Abstract
Background:
MicroRNA-143 (miRNA-143) is frequently down-regulated in colorectal cancer (CRC) and may influence CRC cell proliferation, apoptosis and sensitivity to 5-fluorouracil. mRNA encoded by the KRAS oncogene has been identified as a target of miRNA-143. However, the prognostic significance of miRNA-143 expression and the ability to predict patient response to epidermal growth factor receptor (EGFR)-targeted agents have not yet been explored.
Methods:
We examined 77 CRC patients who were identified by pyrosequencing to have wild-type KRAS and were subsequently treated with EGFR-targeted therapy with the monoclonal antibodies cetuximab or panitumumab. MicroRNA-143 expression was measured in CRC tissue and corresponding non-neoplastic colon tissue by RT–PCR and its expression level was correlated with clinico-pathological characteristics. Univariate and multivariate analyses were used to calculate cancer-specific survival (CSS). The progression-free survival (PFS) and objective response rates on EGFR-targeted therapy were also evaluated.
Results:
Down-regulation of miRNA-143 was observed in 47 out of 77 (61%) tumours. Multivariate Cox regression analysis identified low levels of miRNA-143 expression as an independent prognostic factor with respect to CSS (hazard ratio=1.92, confidence interval=1.1–3.4, P=0.024). A significant difference was also observed with regard to PFS on EGFR-targeted therapy (P=0.031), but there were no significant differences with regard to the objective response rates.
Conclusion:
Our data indicate that miRNA-143 expression levels serve as an independent prognostic biomarker for CRC in KRAS wild-type patients. No role for miRNA-143 expression as a predictive biomarker for EGFR-targeted agents could be identified. Given its negative impact on CSS and PFS, miRNA-143 represents a novel prognosticator and a promising drug target for patients with CRC.
Keywords: microRNA, colorectal cancer, prognosis, KRAS , targeted therapy
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with over 1.2 million new cancer cases and 608 700 deaths estimated to have occurred in 2008 (Jemal et al, 2011). In its early stages, CRC represents a curable disease. Once metastasised, however, for the vast majority of patients only life-prolonging palliative treatment options remain. Since the 1990s, 5-fluorouracil (5-FU)-based chemotherapeutic backbone has been the standard of care for all treatment approaches for metastatic CRC (mCRC), and efficacy has been substantially improved by the introduction of the cytotoxic agents irinotecan and oxaliplatin (Grothey et al, 2004). Advances in deciphering the molecular mechanisms underlying mCRC have led to the development of novel agents that selectively inhibit defined molecular targets. Accordingly, the monoclonal antibodies cetuximab and panitumumab, which selectively target the extracellular domain of the epidermal growth factor receptor (EGFR), have expanded the spectrum of therapeutic options and improved the clinical outcome of patients with mCRC (Cunningham et al, 2004; Douillard et al, 2010; Peeters et al, 2010). Despite the development of these relatively expensive and novel agents, early reports have demonstrated that only 10–20% of unselected patients with mCRC clinically benefited from EGFR-targeted monoclonal antibodies (Cunningham et al, 2004). The subsequent analysis of large clinical trials has demonstrated a clear negative relationship between objective response rates and the occurrence of mutations in the KRAS gene, establishing KRAS mutations as negative predictors for EGFR-targeted therapies in mCRC (Amado et al, 2008; Karapetis et al, 2008; Lievre et al, 2008). However, the occurrence of KRAS mutations only accounts for ∼35–45% of non-responsive patients, and therefore, there is a clear need to identify additional predictive biomarkers to help patients avoid ineffective, toxic and expensive therapies (Bardelli and Siena, 2010). Recently, the classical dogma that protein-coding genes recognised as tumour suppressors and oncogenes are the key factors implicated in carcinogenesis has been expanded by the identification of a class of non-protein-coding RNA molecules known as microRNAs (miRNAs) (Calin and Croce, 2006). MicroRNAs are naturally occurring small RNAs that are 18–25 nucleotides in length (thus termed ‘micro’ RNAs). As a generalised mechanism of action, miRNAs suppress endogenous gene expression by binding to the 3′-untranslated region (3′UTR) of large target mRNAs, leading to either translational repression or the cleavage of their target mRNAs (Grothey et al, 2004). Chen et al (2009) were the first to demonstrate a direct link between a specific miRNA and the RAS/RAF/MAP kinase pathway. They found that miRNA-143 inhibits the translation of KRAS mRNA to alter this RAS signalling network and consequently inhibits tumour cell growth. Recently, Loboda et al (2010) demonstrated that activation of the RAS signalling pathway as determined by the analysis of a RAS pathway gene expression signature can predict resistance to cetuximab in CRC. As miRNA-143 was shown to target KRAS mRNA and therefore influence the KRAS signalling pathway in CRC cells, the regulation of KRAS gene expression by miRNA-143 or other miRNAs might contribute to a resistance against EGFR-targeted agents. The first evidence for this hypothesis comes from two reports showing that a polymorphism in the 3′UTR of the KRAS gene represents a binding site for a particular miRNA (let-7) and that a specific genotype of this miRNA-binding site polymorphism predicts for cetuximab responsiveness in KRAS wild-type patients (Graziano et al, 2010; Zhang et al, 2011). Second, a recently published study has reported that a specific miRNA, miRNA-212, also regulates resistance against cetuximab in head and neck squamous cell carcinoma (Hatakeyama et al, 2010). Given the previously described role for miRNA-143 in CRC tumourigenesis and its known KRAS-interacting properties, we evaluated its potential relation to prognosis as well as its predictive value in CRC patients treated by EGFR-targeted agents.
