Recognition of multiple peptides was observed despite the unknown history of recent influenza virus infections; representative responses for 3 donors (753, 6018, 709) are presented in Fig. 1. Note that for donor 753, a total of 16 epitopes (M1.100, NP.36, NP.186, NP.255, NS1.114, NS2.37, NS2.76, PA.175, PA.457, PB1.216, PB1.408, PB1.446, PB2.411, PB2.435 and PB2.551) were recognized using the ex vivo, version of the assay indicating the presence of circulating activated CD4+ T cells (Fig. 1A). The magnitude of CD4+ T cell responses specific for these epitopes ranged from 160 to 2,040 IFN-γ SFC/106 cells. Following the 7 day peptide expansion step, response to an additional peptide, M1.57, was also observed (Fig. 1B). Responses with somewhat reduced breadth are shown for two other PBMC donors, 6018 and 709, Fig. 2 C, D, E and F.
Fig. 1.
HLA-DR-restricted influenza recall responses from human donors. Human donor 753 panels A and B; donor 6018 panels C and D and donor 709 panels E and F. Panels A, C and E depict recall responses measured in the absence of in vitro peptide expansion while panels B, D and F depict recall responses measured using an in vitro peptide expansion step. CD4+ cells from normal human donors were enriched for assay use. The experimental values are expressed as mean IFN-γ net spots/106 CD4+ lymphocytes ± SEM for each peptide; tests were performed using triplicate wells. Significant responses from the indicated epitopes were defined as responses > than mean of background responses + (2.0 × Std. Dev.). Background responses were determined using supertype HLA-DR binding peptides from pathogens to which donors were not exposed, i.e., HIV, HBV, HCV and Plasmodium falciparum.
Fig. 2.
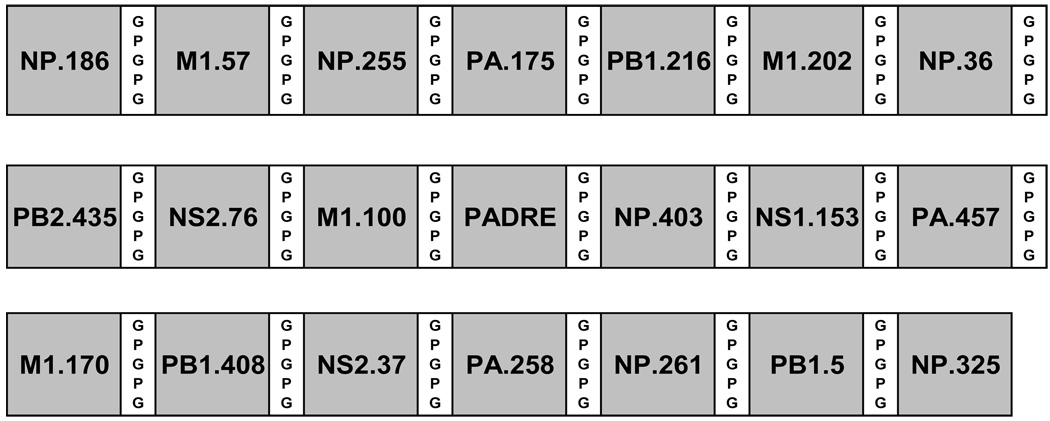
Schematic diagram of the CD4+ 20 epitope DNA insert. The epitope order and amino acid spacer usage are shown. Influenza-derived epitopes were used in addition to the universal helper epitope, PADRE [23].
We believe the observed reactivity is most likely mediated by CD4+ T cells since the PBMC preparations were depleted of CD8+ cells prior to the IFN-γ ELISPOT analysis. Also, the reactivity was detected in donors expressing different HLA Class I alleles. For example, the M1.57 helper epitope has an embedded HLA-A2-restricted CD8+ epitope, indicated by italics and underlined, KGILGFVFTLTVPSE, but the CD4+ epitope was recognized using PBMC from 6 different donors, of which two donors were HLA-A1 and HLA-A3 but not HLA-A2.
Together these data demonstrate the utility for the motif scanning and HLA-DR peptide binding assays for identifying CD4+ T cell epitopes. In total, 37 of the 45 selected peptides were documented to be epitopes based on recognition by T cells from at least 2 donors (Table 2).
