Abstract
Mismatches in DNA occur naturally during replication and as a result of endogenous DNA damaging agents, but the mismatch repair (MMR) pathway acts to correct mismatches before subsequent rounds of replication. Rhodium metalloinsertors bind to DNA mismatches with high affinity and specificity and represent a promising strategy to target mismatches in cells. Here we examine the biological fate of rhodium metalloinsertors bearing dipyridylamine ancillary ligands in cells deficient in MMR versus those that are MMR-proficient. These complexes are shown to exhibit accelerated cellular uptake which permits the observation of various cellular responses, including disruption of the cell cycle, monitored by flow cytometry assays, and induction of necrosis, monitored by dye exclusion and caspase inhibition assays, that occur preferentially in the MMR-deficient cell line. These cellular responses provide insight into the mechanisms underlying the selective activity of this novel class of targeted anti-cancer agents.
Mismatches in DNA arise naturally as a result of replication errors, endogenous DNA damaging agents, and spontaneous processes such as cytosine deamination. The mismatch repair (MMR) pathway acts to correct mismatches before subsequent rounds of replication, reducing the number of DNA mismatches in the human genome from ~1000 to ~1.1 The loss of MMR carries dire consequences, including increased mutation rates2-4, carcinogenesis5-8, and resistance to a variety of clinical anti-cancer agents, such as anti-metabolites, DNA alkylators, and cisplatin.9-16 Furthermore, this resistance to commonly used agents leads to enrichment of MMR-deficient cells; roughly half of secondary leukemias show MMR-deficiency.17 These issues point to the need for a therapeutic agent that specifically targets MMR-deficient cells.
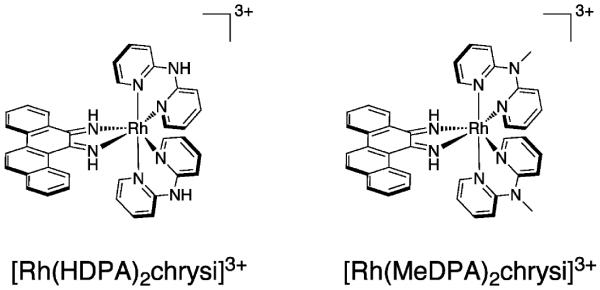
Rhodium metalloinsertors have been developed in our laboratory to target DNA mismatches in vitro (Figure 1).18-22 DNA mismatches, owing to their loss of hydrogen bonding and poor stacking, are destabilized relative to well matched DNA.23 It is this thermodynamic destabilization that our laboratory seeks to exploit as a means of targeting mismatches, since mismatches do not significantly perturb the structure of the B-form DNA duplex.24-28 The width of the expansive aromatic system of the chrysi ligand (chrysi = chrysene-5,6-quinonediimine) exceeds the width of well matched base pairs, 11.3 Å versus 10.8 Å. As a result, complexes bearing the chrysi ligand can be preferentially accommodated by DNA at thermodynamically destabilized mismatch sites.20 In addition to their DNA binding capabilities, these complexes promote single-stranded cleavage of the DNA backbone upon photoactivation. This photocleavage chemistry provides a useful tool to probe DNA binding. Photocleavage titration experiments revealed that the first generation compound, [Rh(bpy)2chrysi]3+, binds 80% of DNA mismatches with typical binding constants of 106 M−1 and remarkable specificity for mismatched DNA; similar experiments also showed single-site targeting of the mismatch in a 2.7 kb DNA fragment.18,19 Subsequent work led to the incorporation of nitrogen atoms into the intercalating ligand and a 50-fold increase in binding affinity for the second compound, [Rh(bpy)2phzi]3+ (phzi = benzo[a] phenazine-5,6-quinonediimine).20
Figure 1.
Structures of rhodium metalloinsertors, [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)2chrysi]3+.
A high resolution crystal structure of [Rh(bpy)2chrysi]3+ bound to single base mismatches within a DNA oligonucleotide duplex reveals a distinctive binding mode at the mismatched site.29 We had previously demonstrated that rhodium intercalators bearing the phi ligand bind to well-matched DNA by partial intercalation of the planar phi ligand from the major groove side into the base pair stack.30 However, binding to the mismatched site involves instead insertion of the more expansive chrysi ligand into the DNA duplex from the minor groove side at the mismatched site with ejection of the mismatched bases out of the DNA stack; the inserted ligand stacks fully with adjacent base pairs. NMR studies confirm this metalloinsertion mode for the complex at mismatched sites in solution.31 Further crystallographic studies revealed the generality of the metalloinsertion binding mode; in independent views of metalloinsertion at four different mismatch sites, the ejected bases superimpose identically.32 It seems likely that such a perturbation of the double helix might have consequences within the cell.
Of course, any potential agent must first reach its target before it may bind. Confocal microscopy and flow cytometry studies with [Ru(L)2dppz]2+ (dppz=dipyrido[3,2-a:2′,3′-c]phenazine) analogues have demonstrated that cellular accumulation occurs through passive diffusion, facilitated by the negative potential difference across the cell membrane.33-34 Taken together, the observations that (i) rhodium metalloinsertors are capable of recognizing mismatches with high selectivity and (ii) analogous ruthenium complexes accumulate within cells and nuclei form a sound basis for the hypothesis that rhodium metalloinsertors recognize mismatched DNA within cells and predict that these complexes should selectively target MMR-deficient cells, which harbor ~1,000 fold more mismatches than MMR-proficient cells.
This predicted selectivity has been demonstrated in the HCT116 cell lines.35,36 The HCT116N/HCT116O pair serve as an isogenic model system for MMR deficiency, differing only in the presence of an active copy of the MLH1 gene, which is essential for MMR.37 Rhodium metalloinsertors were shown to block selectively the growth of the repair-deficient HCT116O cell line versus the repair-proficient HCT116N cell line, as measured by an ELISA for DNA synthesis, which serves as a proxy for cellular proliferation.38,39 Importantly, the difference in activity between the cell lines correlates directly with the binding affinity for DNA mismatches for a series of rhodium metalloinsertors with varying ancillary ligands.36 This observation supports the idea that rhodium metalloinsertors bind to DNA mismatches in cellulo as well as in vitro.
However, the proliferation assay does not distinguish between senescence and cell death, and therefore is insufficient to demonstrate cytotoxicity. Here, we show that rhodium metalloinsertors are preferentially cytotoxic to the MMR-deficient HCT116O cell line. This rhodium-induced death is shown to be necrotic rather than apoptotic, and is preceded by cell cycle arrest at the G1/S-phase boundary. This observation is consistent with activation of the DNA damage response by rhodium metalloinsertors.