Patients and methods
Study population
This retrospective study included data from 77 histologically confirmed CRC patients who underwent treatment between January 2005 and 2011 at the Division of Oncology, Medical University of Graz, Austria. All patients were consecutively selected on the criteria that their tumours were wild-type for KRAS and that they received an EGFR-targeted therapeutic agent. Institutional ethics committees approved this study (No. 23-545 ex 10/11). Patients clinico-pathological data were retrieved from medical records at the same institution. Pathology reports were evaluated for pathological T stage, tumour grade, number of lymph node metastasis (N stage), presence or absence of distant metastasis (M stage), tumour stage (I–IV), levels of the tumour marker carcinoembryonic antigen and the number and characteristics of treatment lines.
First-line and salvage regimens were selected as standard of care regimens and all patients received a combination or monotherapy with the EGFR-targeting monoclonal antibodies cetuximab or panitumumab. The burden of disease was evaluated at baseline and after every four cycles of therapy (8 weeks) during the treatment. Therapeutic response was assessed using the Response Evaluation Criteria in Solid Tumors (RECIST), where disease progression (PD) was defined as the appearance of any new lesion or an increase >20% in the sum of the longest diameter (LD) of target lesions, complete response (CR) was a disappearance of all target lesions, partial response (PR) was a 30% decrease in the sum of the LD of target lesions and stable disease (SD) was defined as small changes that do not meet the above criteria (Trillet-Lenoir et al, 2002).
To test the influence of miRNA-143 expression on objective responses to EGFR-targeted therapies, patients were selected based on evidence that a treatment outcome could be attributable only to the administration of either panitumumab or cetuximab. This included patients who received either cetuximab or panitumumab as monotherapy and patients who experienced progression on previously used chemotherapy, where cetuximab or panitumumab were administered in combination with the prior chemotherapy at the same dose and schedule previously used. In all patients, refractoriness to prior chemotherapy was defined as documented PD after comparison of computed tomography or magnetic resonance imaging scans after 4 cycles (administered for at least 8 weeks) of receiving chemotherapy. Besides the mentioned inclusion criteria, the availability of a tumour block suitable for quantitative molecular analyses was also a requirement for inclusion in the present study.
KRAS mutational testing
To test the samples for KRAS exon 2 mutations, we extracted the DNA from the tumour samples and determined the sequence in codons 12 and 13 by pyrosequencing. Importantly, we used pyrosequencing because its high sensitivity allows for a more accurate assessment of KRAS mutation burden in CRC than reported for conventional DNA-sequencing (Weidlich et al, 2011). Briefly, the histological section of each case were reviewed by two pathologists (GH and SL) and one paraffin block each of tumour and corresponding non-neoplastic colon mucosa were retrieved. An area consisting of at least 50% tumour cells from each sample was selected and genomic DNA was extracted as previously described (Pichler et al, 2009). The quality and concentration of the extracted DNA was determined using a NanoDrop instrument (Thermo Scientific, Wilmington, DE, USA). Pyrosequencing analysis of the samples was performed according to the manufacturer’s recommendations for a PyroMark Q24 KRAS assay (Qiagen, Hilden, Germany) on a PyroMark Q24 instrument with PyroMark Q24 1.0.9 software (Qiagen).