3.2 Testing of the experimental pDNA vaccine in HLA-DR4 transgenic mice
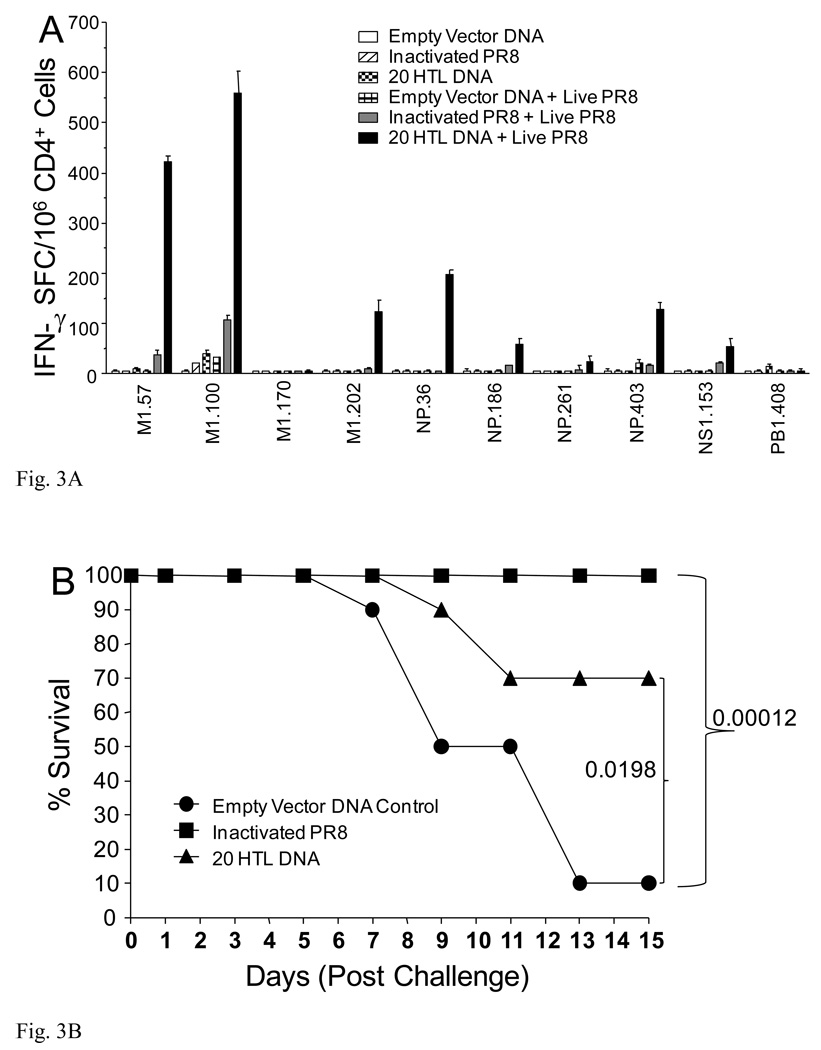
Based on positive recall responses using human PBMC, conservancy among diverse influenza strains and representation of multiple influenza proteins, 20 of the potential 37 epitopes were selected for use in the experimental ‘prototype’ vaccine (Table 2 and Fig 2). HLA-DR4 transgenic mice did not respond to highly significant levels of epitope-specific immunity following immunization with the 20 HTL pDNA vaccine (Fig.3A). However, vaccine immunization followed by live PR8 challenge induced detectable CD4+ T cell responses to multiple epitopes (M1.57, M1.100, M1.202, NP.36, NP.186, NP.403 and NS1.153), ranging from 54 to 559 IFN-γ SFC/106 cells whereas empty vector DNA immunization followed by live PR8 challenge did not induce detectable responses (Fig.3A).
Fig. 3.