EXPERIMENTAL PROCEDURES
Materials
RhCl3 was purchased from Pressure Chemical, Inc. (Pittsburgh, PA). [Rh(NH3)5Cl]Cl2 was obtained from Strem Chemical, Inc. (Newburyport, MA). 2,2′-dipyridylamine (HDPA) and Sephadex ion exchange resin were obtained from Sigma-Aldrich (St. Louis, MO). Sep-Pak C18 solid phase extraction cartridges were purchased from Waters Chemical Co. (Milford, MA). Media and supplements were purchased from Invitrogen (Carlsbad, CA). The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) labeling reagant and acidified lysis buffer (10% SDS in 10 mM HCl) were purchased in kit format from Roche Molecular Biochemicals (Mannheim, Germany). Z-VAD-FMK caspase inhibitor was purchased from Promega. All PARP inhibitors were purchased from Santa Cruz Biotechnology, Inc. All commercial materials were used as received.
Synthesis of Metal Complexes
[Rh(HDPA)2chrysi]Cl3 was prepared according to previously described procedures.36 [Rh(MeDPA)2chrysi]Cl3 was synthesized by refluxing [Rh(NH3)4chrysi]TFA3 with 2 equivalents of MeDPA in 1:1 ethanol:water overnight. The ligand MeDPA was prepared by deprotonation and methylation of HDPA (unpublished results in the laboratory). The reaction mixture was cooled to room temperature, and the solvent was removed under vacuum. The desired product was isolated by cation exchange chromatography with Sephadex CM-25 resin, eluting with 0.03 M MgCl2(aq). Excess magnesium was removed by solid phase extraction with a Sep-pak C18 cartridge and the TFA counterion was exchanged for the chloride by anion exchange chromatography with Sephadex QAE-125 resin. Formation of the desired product was confirmed by UV/vis (H2O, pH 5): 295 nm (55,000 M−1cm−1), 320 nm (39,700 M−1cm−1), 390 nm (14,000 M−1cm−1) and ESI-MS (cation): 727.1 m/z ([M-2H]+), 364.3 m/z ([M-H]2+) obs., 727.2 m/z ([M-2H]+) calc. Purity was confirmed by analytical HPLC (tretention = 13.5 minutes, 10:90:0.1 MeCN:H2O:TFA to 40:60:0.1 MeCN:H2O:TFA over 45 minutes).
Cell Culture
HCT116N and HCT116O cells were grown in RPMI medium 1640 supplemented with: 10% FBS; 2 mM L-glutamine; 0.1 mM nonessential amino acids; 1 mM sodium pyruvate; 100 units/mL penicillin; 100 μg/mL streptomycin; and 400 μg/mL geneticin (G418). Cells were grown in tissue culture flasks and dishes (Corning Costar, Acton, MA) at 37°C under 5% CO2 and humidified atmosphere.
ICP-MS Assay for Cellular Rhodium Levels
Each cell line was treated with 10 μM of [Rh(bpy)2chrysi]3+, [Rh(HDPA)2 chrysi]3+, or [Rh(NH3)4chrysi]3+ for 48 hours. After rhodium incubation, the cells were harvested from adherent culture by trypsinization, washed with cold PBS, and counted by hemacytometer. The samples were pelleted and resuspended in 1% HNO3 (v/v), homogenized by three freeze/thaw cycles in liquid nitrogen, and analyzed for rhodium content on an HP-4500 ICP-MS unit. Rhodium counts were normalized to the number of cells counted in each sample before lysate preparation. Standard errors for three independent experiments are shown.
Cellular Proliferation ELISA
HCT116N and HCT116O cells were plated in 96-well plates at 2,000 cells/well and allowed 24 hours to adhere. The cells were then incubated with rhodium for the durations specified. For incubation less than 72 hours, the Rh-containing media was replaced with fresh media, and the cells were grown for the remainder of the 72 hour period. Cells were labeled with BrdU 24 hours before analysis. The BrdU incorporation was quantified by antibody assay according to established procedures.38,39 Cellular proliferation was expressed as the ratio of the amount of BrdU incorporated by the treated cells to that of the untreated cells.
MTT Cytotoxicity Assay
Cytoxicity assays were performed as described in the literature.40 HCT116N and HCT116O cells were plated in 96-well plates at 50,000 cells/well and incubated with rhodium for the durations specified. After rhodium incubation, cells were labeled with MTT for 4 hours at 37°C under 5% CO2 and humidified atmosphere. The resulting formazan crystals were dissolved with solubilizing reagant purchased from Roche according to the manufacturer’s instructions. The dissolved formazan was quantified as the absorbance at 570 nm minus the background absorbance at 690 nm. Percent viability was determined as the ratio of the amount of formazan in the treated cells to that of the untreated cells. For caspase inhibition assays, Z-VAD-FMK was added to a final concentration of 20 μM. For poly-ADP ribose polymerase (PARP) assays, the inhibitor 3,4-Dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (DPQ) was added to a final concentration of 0, 25, or 50 μM; the inhibitor 3-aminobenzamide (3-AB) was added to a final concentration of 0, 2, or 3 mM; the inhibitor 4-amino-1,8-napthalimide (4-AN) was added to a final concentration of 0, 10, or 20 μM; and the inhibitor 2-((R)-2-Methylpyrrolidin-2-yl)-1H-benzimidazole-4-carboxamide (ABT-888) was added to a final concentration of 0, 5, or 10 μM.
Cell Cycle Distribution Flow Cytometry Assay
Cells were harvested from adherent culture by trypsinization and washed with cold PBS. The resultant pellet was resuspended in PBS (chilled to 4 °C), and ice-cold ethanol was added dropwise to a final concentration of 70% (v/v), with continuous gentle agitation. Cells were fixed at 4°C for 30 minutes and stored for up to one week. Prior to analysis, the fixed cells in 70% ethanol were diluted 1:3 in cold PBS and centrifuged at 1,400 × g for 5 minutes. The resultant pellet was washed twice and resuspended in ice-cold PBS. Ribonuclease was added to a final concentration of 30 μg/mL and the cells were incubated overnight at 4 °C. The next day propidium iodide was added to a final concentration of 20 μg/mL and cells were analyzed by flow cytometry. Data analysis was performed using the FloJo software package (v 8.7.1).
Cell Death Mode Flow Cytometry Assay
Cell death was characterized by a dye exclusion assay.41 After 24, 48, or 72 hour incubation with rhodium, cells were harvested from adherent culture by trypsinization and washed with cold PBS, and centrifuged at 2,000 rpm for 5 minutes. The resultant pellets were resuspended in PBS to a concentration of 106 cells/mL and stained with propidium iodide to a final concentration of 1μg/mL and with YO-PRO-1 to a final concentration of 200 nM for 30 minutes prior to analysis by flow cytometry.