MiRNA-143 expression level
FFPE tumour blocks were reviewed for quality and tumour content, and a single representative tumour block from each patient, containing at least ⩾60% of neoplastic cells, was selected. Corresponding normal adjacent colon mucosa was also used for RNA extraction. The tissue was microdissected, and 2–8 slides of 10-μm thickness were used for RNA extraction. MicroRNAs were isolated using an miRNeasy FFPE Kit 50 (Qiagen) according to the manufacturer’s instructions. cDNA was synthesised from 500 ng of total RNA using a miScript Reverse Transcription Kit (Qiagen). Quantification of miRNAs was performed using a miRscript SYBR Green PCR kit (Qiagen) and a specific miScript Primer Assay: Hs_ miR-143_1 miScript Primer Assay (Qiagen) according to the manufacturer’s recommendations on a Light Cycler 480 real-time PCR device (Roche, Mannheim, Germany). Expression data of mirRNA-143 have been normalised using the geometric mean expression of three suitable and widely used reference genes (RNU6B (Ma et al, 2011), miRNA-16 (Chang et al, 2010) and miRNA-345 (Chang et al, 2010); all specific miScript Primer Assays from Qiagen). Differences in the fold expression with regard to the adjacent normal colon mucosa were calculated from the duplicates of CT values following the 2−ΔΔCt method (Schmittgen and Livak, 2008).
Statistical analyses
All statistical analyses were performed using SPSS (version 19.0) software (SPSS Inc., Chicago, IL, USA). The continuous miRNA-143 expression variable was analysed as a dichotomous variable according to approximately ‘optimal cutpoints’, determined as follows (Atzpodien et al, 2003): The value best discriminating between dead and alive (i.e., which had the most significant P-value on a log-rank test) was found by testing all possible cutoff points near the expression level of 1. The expression value near of 1 is based on the biologically meaningful consideration that the expression level of 1, which means no difference between neoplastic and non-neoplastic tissue, separates the two groups into a high and a low expression group. χ2 test and the Mann–Whitney U-test were used where appropriate to analyse miRNA-143 expression in relation to each clinico-pathological parameter. Cancer-specific survival (CSS) was defined as the time from date of diagnosis to the date of cancer-related death. Cancer-specific survival was assessed using the Kaplan–Meier method and a log-rank test was used to compare the survival curves for individual groups. Univariable and multivariable Cox proportional hazards models, including age, gender, tumour stage (according to the AJCC TNM classification), tumour grade, level of CEA, number of therapies and miRNA-143 expression, were fit to determine the clinico-pathological parameters that were significantly statistically associated with CSS, and the results were reported, including hazard ratios (HRs) and 95% confidence intervals (CIs). The assumption of proportional hazards was checked by LML-plots and residual analyses by Schoenfeld-plots. The objective tumour response rate after 8 weeks of treatment with an EGFR-targeted agent was the end point of our predictive analysis. Qualitative comparisons of objective response to therapy after 8 weeks of treatment and miRNA-143 expression as a predictor were performed by two-tailed Fisher’s exact test to check possible significance. A P-value of <0.05 was considered as statistically significant. Progression-free survival (PFS) was calculated from the date of the start of a therapy with an EGFR-targeted agent to the next documentation of PD according to the RECIST criteria.
Results
We confirmed KRAS wild-type status by pyrosequencing in all 77 patients who underwent subsequent survival and response analyses. Clinico-pathological parameters of the study cohort are summarised in Table 1. The mean age was 59.8 years (s.d.±10.2, range: 31–77). The median follow-up time was 38 months (interquartile range: 25–60). Sixty-five of 77 (84.4%) patients died due to underlying malignant disease during follow-up. MicroRNA-143 was significantly down-regulated in 48 out of 77 (62.3%) of the tumours compared with adjacent non-tumour tissue (median relative expression level: 0.73, range: 0.15–7.78, P<0.05, Mann–Whitney U-test). To analyse the association of miRNA-143 expression with clinico-pathological parameters, CSS and PFS during treatment with an EGFR-targeted therapy, we divided CRC cases into categories of either low or high expression of miRNA-143 using an optimal cutoff value calculated as described in detail in the Patients and methods section. This cutoff value was universally used throughout the study. MicroRNA-143 expression did not significantly correlate with patient age, gender (all P-value >0.05, χ2 test), tumour stage at diagnosis, tumour grade or the level of carcinoembryonic antigen (all P-value >0.05, Mann–Whitney U-test; Table 2). Univariable analysis identified high tumour grade, high tumour stage, high levels of the tumour marker CEA and low expression of miRNA-143 as predictors of poor prognosis for CSS (all P-values <0.05, univariate Cox proportional analysis), whereas age, gender, lines of therapy and tumour location were not significantly associated with CSS (Table 3).