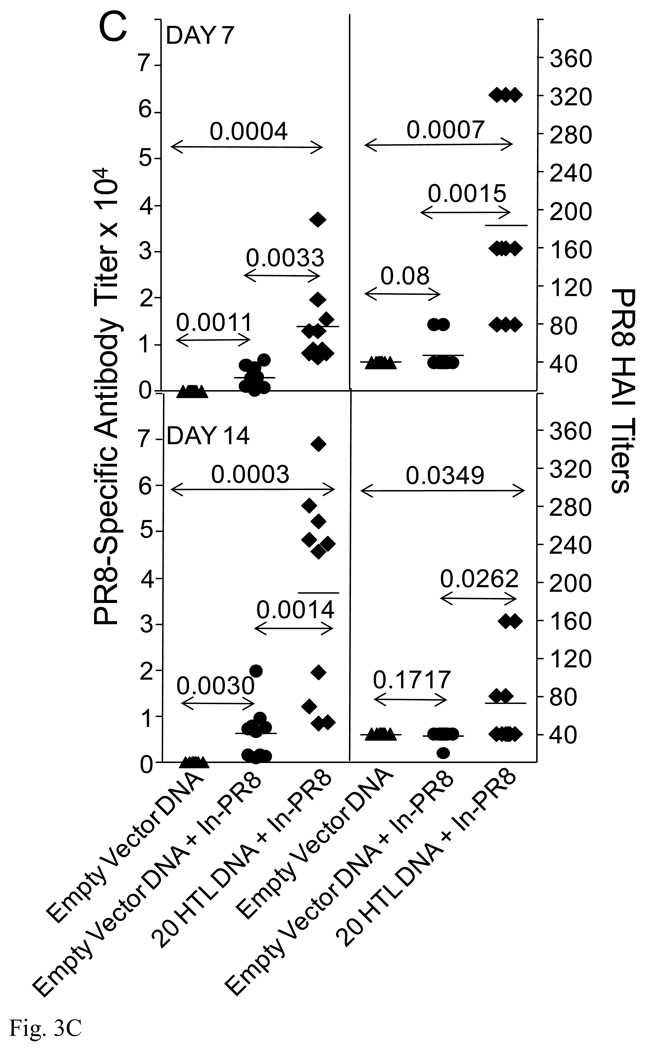
Epitope-specific immune responses, animal survival and augmentation of PR8-specific antibody responses. HLA-DR4 transgenic H-2b mice, (n=14), were immunized with CD4+20 epitope DNA construct or empty vector DNA control using Ichor’s in vivo electroporation device with 50 µg per mouse. Additional, a group of mice was also immunized with inactivated PR8. Two weeks post-immunization, two mice per group were sacrificed and splenocytes prepared for an epitope-specific IFN-γ ELISPOT (A). All 20 CD4+ epitope-specific responses were evaluated. Only positive responses plus three negative responses as comparators are shown. At this same time, the remaining 12 mice were infected with 100 HAU PR8 (4MLD50). Six days following the PR8 infection, two additional mice were sacrificed from each group for a second epitope-specific ELISPOT evaluation (A). The remaining 10 mice were monitored for survival of the animals (B). HLA-DR4 transgenic mice, (n=10) were immunized with CD4+20 epitope DNA vaccine or empty vector DNA control. Two weeks later, mice were immunized with 15 µg inactivated PR8. Seven and 14 days following inactivated PR8 immunization, mice were bled for determination of HA-specific antibody responses (C).
Vaccine induced protection was assessed using PR8 lethal challenge in the HLA-DR4 transgenic mice. In the 20 HTL pDNA vaccinated group, 7 of 10 immunized animals survived the 15 day duration of infection (Fig. 3B) whereas only a single animal in the empty vector DNA control animal survived. Immunization with inactivated PR8 virus induced a higher level of protection as the entire positive control group survived. These results demonstrate that influenza-derived HLA-DR-restricted CD4+ T cell epitopes can induce immune responses that partially protect against influenza infection.
Influenza-derived CD4+ T cell epitopes may afford protection by providing ‘help’ to HA-specific B cells and thus increase the efficiency or rate of antibody production. To evaluate this potential mechanism, we immunized mice with the 20 HTL pDNA vaccine followed by immunization with the inactivated PR8 influenza virus. Seven days following the administration of the virus, significant levels of antibodies were measured in the animals primed with the 20 HTL pDNA vaccine; antibody titers varied from 7,300 to 36,900, mean = 14,000 (Fig. 3C). In contrast, unprimed animals generated significantly lower responses (P=0.0033) with mean antibody titers of 3,000. HAI antibody titers were also significantly higher (P=0.0015) in the 20 HTL pDNA immunized mice; antibody titers in the 80 to 320 range in contrast to a titer of 80 for control animals. Significant differences in antibody responses were also noted day 14 post-inactivated PR8 immunization (Fig. 3C).