RESULTS
ICP-MS of whole cell lysates
Each cell line was treated with 10 μM of [Rh(bpy)2chrysi]3+, [Rh(HDPA)2chrysi]3+, or [Rh(NH3)4chrysi]3+ for 48 hours. Whole cell lysates were analyzed for rhodium levels by ICP-MS (Figure 2). As expected, the HDPA complex exhibits a higher degree of cellular uptake than the other complexes. This supports the notion that the early activity displayed by the complex in the ELISA assay results from accelerated uptake.36 It should be noted that these treatment conditions directly reflect those used previously.
Figure 2.
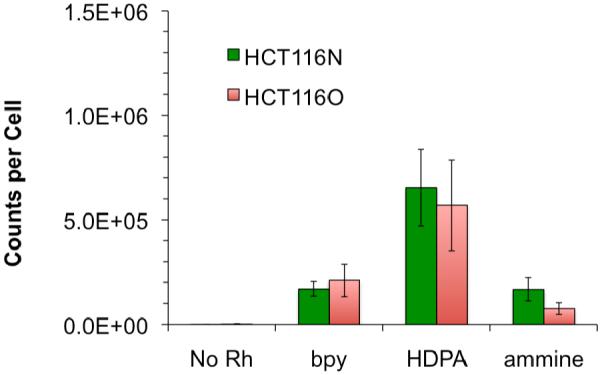
ICP-MS assay for rhodium accumulation. Normalized rhodium counts for whole cell lysates treated with 0 or 10 μM [Rh(bpy)2chrysi]3+, [Rh(HDPA)2chrysi]3+, or [Rh(NH3)4chrysi]3+ for 48 hours. Standard error bars for three trials are shown. Cellular concentrations were determined by counting on hemacytometer prior to lysis. Rhodium counts were normalized to total cells per sample.
MTT cytotoxicity assay
The cytotoxicities of [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)2chrysi]3+ were determined by MTT assay.40 Briefly, reduction of the MTT reagant by mitochondrial enzymes leads to the production of formazan, which can then be dissolved in acidified SDS to produce a characteristic absorbance at 570 nm. This absorbance reflects the relative metabolic activity, which in turn reflects the percentage of viable cells in each sample. HCT116N and HCT116O cells were plated and treated with 0-25 μM of [Rh(bpy)2chrysi]3+, [Rh(HDPA)2chrysi]3+, or [Rh(MeDPA)2chrysi]3+ for 48 or 72 hours. The results are shown in Figure 3. At 48 hours [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)2chrysi]3+ clearly display an enhanced toxicity in the MMR-deficient HCT116O cell line versus the HCT116N cell line. For example, 72 hours after treatment with 20 μM [Rh(HDPA)2chrysi]3+, the number of viable HCT116N cells is 80 ± 5.2 % of untreated controls, whereas the number of viable HCT116O cells is 37 ± 4.4 % of untreated controls. [Rh(MeDPA)2chrysi]3+ also shows differential toxicity against the HCT116O cell line in this assay comparable to that of [Rh(HDPA)2chrysi]3+.
Figure 3.
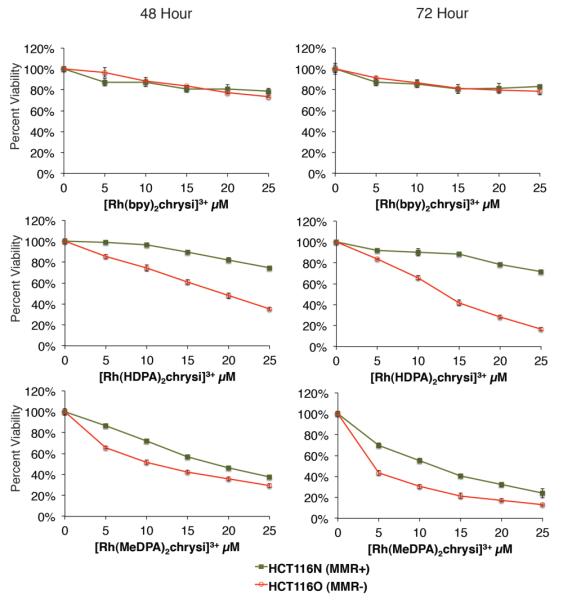
Selective toxicity of [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)2chrysi]3+ in MMR-deficient cells. HCT116N and HCT116O cells were plated in 96-well format at densities of 5 × 104 cells/well and treated with 0 – 25 μM of [Rh(bpy)2chrysi]3+, [Rh(HDPA)2chrysi]3+, or [Rh(MeDPA)2chrysi]3+. After 48–72 hours, the cells were labeled with MTT for 4 hours. While the first generation complex [Rh(bpy)2chrysi]3+ is non-toxic up to 72 hours, the dipyridylamine derivatives [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)2chrysi]3+ exhibit toxicity specifically in the MMR-deficient HCT116O cell line at 48 hours.
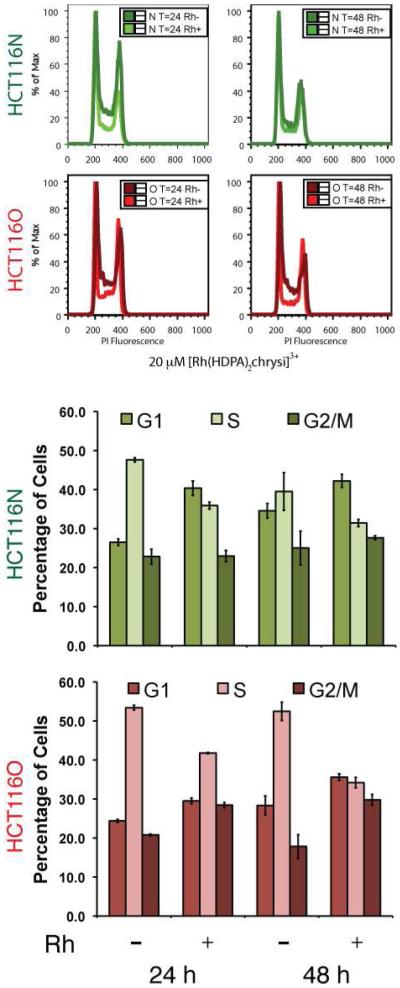
Cell cycle distribution
Given the observation that the complexes inhibit DNA synthesis, a flow cytometry assay was performed to determine if the cytotoxicity of [Rh(HDPA)2chrysi]3+ is accompanied by disruption of the cell cycle.36 MMR-proficient HCT116N and MMR-deficient HCT116O cells were treated with 20 μM [Rh(HDPA)2chrysi]3+ for 24 or 48 h. After treatment, cells were stained with propidium iodide (PI) and analyzed by flow cytometry. The PI fluorescence reports the amount of DNA in each cell and follows a bimodal distribution, where the first peak contains cells with one copy of the genome, i.e. cells in G0/G1-phase, and the second peak contains cells with two copies of the genome, i.e. cells in G2- or M-phase. Cells in S-phase occupy the region between the two peaks. Figure 4 shows these distributions for both cell lines, with or without rhodium treatment. In both cases, a decrease in the area under the curve between the peaks can be seen, indicating a depletion of the S-phase populations. Fitting the raw distributions to G1-, S-, or G2/M-phases confirms the depletion of the S-phase population, concomitant with an increase in the G1-phase population. Notably, at this concentration of [Rh(HDPA)2chrysi]3+ large differentials in both the ELISA and MTT assay were observed. The changes to the cell cycle are more pronounced in the mismatch repair deficient HCT116O cell line, which continues to grow aggressively at 48 hours (> 50% S-phase) in the absence of rhodium treatment, whereas growth of the HCT116N cell line slows slightly as the density of the culture increases. Furthermore, the HCT116O cell line also shows a significant increase in the G2/M population as well as the G1 population.
