Tumor metastasis is the primary cause of death in cancer patients, but the molecular mechanisms driving the evolution of the phenotype toward a specific organ is one of its less understood aspects. The scaffolding protein NHERF1 reprograms the metastatic phenotype and organotropism via the differential function of its PDZ domains.
Abstract
Metastatic cells are highly plastic for differential expression of tumor phenotype hallmarks and metastatic organotropism. The signaling proteins orchestrating the shift of one cell phenotype and organ pattern to another are little known. Na+/H+ exchanger regulatory factor (NHERF1) is a molecular pathway organizer, PDZ-domain protein that recruits membrane, cytoplasmic, and cytoskeletal signaling proteins into functional complexes. To gain insight into the role of NHERF1 in metastatic progression, we stably transfected a metastatic breast cell line, MDA-MB-231, with an empty vector, with wild-type NHERF1, or with NHERF1 mutated in either the PDZ1- or PDZ2-binding domains to block their binding activities. We observed that NHERF1 differentially regulates the expression of two phenotypic programs through its PDZ domains, and these programs form the mechanistic basis for metastatic organotropism. The PDZ2 domain promotes visceral metastases via increased invadopodia-dependent invasion and anchorage-independent growth, as well as by inhibition of apoptosis, whereas the PDZ1 domain promotes bone metastases by stimulating podosome nucleation, motility, neoangiogenesis, vasculogenic mimicry, and osteoclastogenesis in the absence of increased growth or invasion. Collectively, these findings identify NHERF1 as an important signaling nexus for coordinating cell structure with metastatic behavior and identifies the “mesenchymal-to-vasculogenic” phenotypic transition as an essential step in metastatic progression.
INTRODUCTION
Metastatic disease is the primary cause of death among cancer patients, remaining largely unmanageable due to a poor understanding of its underlying molecular mechanisms. Although the biological properties of cancer cells to progress toward overt metastases (i.e., local invasion, intravasation, survival in circulation, extravasation, and organ colonization) have been identified (Geiger and Peeper, 2009), some fundamental aspects are unresolved. Recent data suggest that specific tumor cells might be imprinted intrinsically ab initio with metastatic competence but that the final metastatic organotropism (i.e., specific organ colonization) might depend on additional independent, organ-specific gene signatures (Gupta and Massague, 2006; Chaffer and Weinberg 2011). However, although clear organospecific metastasis signatures have been reported (Hunter and Alsarraj, 2009), the mechanisms underlying these processes are unknown. Indeed, the molecular signaling systems orchestrating both the determination of a particular malignant phenotype and a specific pattern of selected organs are poorly understood.
The emerging vision of signal transduction is that the building of the signaling complexes and the “controlled” shift from one signal complex to another during neoplastic progression are finely coordinated by a class of proteins called scaffolding proteins (Jin et al., 2009). These proteins create tightly controlled complexes integrating diverse signal functions from distinct receptors or microenvironmental conditions into specific phenotypic outcomes. Changes in the scaffolding protein concentrations, cellular localization, and/or binding specificities of their consensus sequence can radically alter the composition of the signaling modules and, as a consequence, the final biochemical/physiological phenotype. This ability to forge new interactions and reprogram cellular behavior could be a strategy adapted by tumor cells to subvert the cellular machinery regulating the normal constraints on cell growth, survival, migration, and invasion.
Recent studies identified a central role for the scaffolding protein Na+/H+ exchanger regulatory factor (NHERF1, EBP50, SLC9A3R1) in multiple malignancies. NHERF1 contains two tandem PDZ domains and recruits membrane receptors/transporters, cytoplasmic signaling proteins, and transcriptional coactivators into functional complexes that regulate many processes (Shenolikar et al., 2004), including cell proliferation (Pan et al., 2006), survival (Molina et al., 2012), apoptosis (Zheng et al., 2010), and migration and invasion (Cardone et al., 2007; Kislin et al., 2009), and is overexpressed in diverse cancers, where it correlates with aggressive stage and poor prognosis (Cardone et al., 2007; Song et al., 2007; Georgescu et al., 2008; Bellizzi et al., 2010; Tabrizi et al. 2010; Karn et al., 2011). However, its effective role and, in particular, its mechanism and the specific roles of the two PDZ domains in regulating cancer development and metastatic progression are undefined.
To gain further insights into how NHERF1 regulates a range of neoplastic phenotypes and metastatic organotropism, we stably transfected the most commonly used cell line to study metastasis, the human metastatic breast cell line MDA-MB-231, with 1) the pcDNA 3.1/Higro empty vector, 2) wild-type (WT) NHERF1, or NHERF1 mutated in 3) the PDZ1 domain (PDZ1mut) or 4) the PDZ2 (PDZ2mut) domain (Supplemental Figure S1). The distinct metastatic properties of four cell lines that share a very closely related genetic background allowed us to comparatively investigate the contribution of NHERF1 to tumor growth and metastasis.