4. Discussion
Currently, influenza vaccines are not capable of conferring protection against emerging variants of even a single influenza virus subtype. Herein, we explored the feasibility of an influenza DNA vaccine encoding HLA-DR-restricted CD4+ T cell epitopes derived from highly conserved regions of influenza virus proteins from past, current and potential emerging viral strains. Our vaccine differs from that of standard commercial influenza vaccines in that the vaccine is specifically designed to induce T helper cell activity and its utility would remain effective against both circulating or newly emerging influenza virus because conserved epitopes are targeted.
Influenza A-derived CD4+ T cell epitopes were identified using a well established process allowing us to identify 45 conserved CD4+ T cell epitopes, 20 which were selected for inclusion in a prototype DNA vaccine. Interestingly, 10 of these epitopes were also identified in independent studies (see Table 2 legend) but 27 are described herein for the first time.
We focused our analyses on the internal proteins of influenza A, specifically; M1, M2, NP, PB1, PB2, PA, NS1 and NS2. This bias was based on data from studies using animal models indicating that protective cellular responses were induced by internal proteins. Numerous studies using the nucleoprotein (NP) and the matrix (M) proteins as vaccine immunogens have been published [10,27,28]. Our results expanded the antigen sources, with a test package consisting of 20 epitopes, 10 of which derived from antigens other than M and NP.
Data from multiple studies in mice have demonstrated the contribution of cellular immune responses in vaccine-induced protection against disease and death; using various vaccine formats [10,27,29–32] and adoptive transfer of CD4+ T cells [16]. In our study, a DNA construct encoding the CD4+ T cell helper epitopes was produced because this format provides an effective means to deliver multiple epitopes [26]. It is interesting to note that immunization of HLA-DR4 transgenic mice, using the DNA vaccine and the in vivo electroporation device, induced only minimal primary responses (Fig. 3). However, this treatment effectively primed CD4+ T cell responses; which was demonstrated by the rapid and high magnitude responses observed after infection with the PR8 virus. The vaccine induced priming specific for these peptides in HLA-DR4 transgenic mice which is consistent with their measured HLA-DRB1*0401 (DR4) binding affinities. Five of the 7 peptides exhibited high binding affinity to DR4 of 53nM or less, while the others were characterized with somewhat reduced levels of binding. In contrast, similar responses were not boosted using inactivated PR8 virus in place of the infection; perhaps because the amount of antigen entering the Class II pathway was higher for live viral replication.
To test benefit of these CD4+ T cells, mice were first immunized with the DNA vaccine, or control empty vector, and the challenged with a lethal dose of the PR8 virus. A majority of the immunized animals (7 of 10) survived whereas only 1 of the 10 control animals survived. Thus the benefit of primed helper T cell epitope responses was established although the level of protection was not absolute and less than achieved using inactivated PR8 virus. We propose that the vaccine would be ‘universal’ and thus protective against various strains of influenza representing relevant epidemic and potential pandemic strains. However, it should be noted that this assumption is based solely on the fact that the epitopes encoded within the vaccine are conserved among multiple influenza strains and not that multiple strains of influenza were used as challenge virus in this study.
The mechanism of protection mediated by CD4+ T cells appeared most likely to be indirect since the survival rate was not as high as that achieved using the inactivated PR8 virus. More specifically, the effect could be related to the action of antibodies and the CD4+ T cells provided support for antibody production. To test this hypothesis, mice were primed with the DNA vaccine and boosted with inactivated PR8 and significantly higher PR8- and HA-specific antibody responses were observed; relative to mice immunized only with the empty vector DNA control. Thus the benefit of the DNA vaccine was likely the positive effect on antibody production. The effect of the vaccine on augmenting cytotoxic T-cell activity was not evaluated but should be considered as a possible explanation for the protective effects of the vaccine.