Table 1. Clinico-pathologic characteristics of the colorectal cancer patients included in this study.
| Clinico-pathologic parameters | Patients ( n =77) | Proportion |
|---|---|---|
| Gender | ||
| Male | 52 | 67.5% |
| Female | 25 | 32.5% |
| Age at diagnosis (years) | ||
| <60 | 35 | 45.5% |
| ⩾60 | 42 | 54.5% |
| Tumour location | ||
| Colon | 49 | 63.6% |
| Rectum | 28 | 36.4% |
| Tumour grade | ||
| G1 | 2 | 2.6% |
| G2 | 55 | 71.4% |
| G3 | 20 | 26% |
| Stage at diagnosis | ||
| II–III | 32 | 41.6% |
| IV | 45 | 58.4% |
Table 2. Correlations between miRNA-143 expression and clinico-pathological parameters.
|
miRNA-143 expression
|
|||
|---|---|---|---|
| Clinico-pathological parameter | miRNA-143 low ( n =41) | miRNA-143 high ( n =36) | P -value |
| Age (years) | |||
| <60 | 17 (40.4%) | 18 (50%) | 0.453 |
| ⩾60 | 24 (59.6%) | 18 (50%) | |
| Gender | |||
| Male | 30 (73.2%) | 22 (61.1%) | 0.259 |
| Female | 11 (26.8%) | 14 (38.9%) | |
| Tumour location | |||
| Colon | 25 (61%) | 24 (66.7%) | 0.604 |
| Rectum | 16 (39%) | 12 (33.3%) | |
| Tumour grade | |||
| G1 | 1 (2.4%) | 1 (2.8%) | 0.495 |
| G2 | 28 (68.3%) | 27 (75%) | |
| G3 | 12 (29.3%) | 8 (22.2%) | |
| Stage | |||
| II–III | 14 (34.1%) | 18 (50%) | 0.137 |
| IV | 27 (65.9%) | 18 (50%) | |
| Levels of tumour marker (median ng ml−1) | |||
| CEA | 4 | 3 | 0.606 |
Abbreviations: CEA=carcinoembryonic antigen; miRNA=microRNA.
Table 3. Univariable analysis of clinico-pathological parameters for the prediction of cancer-specific survival in patients with colorectal cancer.
|
Cancer-specific survival
|
||
|---|---|---|
| Parameter | HR (95% CI) | P -value |
| Age at diagnosis (years) | ||
| <60 | 1 (Reference) | 0.526 |
| ⩾60 | 1.18 (0.71–1.95) | |
| Gender | ||
| Male | 1 (Reference) | 0.956 |
| Female | 1.02 (0.59–1.74) | |
| Tumour location | ||
| Colon | 1 (Reference) | 0.597 |
| Rectum | 0.87 (0.53–1.44) | |
| Tumour grade | ||
| G1+G2 | 1 (Reference) | <0.001 |
| G3 | 2.5 (1.41–4.4) | |
| Tumour stage | ||
| II+III | 1 (Reference) | 0.015 |
| IV | 1.9 (1.13–3.2) | |
| Lines of therapy | ||
| ⩽3 | 1 (Reference) | 0.052 |
| >3 | 0.59 (0.34–1.01) | |
| Level of CEA | ||
| Normal | 1 (Reference) | 0.003 |
| Increased | 2.25 (1.33–3.81) | |
| miRNA-143 expression | ||
| High | 1 (Reference) | 0.007 |
| Low | 2.09 (1.22–3.58) | |
Abbreviations: CEA=carcinoembryonic antigen; CI=confidence interval; HR=hazard ratio; miRNA=microRNA.