Figure 4.
Cell cycle distribution assay. HCT116N (green) and HCT116O (red) cells were treated with 20 μM [Rh(HDPA)2chrysi]3+ for 24 or 48 hours. After fixation and staining with PI, cells were analyzed by flow cytometry. The raw distributions were analyzed for cell cycle phase using commercially available software. Upon rhodium treatment, the S-phase population is depleted, with a concomitant increase in the G1-phase population.
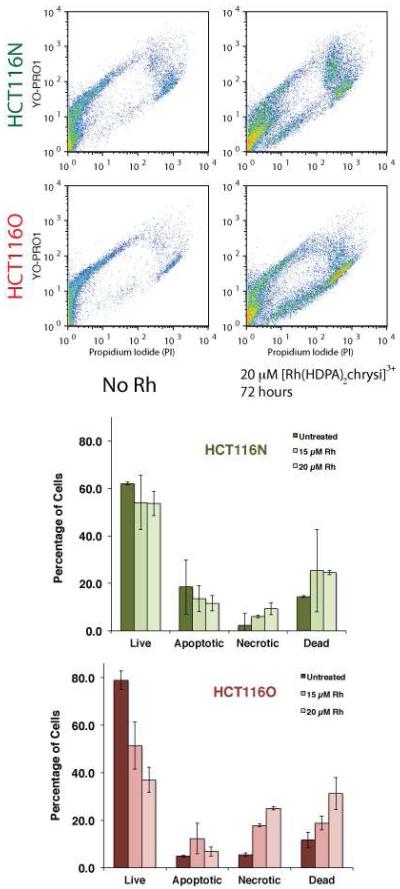
Mode of cell death
To characterize the cell death occurring in response to rhodium treatment, a dye exclusion flow cytometry assay was employed.41 The assay differentiates between live cells, dead cells, and cells undergoing apoptosis or necrosis through concurrent staining with propidium iodide (a dead cell permeable dye) and YO-PRO-1 (an apoptotic cell permeable dye). By plotting the fluorescence of the YO-PRO-1 channel against the PI channel, a pattern emerges. Healthy cells are seen in the lower lefthand corner of the plot. Apoptotic cells exhibit higher YO-PRO-1 fluorescence, but still exclude propidium iodide, placing them in the upper lefthand quadrant of the pattern. Dead cells admit both dyes and are therefore seen in the upper righthand quadrant of the image. Upon flow cytometry analysis, cells can be classified as live, apoptotic, necrotic, or dead by defining regions in the fluorescence plane corresponding to each category.
The HCT116N and HCT116O cell lines were incubated with 0–25 μM of [Rh(HDPA)2chrysi]3+ for 24–72 hours. After harvesting the cells and staining with both PI and YO-PRO-1, the cells were analyzed by flow cytometry to obtain raw fluorescence data. Representative data for 20 μM rhodium treatment for 72 hours are shown in Figure 5. YO-PRO-1 fluorescence is shown on the y-axis, and PI fluorescence is shown on the x-axis. The color scale represents the number of cells, with blue indicating fewer cells at a given pair of fluorescence levels, and orange representing a greater number of cells at a given pair of fluorescence levels. The raw data were analyzed by gating the fluorescence events into one of four categories, depending on the fluorescence levels of the two dyes. Figure 5 also shows histograms of live, apoptotic, necrotic, and dead cells for the HCT116N and HCT116O cell lines based on the flow cytometry. Rhodium treatment was either 15 or 20 μM [Rh(HDPA)2chrysi]3+ for 72 hours. As before, rhodium treatment alone induces necrosis preferentially in the MMR-deficient HCT116O cell line; there is no significant change in the percentage of cells in the apoptotic region in either cell line. The effect is significantly more pronounced in the MMR-deficient HCT116O cell line, which drops from 79 ± 3.8 % to 37 ± 5.3 % after treatment with 20 μM [Rh(HDPA)2chrysi]3+, versus the MMR-proficient HCT116N cell line, which shows a minimal decrease in live cells from 62 ± 0.6 % to 54 ± 5.1 % after treatment with 20 μM [Rh(HDPA)2chrysi]3+.
Figure 5.
Flow cytometry assay of cell death. (Top) HCT116N and HCT116O cells were treated with 20 μM [Rh(HDPA)2chrysi]3+ for 72 hours. Rhodium treatment causes cells to move away from the origin, along the necrotic pathway (lower branch of pattern). The effect is more pronounced in the HCT116O cell line. (Bottom) HCT116N and HCT116O cells were treated with 15 μM or 20 μM [Rh(HDPA)2chrysi]3+ for 24 – 72 hours. Rhodium treatment causes a sharp decrease in the live population of the HCT116O cell line with a corresponding increase in the necrotic and dead cell populations. Minimal effect is seen in the HCT116N cell line. Thus [Rh(HDPA)2chrysi]3+ preferentially induces necrosis in the MMR-deficient HCT116O cell line.