Our belief is that primed CD4+ T cells specific to the internal proteins were able to support B cell differentiation and maturation resulting in enhanced influenza-specific antibody response kinetics. Several investigators have generated data that support this hypothesis using active vaccination experiments [18,19] and the passive transfer of CD4+ effector T-cells [16]. As such, our finding does not appear to be unusual.
Several studies, including ours, demonstrated the presence of effector and memory CD4+ T cells in humans specific for highly conserved epitopes; epitopes that occur in both human season influenza virus strains and the avian H5N1 virus [33,34]. It is tempting to speculate that expanding these reactive CD4+ T cells in the human population by deliberate vaccination might, at least to some degree, afford increased protection against influenza infection in vulnerable subpopulations, as well as provide protection against new seasonal variants and possibly even new pandemic strains and this could be the basis for a new approach to augmenting the efficacy of current influenza virus vaccines.
The HLA system is highly polymorphic and the use of individual epitopes to design a vaccine could result in a product that fails to provide the desired efficacy in diverse ethnicities. However, we reasoned that the use of multiple epitopes, each binding multiple HLA molecules, would enable the development of a vaccine that would be sufficiently immunogenic in most major ethnic populations. The potential population coverage was calculated using peptide-HLA-DR binding affinities and reported gene frequencies for HLA-DR types to estimate the fraction of individuals with the genetic potential to respond to the vaccine encoded epitopes [35]. Based on the 20 epitopes used in this study and the 13 HLA-DR allelic products used in the binding analyses (Table 2) we calculated a theoretical minimal population coverage of 89%. Thus, this approach appears well suited for use in the design new influenza virus vaccine product.
Acknowledgements
We thank Barbara Stewart, Kerin Arora, Barry Ellefsen, John Sidney, Jason Mendy and Xin Dong for technical assistance and data analysis. This work was partially supported by NIH-NIAID contract number N01 AI-30039.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. Influenza Fact Sheet No. 211. http://www.who.int/mediacentre/factsheets/fs211/en/.
- 2.Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165:265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 3.Beveridge WI. Webster R, Bean W., Jr . Evolution and ecology of human influenza viruses. London: Heinemann; 1977. Influenza: the last great plague. [Google Scholar]
- 4.Oxford JS. Influenza A pandemics of the 20th century. Rev Med Virol. 2000;10:119–133. doi: 10.1002/(sici)1099-1654(200003/04)10:2<119::aid-rmv272>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 5.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci USA. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, K Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webster RG, Peiris M, Chen MH, Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12:3–8. doi: 10.3201/eid1201.051024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichelberger M, Golding H, Hess M, Weir J, Subbarao K, Luke CJ, Friede M, Wood D. FDA/NIH/WHO public workshop on immune correlates of protection against influenza A viruses in support of pandemic vaccine development. Vaccine. 2008;26:4299–4303. doi: 10.1016/j.vaccine.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Rimmelzwaan GF, McElhaney JE. Correlates of protection: Novel generations of influenza vaccines. Vaccine. 2008;26S:D41–D44. doi: 10.1016/j.vaccine.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 10.Epstein SL, Stack A, Misplon JA, Lo C-Y, Mostowski H, Bennink J, Subbarao K. Vaccination with DNA encoding internal proteins of influenza virus does not require CD8+ cytotoxic T lymphocytes: either CD4+ or CD8+ T cells can promote survival and recovery after challenge. International Immunol. 2000;12:91–101. doi: 10.1093/intimm/12.1.91. [DOI] [PubMed] [Google Scholar]
- 11.Crowe SR, Miller SC, Brown DM, Adams PS, Dutton RW, Harmsen AG, Lund FE, Randall TD, Swain SL, Woodland DL. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24:457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 12.McMichael AJ, F Gotch FM, Noble GR, Beare PAS. CTL immunity to influenza. N Engl J Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 13.Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. Cross-subtype protection in humans during sequential, overlapping, or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. J Infect Dis. 1985;151:81–88. doi: 10.1093/infdis/151.1.81. [DOI] [PubMed] [Google Scholar]