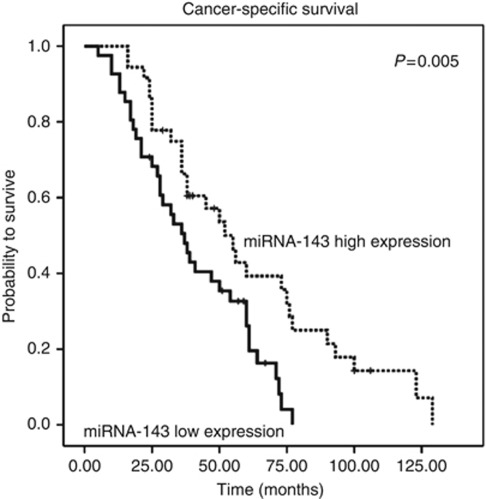
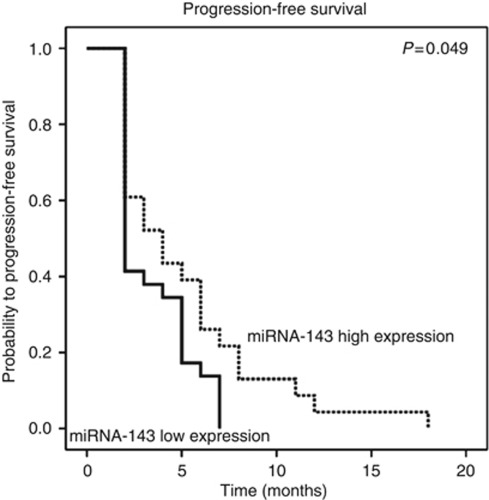
The median CSS for patients with high expression of miRNA-143 was 42.5 months, whereas the median CSS for patients with low expression of miRNA-143 was 36 months (P=0.035, Mann–Whitney U-test). Figure 1 shows the Kaplan–Meier curve for CSS and reveals that low miRNA-143 expression is a factor for poor prognosis in CRC (P=0.005, log-rank test). To test whether the prognostic value of low miRNA-143 expression was independent of other risk factors for poor survival, a multivariate analysis was performed using a Cox proportional hazards model. In multivariate analyses that included age and all other factors significant in univariate analysis (tumour stage, tumour grade, level of CEA and level of miRNA-143 expression), low miRNA-143 expression was identified as an independent predictor for CSS in wild-type KRAS CRC patients (HR=1.86, CI=1.06–3.25, P<0.031, multivariate Cox proportional analysis). Statistically significant results were also obtained for patients with a high tumour grade (P=0.003) and elevated CEA levels (P=0.023), whereas age and tumour stage were not significantly associated with CSS (Table 4). To analyse whether miRNA-143 expression may impact the efficacy of EGFR-targeted therapy and could serve to predict therapeutic response to these drugs, we analysed cases where treatment outcome could be attributable only to the administration of either panitumumab or cetuximab. Applying the criteria detailed in the Patients and Methods section, 52 patients could be analysed accordingly. The other 25 patients who were excluded for an evaluation of the predictive value of miRNA expression received an EGFR-targeted agent in combination with a first- or second-line chemotherapy so that the therapeutic response was not solely attributable to the EGFR-targeted therapy. Overall, 27 patients were treated with a cetuximab-based therapy, and 25 were treated with a panitumumab-based therapy. Sixteen patients received monotherapy either with panitumumab (n=14) or with cetuximab (n=2), whereas 36 patients who were refractory to previous chemotherapeutical regimens received the monoclonal antibodies in combination with prior chemotherapy. Overall, there were 25 out of 52 (48%) patients with progressive disease, 17 out of 52 (33%) with SD, 9 out of 52 (17%) with partial remission and one patient out of 52 (2%) with complete remission after receiving four cycles of an EGFR-targeted therapy according to the international established RECIST criteria. Mean PFS time for the 52 patients was 4.2 months (range: 2–18 months). Dividing the patients into groups according to low or high miRNA-143 expression levels resulted in 29 patients with low miRNA-143 expression and 23 patients with high miRNA-143 expression. There were no significant differences found regarding therapeutic response in the 52 evaluable patients with an EGFR-targeting agent (P=0.288, χ2 test; Table 5). Median PFS in the high miRNA-143 expression group was 4 months compared with a median PFS of 2 months for the group with low miRNA-143 expression. Progression-free survival was significantly lower among patients whose tumours showed low expression of miRNA-143 compared with patients with miRNA-143 high expression (P=0.049, log-rank test; Figure 2).
Figure 1.
Kaplan–Meier plot for CSS in KRAS wild-type CRC patients (n=77) stratified by miRNA-143 expression level.