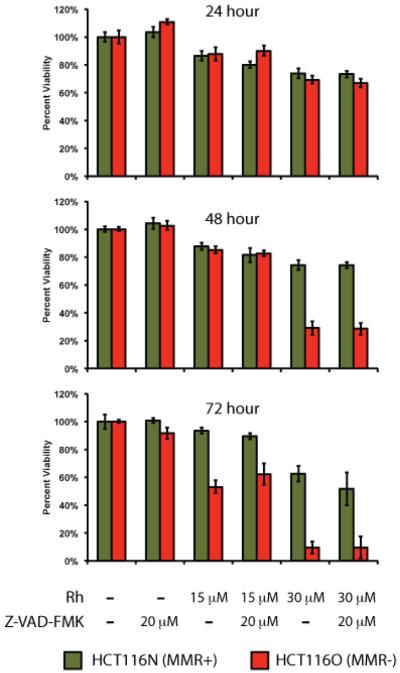
Caspase inhibition
As a complement to the dye exclusion flow cytometry assay, the MTT cytotoxicity assay was repeated in the absence and presence of the pan-caspase inhibitor Z-VAD-FMK.42 This inhibitor works by irreversibly binding to the active site of caspases. As before, the HCT116N and HCT116O cell lines were treated with 0 – 30 μM of the [Rh(HDPA)2chrysi]3+ complex for 24 – 72 hours. In addition, each treatment was also combined with the inhibitor at a final concentration of 20 μM (Figure 6). As before, the rhodium complex exhibited selective toxicity in the repair-deficient HCT116O cell line, with cell viability dropping to 9.7 ± 4.4 % after treatment with 30 μM metal complex for 72 hours, versus 63 ± 5.7 % viability in the repair-proficient HCT116N cell line. Addition of the caspase inhibitor at 20 μM offered no protection from rhodium to the HCT116N cell line (63 ± 5.7 % without inhibitor, 52 ± 9.8 % with inhibitor) or to the HCT116O cell line (9.7 ± 4.4 % without inhibitor, 9.8 ± 7.8 % with inhibitor). At a final concentration of 40 μM, the caspase inhibitor provided some protection from rhodium to the HCT116O cell line (16 ± 10 % without inhibitor, 28 ± 3.7 % with inhibitor), but this difference was small in relation to the differential between the HCT116N and HCT116O cell lines and roughly within error (data not shown).
Figure 6.
Caspase inhibition assay. HCT116N and HCT116O cells were plated in 96-well format at densities of 5 × 104 cells/well and treated with 0 – 30 μM of [Rh(HDPA)2chrysi]3+ with or without 20 μM of the pan-caspase inhibitor Z-VAD-FMK. After 24 – 72 hours, the cells were labeled with MTT for 4 hours. The resulting formazan crystals were dissolved by addition of 10% SDS acidified with 10 mM HCl, and absorbance was measured at 570 nm. The caspase inhibitor confers no protection from rhodium-induced toxicity in either cell line.
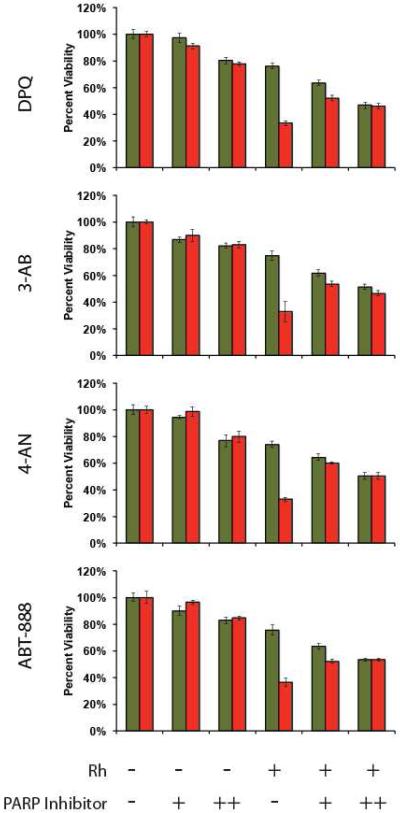
PARP inhibition
The MTT cytotoxicity assay was also repeated in conjunction with a panel of poly-ADP ribose polymerase (PARP) inhibitors: DPQ, 3-AB, 4-AN, and ABT-888.43-46 As before, the HCT116N and HCT116O cell lines were treated with 0 or 20 μM of the [Rh(HDPA)2chrysi]3+ complex for 72 hours, with or without one of the four inhibitors. In each case, treatment with the inhibitor completely abolished the selective MMR-dependent effects of the rhodium compound (Figure 7), as determined by the difference between the percentage of viable cells in the HCT116N cell line and the percentage of viable cells in the HCT116O cell line. For example, in the case of the compound DPQ, this difference was 43 ± 2.7 % without inhibitor and 0.6 ± 3.0 % with inhibitor. Similar results are seen with each of the other three compounds as well; taken together, these data implicate PARP in the MMR-dependent response to [Rh(HDPA)2chrysi]3+.
Figure 7.
PARP inhibition assay. HCT116N and HCT116O cells were plated in 96-well format at densities of 5 × 104 cells/well and treated with 0 μM (“−”) or 20 μM (“+”) of [Rh(HDPA)2chrysi]3+ with or without PARP inhibitor (“-” = 0 μM DPQ, 3-AB, 4-AN, or ABT-888; “+” = 25 μM DPQ, 2 mM 3-AB, 10 μM 4-AN, or 5 μM ABT-888; “++” = 50 μM DPQ, 3 mM 3-AB, 20 μM 4-AN, or 10 μM ABT-888). After 72 hours, the cells were labeled with MTT for 4 hours. The resulting formazan crystals were dissolved by addition of 10% SDS acidified with 10 mM HCl, and absorbance was measured at 570 nm. All four PARP inhibitors confer protection from rhodium-induced MMR-dependent toxicity, reducing the differential activity significantly.
DISCUSSION
The direct correlation between the binding affinity of rhodium metalloinsertors for DNA mismatches and the differential in their activity between the HCT116N and HCT116O cell lines fails to predict the high selective activity of [Rh(HDPA)2chrysi]3+ . In light of the shorter incubations required for this complex, it seemed likely that accelerated cellular uptake is a contributing factor in the increased cellular response. As [Rh(HDPA)2chrysi]3+ is not luminescent, another method was needed to examine its cellular uptake. The high atomic mass and monoisotopic distribution of rhodium make the cellular accumulation of rhodium metalloinsertors well suited to analysis by inductively coupled plasma – mass spectrometry (ICP-MS).47-49 ICP-MS affords a direct comparison of the cellular accumulation of a series of rhodium metalloinsertors after treatments similar to those applied in ELISA assays for activity. The highest levels of cellular accumulation are clearly seen for the complex bearing the HDPA ligand, which was expected in light of the previous observation that [Rh(HDPA)2chrysi]3+ exhibits increased anti-proliferative activity against the HCT116 cell lines at shorter incubation times than other rhodium metalloinsertors. Significant activity is seen with [Rh(HDPA)2chrysi]3+ in as little as 12 hours, while [Rh(bpy)2chrysi]3+ and [Rh(NH3)4chrysi]3+ display no activity at this incubation time.