- 14.McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, Ewen C, Kane KP, Bleackley RC. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 15.Forrest BD, Pride MW, Dunning AJ, Capeding MRZ, Chotpitayasunondh T, Tam JS, Rappaport R, Eldridge JH, Gruber WC. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin Vaccine Immunol. 2008;15:1042–1053. doi: 10.1128/CVI.00397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 17.Marshall D, Sealy R, Sangster M, Coleclough C. TH cells primed during influenza virus infection provide help for qualitatively distinct antibody responses to subsequent immunization. J Immunol. 1999;163:4673–4682. [PubMed] [Google Scholar]
- 18.Russell SM, Liew FY. T cells primed by influenza virion internal components can cooperate in the antibody response to haemagglutinin. Nature. 1979;280:147–148. doi: 10.1038/280147a0. [DOI] [PubMed] [Google Scholar]
- 19.Scherle PA, Gerhard W. Functional analysis of influenza-specific helper T cell clones in vivo. T cells specific for internal viral proteins provide cognate help for B cell responses to hemagglutinin. J Exp Med. 1986;164:1114–1128. doi: 10.1084/jem.164.4.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun JC, M Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 22.Tangri S, Mothé BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, Zinckgraf J, Bilsel P, Newman M, Chesnut R, LiCalsi C, Sette A. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174:3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]
- 23.Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A, Grey HM. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 24.Livingston B, Crimi C, Newman M, Higashimoto Y, Appella E, Sidney J, Sette A. A rational strategy to design multiepitope immunogenes based on multiple TH lymphocyte epitopes. J Immunol. 2002;168:5499–5506. doi: 10.4049/jimmunol.168.11.5499. [DOI] [PubMed] [Google Scholar]
- 25.Luxembourg A, Evans CF, Hannaman D. Electroporation-based DNA immunization: translation to the clinic. Expert Opin Biol Ther. 2007;7:1647–1664. doi: 10.1517/14712598.7.11.1647. [DOI] [PubMed] [Google Scholar]
- 26.Depla E, Van der Aa A, Livingston BD, Crimi C, Allosery K, De Brabandere V, Krakover J, Murthy S, Huang M, Power S, Babé L, Dahlberg C, McKinney D, Sette A, Southwood S, Philip R, Newman MJ, Meheus L. Rational design of a multiepitope vaccine encoding T-lymphocyte epitopes for treatment of chronic hepatitis B virus infections. J Virol. 2008;82:435–450. doi: 10.1128/JVI.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altstein AD, Gitelman AK, Smirnov YA, Piskareva LM, Zakharova LG, Pashvykina GV, Shmarov MM, Zhirnov OP, Varich NP, Ilyinskii PO, Shneider AM. Immunization with influenza A NP-expressing vaccinia virus recombinant protects mice against experimental infection with human and avian influenza viruses. Arch Virol. 2006;151:921–931. doi: 10.1007/s00705-005-0676-9. [DOI] [PubMed] [Google Scholar]
- 28.Mbawuike IN, Zhand Y, Couch RB. Control of mucosal virus infection by influenza nucleoprotein-specific CD8+ cytotoxic T lymphocytes. Respiratory Research. 2007;8:44. doi: 10.1186/1465-9921-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoloff GA, Caparros-Wanderley W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur J Immunol. 2007;37:2441–2449. doi: 10.1002/eji.200737254. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Yedidia T, Arnon R. Epitope-based vaccine against influenza. Expert Rev Vaccines. 2007;6:939–948. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]
- 31.Kreijtz JHCM, Bodewes R, van Amerongen G, Kuiken T, Fouchier RAM, Osterhaus ADME, Rimmelzwaan GF. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine. 2007;25:612–620. doi: 10.1016/j.vaccine.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 32.Nagata T, Toyota T, Ishigaki H, Ichihashi T, Kajino K, Kashima Y, Itoh Y, Mori M, Oda H, Yamamura H, Taneichi M, Uchida T, Ogasawara K. Peptides coupled to the surface of a kind of liposome protect infection of influenza viruses. Vaccine. 2007;25:4914–4921. doi: 10.1016/j.vaccine.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Lee LY, Ha do LA, Simmons C, Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend ARM, Askonas BA, Rowland-Jones S, Dong T. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118:3273–3275. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008;180:1758–1768. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bui H-H, Sidney J, Ding K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, James EA, Huston L, Danke NA, A Liu AW, Kwok WW. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clinical Immunol. 2006;120:21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Assarsson E, Bui H-H, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol. 2008;82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]