Table 4. Multivariable analysis of clinico-pathological parameters for the prognosis of cancer-specific survival in patients with KRAS wild-type colorectal cancer.
|
Cancer-specific survival
|
||
|---|---|---|
| Parameter | HR (95% CI) | P -value |
| Age at diagnosis (years) | ||
| <60 | 1 (Reference) | 0.859 |
| ⩾60 | 1.05 (0.61–1.81) | |
| Tumour grade | ||
| G1+G2 | 1 (Reference) | 0.003 |
| G3 | 2.47 (1.36–4.51) | |
| Tumour stage | ||
| II+III | 1 (Reference) | |
| IV | 1.38 (0.78–2.46) | 0.270 |
| Level of CEA | ||
| Normal | 1 (Reference) | 0.023 |
| Increased | 1.92 (1.09–3.37) | |
| miRNA-143 expression | ||
| High | 1 (Reference) | 0.031 |
| Low | 1.86 (1.06–3.25) | |
Abbreviations: CEA=carcinoembryonic antigen; CI=confidence interval; HR=hazard ratio; miRNA=microRNA.
Table 5. Therapy response in KRAS wild-type CRC patients treated by an EGFR-targeting agent.
|
miRNA-143 expression
|
||
|---|---|---|
| RECIST criteria | Low expression, no. (%) | High expression, no. (%) |
| Complete remission | 0 (0%) | 1 (4.3%) |
| Partial remission | 4 (13.8%) | 5 (21.7%) |
| Stable disease | 8 (27.6%) | 9 (39.1%) |
| Progressive disease | 17 (58.6%) | 8 (34.8%) |
Abbreviations: CRC=colorectal cancer; EGFR=epidermal growth factor receptor; RECIST=Response Evaluation Criteria in Solid Tumors; miRNA=microRNA.
Figure 2.
Kaplan–Meier plot for PFS in KRAS wild-type CRC patients (n=52) treated by an EGFR-targeted agent.
Discussion
Several factors influence the survival of patients with mCRC. Although novel drugs that target the EGFR signalling pathway in CRC have been approved for routine clinical use, data from large clinical trials suggest that only a small percentage of patients truly benefit from these expensive agents (Bardelli and Siena, 2010). Novel molecular biomarkers that facilitate the prediction of survival in CRC patients could, therefore, provide a tool to assess a patient’s individual risk of progression and identify patients who are candidates for more aggressive initial therapies. Moreover, biomarkers that can predict the response to novel EGFR-targeted agents would enable a better patient selection as well as an appropriate treatment protocol. In the last several years, the most useful biomarker for selecting patients with mCRC has been KRAS mutations, which serve as a negative predictor for response to EGFR-targeted agents (Amado et al, 2008; Lievre et al, 2008). Although this genetic alteration leads to the immediate exclusion of 35–40% CRC patients with KRAS mutations, there remains a great demand to discover other predictive biomarkers to further identify additional non-responsive patients in the remaining 60% of KRAS wild-type CRC patients. Notably, other alterations in the EGFR/RAS/RAF signalling pathway have also been linked with resistance to EGFR-targeted agents, but these findings have been not yet translated to clinical practice (Bardelli and Siena, 2010). To date, several lines of evidence suggest a strong role for miRNAs in the development, diagnosis and prognosis of CRC (Valeri et al, 2009; Dong et al, 2011). Specifically, miRNA-143 has been described to be down-regulated in CRC (Michael et al, 2003). More recently, miRNA-143 has also been found to be involved in targeting KRAS mRNA to influence CRC cell proliferation and survival (Chen et al, 2009). However, the prognostic significance or the predictive value to assess response to EGFR-targeted agents in CRC patients has not yet been explored. There are three main findings in this study regarding miRNA-143 and CRC: (a) low levels of miRNA-143 expression are an independent negative prognostic factor for CSS; (b) low levels of miRNA-143 expression are associated with a shorter PFS in patients with KRAS wild-type CRC who are treated with EGFR-targeted agents; and (c) miRNA-143 is not useful as a predictive biomarker in KRAS wild-type CRC patients treated with EGFR-targeting agents. In detail, we found that >60% of the primary tumours included in our study showed down-regulation of miRNA-143 compared with normal adjacent tissue. This finding is not novel but nonetheless confirms previously published reports showing a down-regulation of miRNA-143 in CRC patients (Michael et al, 2003; Slaby et al, 2007; Kulda et al, 2010). Patients expressing low levels of miRNA-143 had a significantly lower CSS. To our knowledge, only a few other studies have addressed the clinical relevance of miRNA-143 down-regulation in CRC, but none of them evaluated its prognostic