Rhodium metalloinsertors have shown differential anti-proliferative activity in an ELISA assay for DNA synthesis. This assay directly reports on the amount of BrdU label incorporated during DNA replication, and as such, does not distinguish between cells that are viable but not replicating, e.g. G0 cells that have exited the cell cycle, and cells that are inviable, or dead. Therefore, this assay can be used to determine inhibitory, but not cytotoxic activity. In contrast, the MTT assay reports directly on cell viability as measured by metabolic activity, with the action of mitochondrial reductases catalyzing the cleavage of the labeling agent MTT. Here cells that are viable still produce signal, i.e. formazan absorbance, whether or not they are actively dividing. Thus, this assay can distinguish between senescence and true cell death, and the effects observed in response to rhodium treatment are truly cytotoxic. Importantly, the concentration ranges and incubation times of the treatments applied in the MTT assays for [Rh(HDPA)2chrysi]3+ and the closely related complex [Rh(MeDPA)2chrysi]3+ (0 – 25 μM, 24 – 72 hours) are identical to those that inhibit DNA synthesis as seen by ELISA.
Accordingly, the result that [Rh(HDPA)2chrysi]3+ and [Rh(MeDPA)23+chrysi] trigger cell death selectively in the MMR-deficient HCT116O cell line versus the MMR-proficient HCT116N cell line as measured by MTT assay represents a significant advance in the development of these complexes as anti-cancer agents; clearly, these agents are more potent than previously considered. Although [Rh(bpy)2chrysi]3+ does not appear to be selectively toxic at these concentrations, it is likely that this is due to differences in the kinetics of cellular uptake, rather than fundamental differences in its mode of action as compared to [Rh(HDPA)2chrysi]3+, since both bind DNA mismatches with equal affinity.36 For either activity assay, the complex must first accumulate within the cell, and then cellular response must be triggered. While [Rh(HDPA)2chrysi]3+ displays activity in ELISA after 12 h incubations, [Rh(bpy)2chrysi]3+ requires 48 h or more to show significant differential activity. In light of the higher levels of rhodium accumulation seen by ICP-MS after treatment with the HDPA complex versus the bpy complex, it seems likely that accumulation of the bpy complex is delayed by ~36 h relative to the HDPA complex. Cellular responses that occur quickly after accumulation, such as the inhibition of DNA synthesis, can still be observed within the 72 h timeframe of the ELISA assay, and both complexes display activity. However, a lag time will exist between the inhibition of DNA synthesis and the onset of cell death, and when combined with the slow uptake of [Rh(bpy)2chrysi]3+, (t > 48 h), cell death is delayed accordingly and cannot be observed within the timeframe of the MTT assay (also 72 h).
The fact that the [Rh(MeDPA)2chrysi]3+ complex displays differential toxicity against the HCT116O cell lines comparable to that of the [Rh(HDPA)2chrysi]3+ complex suggests that it shares the accelerated uptake of the HDPA complex. This was certainly expected, as the complexes are almost identical, but does address the question of uptake mechanism, and clearly refutes the hypothesis that accelerated uptake requires a hydrogen bonding interaction with the bridging secondary amine of the HDPA ligand. More importantly, this suggests that selective toxicity as a function of MMR-competency is a general property of rhodium metalloinsertors with dipyridylamine ancillary ligands, and establishes these ligands as the basis for the development of the next generation of complexes.
Flow cytometry analysis reveals that cell death is preceded by disruption of the cell cycle. Treatment with 20 μM [Rh(HDPA)2chrysi]3+ for 24 hours leads to a marked depletion of the S-phase population with a concomitant increase in the G1 population. These data suggest that the G1/S-phase DNA damage checkpoint may be activated in response to rhodium treatment. In the case of the HCT116O line, the G2/M-phase population also increases in response to rhodium treatment. This could represent a secondary checkpoint activation occurring at the G2/M-phase transition, and might signal a “two-alarm fire” that accompanies the enhanced activity against this cell line.
The biochemical events associated with apoptosis have been extensively studied.50 An extensive network of regulatory proteins controls the initiation of this process in response to both internal and external signals.51 Upon activation, the cascade of initiator and effector caspases cleaves a variety of substrates to bring about the morphological changes associated with this mode of cell death, including nuclear condensation and fragmentation, plasma membrane blebbing, decomposition of the cell into apoptotic bodies, and ultimately, the engulfment of these bodies by neighboring cells through phagocytosis.52 Importantly, apoptotic cells retain their membrane integrity until the very last stages of this process, preventing release of cytokines and thus avoiding inflammation. By comparison, both the causes and the progression of necrosis are much less defined at the molecular level, and this mode of cell death is most frequently characterized by morphological criteria.52-54 Perhaps the most reliable marker of necrosis then, is the early rupture of the plasma membrane, in direct contrast to apoptosis. These differences in membrane integrity enable the facile characterization of cell death by flow cytometry. The admission of the dead cell stain propidium iodide by the HCT116 cell lines upon rhodium treatment reveals that cell death proceeds through a necrotic, rather than apoptotic pathway. This conclusion is supported by the observation that the caspase inhibitor Z-VAD-FMK is unable to block rhodium-induced toxicity in the MTT assay, indicating that death occurs in a caspase-independent fashion.
It is interesting to consider that rhodium metalloinsertors selectively induce necrosis in MMR-deficient cells. Traditionally, necrotic cell death has been considered to be “accidental”, occurring mainly in response to non-physiological insults.55,56 More recently however, the notion that necrosis may in fact be an ancestral mode of programmed cell death has gained attention in the literature and community of cell death research.52,53,57 Hitomi et al screened an siRNA library covering the mouse genome and identified 432 gene knockdowns that blocked the induction of necrosis.58 Chan et al found anti-necrotic proteins encoded in the genomes of several viruses.59 While these studies present some of the most compelling evidence for the notion that necrosis is in fact a regulated process, several other groups are also working to provide a molecular definition for the process as has been done for apoptosis.56,57
Thompson and co-workers have reported that the DNA repair protein poly ADP-ribose polymerase-1 (PARP-1) mediates the induction of necrosis in response to DNA damage by the alkylator MNNG.60 Upon activation at sites of DNA damage, PARP-1 covalently modifies itself with long chains of ADP-ribose polymers in order to recruit downstream components of the repair machinery.61 As a result, PARP-1 along with PARP-2 has attracted much attention as a therapeutic target recently, and several specific and potent inhibitors of the enzyme are currently in clinical trials.62 Here, treatment with a panel of PARP-1 inhibitors confers protection from MMR-dependent toxicity, abolishing differential activity between the HCT116 cell lines by Rh. This result strongly suggests that PARP activation is required for the differential biological effects of rhodium metalloinsertors. Importantly, the fact that each of the four PARP inhibitors on the panel abolishes differential activity argues against the notion that off-target effects of any particular inhibitor are responsible for blocking rhodium activity. Although the role of PARP in regulating necrosis is still emerging, the requirement for PARP activation is consistent with the notion that these rhodium complexes elicit a damage response upon binding to DNA mismatches. This is supported by flow cytometry analysis that reveals cell death is preceded by disruption of the cell cycle. Upon rhodium treatment, there is a sharp decline in the population of cells that are actively synthesizing DNA. This observation suggests that the G1/S-phase DNA damage checkpoint is activated.
As necrosis triggers a pro-inflammatory response by releasing cytokines, selective induction of necrosis in cancerous tissues may be a way to activate the immune system against cancer cells and ultimately improve efficacy of a therapeutic agent.63 One accepted trigger of necrosis is bioenergetic catastrophe, i.e. severe ATP depletion.55,56 Rhodium metalloinsertors might trigger such a catastrophe by targeting mitochondrial DNA. Previously, it was thought that damaged mitochondrial genomes simply would be degraded and replaced by the replication of undamaged DNA.64 However, emerging research has uncovered DNA repair processes in mitochondria, including mismatch repair capability.64,65 As lipophilic cations, rhodium metalloinsertors likely accumulate in mitochondria66-71, as we have seen with analogous ruthenium complexes in our laboratory.33 If mitochondrial DNA mismatches are in fact a target for rhodium metalloinsertors, then the resultant disruption of mitochondrial function would lead to energy depletion and necrosis. Future mechanistic work must explore the biochemical consequences of mismatch binding in nuclear DNA versus mitochondrial DNA. It should be noted that these mechanisms are not mutually exclusive; as such, both might contribute to the biological response to rhodium metalloinsertors.
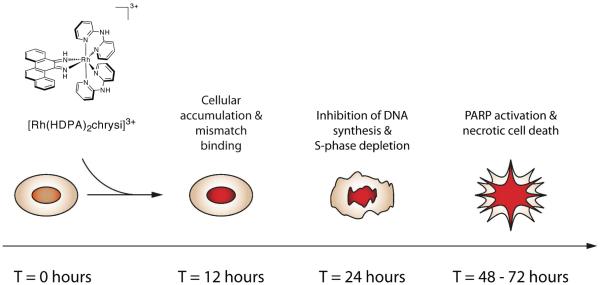
These studies support the notion that rhodium metalloinsertors bearing HDPA ligands benefit from increased cellular accumulation, and thus provide an explanation for the observation that [Rh(HDPA)2chrysi]3+ exceeds the activity predicted by its binding affinity for DNA mismatches. This increased uptake allows us to observe additional cellular responses to these agents, and, as a whole, a picture of the biological response to rhodium metalloinsertors, exemplified by [Rh(HDPA)2chrysi]3+ in particular, is beginning to emerge (Figure 8). Over the course of the first 12 hours, the rhodium complex accumulates in cells, binding to either mitochondrial or genomic DNA mismatches. Within 24 hours, DNA synthesis is inhibited, and cells accumulate in G1-phase. Over the next 24–48 hours, the DNA damage response is likely activated, and ultimately leads to cell death by a caspase-independent, necrotic mechanism. These biological effects are more pronounced at each stage of the response in the MMR-deficient HCT116O cell line relative to the MMR-proficient HCT116N cell line, strongly suggesting that DNA mismatches are in fact the cellular target of rhodium metalloinsertors.
Figure 8.
Model for the cellular response to rhodium metalloinsertors.
This work shows that dipyridylamine ancillary ligands serve to accelerate the cellular uptake of rhodium metalloinsertors, triggering a selective cytotoxic effect as a function of MMR status and forming the foundation for the next generation of complex development. This new class of agents is significantly more potent than previously considered, and the work begun here on understanding their mechanism of action advances their development as novel anti-cancer agents.
Footnotes
Financial support for this work from the NIH (GM33309) is gratefully acknowledged. We also thank Amgen and the Parsons foundation for fellowship support to R.J.E. and NSF for a predoctoral fellowship to A.C.K.
REFERENCES
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 3.Bhattacharya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc. Natl. Acad. Sci. USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss BS. Frameshift mutation, microsatellites and mismatch repair. Mutation Res. 1999;437:195–203. doi: 10.1016/s1383-5742(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Human Mutation. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Human Mol. Gen. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 7.Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. European J. of Surgical Oncology. 2003;29:201–212. doi: 10.1053/ejso.2002.1399. [DOI] [PubMed] [Google Scholar]
- 8.Arzimanoglou II, Gilbert F, Barber HRK. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 9.Pors K, Patterson LH. DNA mismatch repair deficiency, resistance to cancer chemotherapy and the development of hypersensitive agents. Current Topics in Med. Chem. 2005;5:1133–1149. doi: 10.2174/156802605774370883. [DOI] [PubMed] [Google Scholar]
- 10.Valentini AM, Armentano M, Pirrelli M, Caruso ML. Chemotherapeutic agents for colorectal cancer with a defective mismatch repair system: the state of the art. Cancer Treatment Reviews. 2006;32:607–618. doi: 10.1016/j.ctrv.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Carethers JM, Hawn MT, Chauhan DP, Luce MC, Marra G, Koi M, Boland CR. Competency in mismatch repair prohibits clonal expansion of cancer cells treated with N-methyl-N’-nitro-N-nitrosoguanidine. J. of Clinical Investigation. 1996;98:199–206. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 13.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aebi S, Fink D, Gordon R, Kim HK, Zheng H, Fink JL, Howell SB. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clinical Cancer Res. 1997;3:1763–1767. [PubMed] [Google Scholar]
- 15.Fedier A, Schwarz VA, Walt H, Carpini RD, Haller U, Fink D. Resistance to topoisomerase poisons due to loss of DNA mismatch repair. International J. of Cancer. 2001;93:571–576. doi: 10.1002/ijc.1356. [DOI] [PubMed] [Google Scholar]
- 16.Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clinical Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 17.Karran P, Offman J, Bignami M. Human mismatch repair, drug-induced DNA damage, and secondary cancer. Biochimie. 2003;85:1149–1160. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J. Am. Chem. Soc. 1997;119:12986–12987. [Google Scholar]
- 19.Jackson BA, Barton JK. Recognition of base mismatches in DNA by 5,6-chrysenequinone diimine complexes of rhodium(III): a proposed mechanism for preferential binding in destabilized regions of the double helix. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 20.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch I, Barton JK. A rhodium(III) complex for high-affinity DNA base-pair mismatch recognition. Proc. Natl. Acad. Sci. USA. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeglis BM, Pierre VP, Barton JK. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007;44:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeglis BM, Barton JK. DNA base mismatch detection with bulky rhodium intercalators: synthesis and applications. Nature Protocols. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 24.Brown T, Hunter WN, Kneale G, Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc. Natl. Acad. Sci. USA. 1996;83:2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter WN, Brown T, Kennard O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Research. 1987;15:6589–6606. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter WN, Brown T, Kneale G, Anand NN, Rabinovich D, Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-Å resolution. J. of Biological Chemistry. 1987;262:9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 27.Skelly JV, Edwards KJ, Jenkins TC, Neidle S. Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl. Acad. Sci. USA. 1993;90:804–808. doi: 10.1073/pnas.90.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drew HR, Wing RM, Takano R, Broa C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl. Acad. Sci. USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Structure of a photoactive rhodium complex intercalated into DNA. Nature Structural Biology. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 31.Cordier C, Pierre VC, Barton JK. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: an NMR solution study. J. Am. Chem. Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode. Biochemistry. 2009;20:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puckett CA, Barton JK. Methods to explore cellular uptake of ruthenium complexes. J. Am. Chem. Soc. 2007;129:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puckett CA, Barton JK. Mechanism of cellular uptake of a ruthenium polypyridyl complex. Biochemistry. 2008;47:11711–11716. doi: 10.1021/bi800856t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. DNA mismatch-specific targeting and hypersensitivity of mismatch-repair-deficient cells to bulky rhodium(III) intercalators. Proc. Natl. Acad. Sci. USA. 2006;103(42):15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst RJ, Song HG, Barton JK. DNA mismatch binding and antiproliferative activity of rhodium metalloinsertors. J. Am. Chem. Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N’-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 38.Reitmar AH, Risley R, Bristow RG, Wilson T, Ganesh A, Jang A, Peacock J, Benchimol S, Hill RP, et al. Mutator phenotype in Msh2-deficient murine embryonic fibroblasts. Cancer Res. 1997;57:3765–3771. [PubMed] [Google Scholar]
- 39.Gratzner HG. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 40.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 41.Idziorek T, Estaquier J, DeBels F, Ameisen J-C. YOPRO-1 permits cytofluorometric analysis of programmed cell death (apoptosis) without interfering with cell viability. J. Immunol. Methods. 1995;185:249–258. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 42.Vandenabeele P, Vanden Berghe T, Festjens N. Caspase inhibitors promote alternative death pathways. Science STKE. 2006;358:pe44. doi: 10.1126/stke.3582006pe44. [DOI] [PubMed] [Google Scholar]
- 43.Costantino G, Macchiarulo A, Camaioni E, Pellicciari R. Modeling of poly(ADP-ribose)polymerase (PARP) inhibitors. Docking of ligands and quantitative structure-activity relationship analysis. J. Med. Chem. 2001;44:3786–3794. doi: 10.1021/jm010116l. [DOI] [PubMed] [Google Scholar]
- 44.Purnell MR, Whish WJD. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem. J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J. Biol. Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 46.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LW, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 47.Egger E, Rappel C, Jakupec MA, Hartinger CG, Heffeter P, Keppler BK. Development of an experimental protocol for uptake studies of metal compounds in adherent tumor cells. J. Anal. At. Spectrom. 2009;24:51–61. doi: 10.1039/B810481F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghezzi A, Aceto M, Cassino C, Gabano E, Osella D. Uptake of antitumor platinum(II) complexes by cancer cells, assayed by inductively coupled plasma mass spectrometry (ICP-MS) J. Inorg. Biochem. 2004;98:73–78. doi: 10.1016/j.jinorgbio.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 49.Kirin SI, Ott I, Gust R, Mier W, Weyhermüller T, Metzler-Nolte N. Cellular uptake quantification of metalated peptide and peptide nuclei acid bioconjugates by atomic absorption spectroscopy. Angew. Chem. Int. Ed. 2008;47:955–959. doi: 10.1002/anie.200703994. [DOI] [PubMed] [Google Scholar]
- 50.Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 52.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr. Op. in Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, Zhivotovsky B, Blagosklonny MV, Malorni W, Knight RA, Piacentini M, Nagata S, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death and Differentiation. 2005;12:1463–1467. doi: 10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 54.Kroemer G, Galuzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death and Differentiation. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends in Biochemical Sciences. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 56.McCall K. Genetic control of necrosis- another type of programmed cell death. Curr. Op. Cell Biology. 2010;22:882–888. doi: 10.1016/j.ceb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galluzzi L, Kroemer G. Genetic control of necrosis - another type of programmed cell death. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J of Biol. Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 60.Zong WX, Ditsworth D, Bauer DE, Wang Z-Q, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes and Development. 2004;18:1272–1282. doi: 10.1101/gad.1199904. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. and Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 62.Annunziata CM, O’Shaughnessy J. Poly (ADP-ribose) polymerase as a novel therapeutic target in cancer. Clin. Cancer Res. 2010;16:4517–4526. doi: 10.1158/1078-0432.CCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ricci MS, Zong WX. Chemotherapeutic approaches for targeting cell death pathways. The Oncologist. 2006;11:342–357. doi: 10.1634/theoncologist.11-4-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochimica et Biophysica Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 65.de Souze-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Napolitano SM, Aprille JR. Delocalized lipophilic cations selectively target the mitochondria of carcinoma cells. Adv. Drug Delivery Rev. 2001;49:63–70. doi: 10.1016/s0169-409x(01)00125-9. [DOI] [PubMed] [Google Scholar]
- 67.Murphy MP, Smith RAJ. Drug delivery to mitochondria: the key to mitochondrial medicine. Adv. Drug Delivery Rev. 2000;41:235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 68.Rackham O, Nichols SJ, Leedman PJ, Berners-Price SJ, Filipovska A. A gold(I) phosphine complex selectively induces apoptosis in breast cancer cells: Implications for anticancer therapeutics targeted to mitochondria. Biochem. Pharmacol. 2007;74:992–1002. doi: 10.1016/j.bcp.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 69.Liu JJ, Galettis P, Farr A, Maharaj L, Samarasinha H, McGechan AC, Baguley BC, Bowen RJ, Berners-Price SJ, McKeage MJ. In vitro antitumor and hepatotoxicity profiles of Au(I) and Ag(I) bidentate pyridyl phosphine complexes and relationships to cellular uptake. J. Inorg. Biochem. 2008;102:303–310. doi: 10.1016/j.jinorgbio.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Liberman EA, Topali VP, Tsofina LM, Jasaitis AA, Skulachev VP. Mechanism of coupling of oxidative phosphorylation and the membrane potential of mitochondria. Nature. 1969;222:1076–1078. doi: 10.1038/2221076a0. [DOI] [PubMed] [Google Scholar]
- 71.Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. U.S.A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]