significance through multivariate analyses. In an earlier study, Slaby et al (2007) reported that down-regulation of miRNA-143 was associated with a larger tumour size in 29 patients with CRC. In another study of 98 patients, Wang et al (2009) reported a down-regulation of miRNA-143 in 57% of CRC patients. Similar to our study, these authors found no correlation between miRNA-143 expression and any of the examined clinico-pathological variables. In a smaller study consisting of 46 CRC patients, Kulda et al (2010) showed that overexpression of miRNA-143 was associated with shorter disease-free survival, which is in contrast to the putative tumour suppressive role ascribed to miRNA-143. However, their study included only 44 patients for outcome analysis, showed no data about multivariable analysis and concluded that their findings did not logically correspond with differences in miRNA-143 levels between tumour tissue and normal tissue. Our study shows for the first time using a larger cohort of patients that low miRNA-143 expression in tumour tissue is significantly associated with a decreased CSS in wild-type KRAS patients. The reasons for the negative prognostic role of miRNA-143 expression remain to be more clearly elucidated, but the results of these findings fit well with previously published experimental data that led to the hypothesis that miRNA-143 represents a tumour suppressor in CRC. Along this line of argument, Chen et al (2009) described that inhibition of miRNA-143 stimulated cell proliferation, whereas miRNA-143 overexpression had an opposite effect and that inhibition of KRAS expression by miRNA-143 inhibits constitutive phosphorylation of ERK1/2. Ng et al (2009) demonstrated that restoration of miRNA-143 expression in CRC cell lines decreased tumour cell growth and soft-agar colony formation. They also identified DNA methyltransferase 3A as a potential target of miRNA-143. Importantly, Borralho et al (2009) demonstrated that increased stable expression of miRNA-143 in CRC cell lines was associated with a decreased viability and an increase of cell death after exposure to 5-FU, a drug that every patient in our study received. The data by Chen et al (2009) which suggested a KRAS mRNA-targeting role of miRNA-143, prompted us to investigate whether the expression level of miRNA-143 correlates with response to EGFR-targeted agents. Although we could not find significant differences to predict the response to these agents, a significant association for PFS in patients with low miRNA-143 expression was identified. These data are in line with the findings of reduced CSS in patients with low miRNA-143 expression. The observed shorter PFS might in part be explained by the experimental data of decreased 5-FU sensitivity in those patients with miRNA-143 low expression (Borralho et al, 2009). Interestingly, beyond the role of miRNA-143 as a prognostic biomarker, miRNA-143 might also represent a novel drug target in CRC. Data that support this assumption come from a recently published study, which demonstrated that overexpression of miRNA-143 impairs growth of human CRC xenografts in mice through an induction of apoptosis and an inhibition of proliferation (Borralho et al, 2011). Notably, the first pre-clinical studies with chemically modified miRNA-143 showed promising anti-tumour effects in CRC xenografts (Kitade and Akao, 2010). Recently, Pramanik et al (2011) introduced a lipid-based nanoparticle delivery system for miRNA-143 in a pancreatic cancer xenograft model. Accordingly, these chemically modified miRNA-based drugs and novel delivery systems may represent promising therapeutic potentials for miRNA-143-deficient CRC. The limitations of our study are its retrospective nature and the relative small sample size when analysing the predictive value of miRNA-143.
In conclusion, our present study demonstrates that miRNA-143 is down-regulated in the majority of CRC cases and that low expression of miRNA-143 in tumour tissue is an independent negative prognostic factor for CSS in CRC KRAS wild-type patients. Although associated with shorter PFS in patients treated with EGFR-targeted agents, low miRNA-143 expression seems not to be predictive for the response to these drugs. Further pre-clinical studies are warranted for the validation of miRNA-143 as a novel drug target in CRC.
Acknowledgments
This project was supported in part by the funding of the START Funding program of the Medical University of Graz, Austria (to MP).
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26(10): 1626–1634 [DOI] [PubMed] [Google Scholar]
- Atzpodien J, Royston P, Wandert T, Reitz M (2003) Metastatic renal carcinoma comprehensive prognostic system. Br J Cancer 88(3): 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A, Siena S (2010) Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 28(7): 1254–1261 [DOI] [PubMed] [Google Scholar]
- Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM (2009) MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J 276(22): 6689–6700 [DOI] [PubMed] [Google Scholar]
- Borralho PM, Simoes AE, Gomes SE, Lima RT, Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE, Rodrigues CM (2011) miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS One 6(8): e23787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866 [DOI] [PubMed] [Google Scholar]
- Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N (2010) MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer 10: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang CY (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28(10): 1385–1392 [DOI] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351(4): 337–345 [DOI] [PubMed] [Google Scholar]
- Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS (2011) MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer 104(6): 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, Blasinska-Morawiec M, Smakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J (2010) Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28(31): 4697–4705 [DOI] [PubMed] [Google Scholar]
- Graziano F, Canestrari E, Loupakis F, Ruzzo A, Galluccio N, Santini D, Rocchi M, Vincenzi B, Salvatore L, Cremolini C, Spoto C, Catalano V, D’Emidio S, Giordani P, Tonini G, Falcone A, Magnani M (2010) Genetic modulation of the Let-7 microRNA binding to KRAS 3’-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J 10(5): 458–464 [DOI] [PubMed] [Google Scholar]
- Grothey A, Sargent D, Goldberg RM, Schmoll HJ (2004) Survival of patients with advanced colorectal cancer improves with the availability of fluorouracil-leucovorin, irinotecan, and oxaliplatin in the course of treatment. J Clin Oncol 22(7): 1209–1214 [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough WG, Wheeler DL, Harari PM, Guo Y, Shyr Y, Slebos RJ, Chung CH (2010) Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One 5(9): e12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359(17): 1757–1765 [DOI] [PubMed] [Google Scholar]
- Kitade Y, Akao Y (2010) MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR-143 and -145, function as anti-oncomirs and the application of chemically modified miR-143 as an anti-cancer drug. J Pharmacol Sci 114(3): 276–280 [DOI] [PubMed] [Google Scholar]
- Kulda V, Pesta M, Topolcan O, Liska V, Treska V, Sutnar A, Rupert K, Ludvikova M, Babuska V, Holubec L, Cerny R (2010) Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200(2): 154–160 [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 26(3): 374–379 [DOI] [PubMed] [Google Scholar]
- Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, Roberts B, Zhang T, Chenard M, Haines B, Andersen J, Nagashima K, Paweletz C, Lynch B, Feldman I, Dai H, Huang P, Watters J (2010) A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics 3: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H (2011) miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut; e-pub ahead of print 3 November 2011 [DOI] [PubMed]
- Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ (2003) Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1(12): 882–891 [PubMed] [Google Scholar]
- Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, Rocken C, Ebert MP, Kwok TT, Sung JJ (2009) MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 101(4): 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, Andre T, Chan E, Lordick F, Punt CJ, Strickland AH, Wilson G, Ciuleanu TE, Roman L, Van Cutsem E, Tzekova V, Collins S, Oliner KS, Rong A, Gansert J (2010) Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 28(31): 4706–4713 [DOI] [PubMed] [Google Scholar]
- Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, Guelly C, Bauernhofer T, Samonigg H, Hoefler G, Dandachi N (2009) Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagnost 11(2): 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A (2011) Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther 10(8): 1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protocols 3(6): 1101–1108 [DOI] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R (2007) Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72(5–6): 397–402 [DOI] [PubMed] [Google Scholar]
- Trillet-Lenoir V, Freyer G, Kaemmerlen P, Fond A, Pellet O, Lombard-Bohas C, Gaudin JL, Lledo G, Mackiewicz R, Gouttebel MC, Moindrot H, Boyer JD, Chassignol L, Stremsdoerfer N, Desseigne F, Moreau JM, Hedelius F, Moraillon A, Chapuis F, Bleuse JP, Barbier Y, Heilmann MO, Valette PJ (2002) Assessment of tumour response to chemotherapy for metastatic colorectal cancer: accuracy of the RECIST criteria. Br J Radiol 75(899): 903–908 [DOI] [PubMed] [Google Scholar]
- Valeri N, Croce CM, Fabbri M (2009) Pathogenetic and clinical relevance of microRNAs in colorectal cancer. Cancer Genomics Proteomics 6(4): 195–204 [PubMed] [Google Scholar]
- Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF (2009) Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Disease Markers 26(1): 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidlich S, Walsh K, Crowther D, Burczynski ME, Feuerstein G, Carey FA, Steele RJ, Wolf CR, Miele G, Smith G (2011) Pyrosequencing-based methods reveal marked inter-individual differences in oncogene mutation burden in human colorectal tumours. Br J Cancer 105(2): 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, Hu-Lieskovan S, Mauro DJ, Langer C, Rowinsky EK, Lenz HJ (2011) A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol 22(1): 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]