RESULTS
NHERF1 regulates anchorage-independent growth and xenograft tumorigenicity primarily through the PDZ1 domain
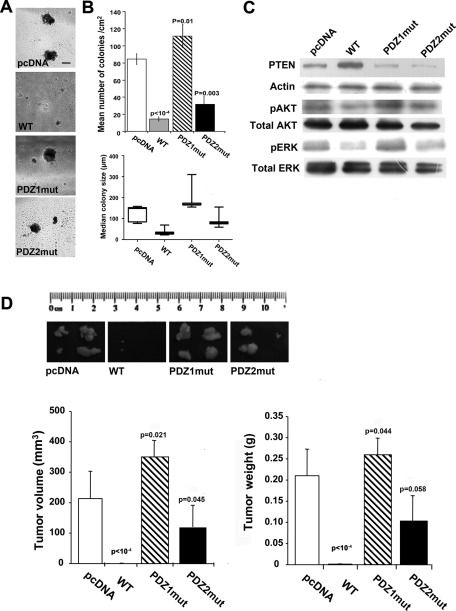
The ability of cells to grow in an anchorage-independent manner in semisolid media is often regarded as one of the hallmarks of tumorigenicity and metastatic potential (Fiebig et al., 2004). We tested our panel of MDA-MB-231 breast cancer cells for their ability to grow in soft agar (Figures 1, A and B), the signaling pathways involved (Figure 1C), and their tumorigenic potential in nude mice (Figure 1D). When only colonies >50 μm in diameter were scored (Figure 1B, top), the WT cells, which overexpress WT NHERF1, formed very few colonies compared with the parental, pcDNA cells, suggesting that NHERF1 overexpression suppresses anchorage-independent growth, whereas cells overexpressing the mutated PDZ1 domain (PDZ1mut) formed more colonies, and cells expressing the mutated PDZ2 domain (PDZ2mut) formed an intermediate number of colonies. The relative median size of the soft-agar colonies (Figure 1B, bottom) displayed significant differences (p < 0.05 in one-way analysis of variance [ANOVA], Kruskal–Wallis test); PDZ1mut had the largest median colony size, followed by the pcDNA and the PDZ2mut cells. Moreover, whereas the WT cells did not form large colonies (Figure 1A), they formed a large number of microcolonies that were <50 μm (Figure 1B, bottom). Taken together, these data indicate that the loss of anchorage-independent growth by overexpression of WT NHERF1 occurs primarily through the PDZ1 domain. NHERF1 has been shown to regulate survival through the PTEN-dependent inhibition of PI3K-phospho-AKT–mediated signals (Molina et al., 2012) and proliferation through the ERK pathway (Zheng et al., 2010). To examine whether these signaling pathways are differentially modulated in the four cell lines, we analyzed the level of PTEN and the phosphorylation status of ERK and AKT by Western blotting (Figure 1C). Indeed, NHERF1-WT–overexpressing cells had higher expression levels of PTEN, whereas both PDZ1-mutated (PDZ1mut) and PDZ2-mutated (PDZ2mut) cells had slightly lower levels of PTEN than the parental, pcDNA cells. In line with these data, WT-NHERF1–overexpressing cells showed a reduced level of p-AKT compared with pcDNA cells, whereas the level of p-AKT was markedly higher in PDZ1-mutated (PDZ1mut) cells and little changed in PDZ2-mutated cells (PDZ2mut cells) in comparison with that of pcDNA cells. Total AKT levels remained unchanged in the four cell lines. Last, NHERF1-WT–overexpressing cells had greatly reduced p-ERK respect to pcDNA cells, whereas PDZ1mut cells expressed much higher p-ERK levels and PDZ2mut cells a less pronounced increase in p-ERK compared with pcDNA cells. These results confirm that NHERF1 growth-suppression function requires a functional PDZ1 domain to suppress the PTEN/AKT-mediated survival signal and increase the p-ERK–driven proliferation pathway.
FIGURE 1:
Increased expression of NHERF1 impairs tumor growth mainly via the PDZ1 domain. (A–C) MDA-MB-231 cells transfected with empty vector (pcDNA), WT NHERF1 (WT), and the PDZ1 (PDZ1mut) and PDZ2 (PDZ2mut) mutated constructs were plated onto soft agar. After 4 wk, colonies were counted, and colony size was assessed by micrometer. (A) Representative photomicrographs of colonies for each cell variant. Results are represented as mean ± SEM; the values of p are vs. pcDNA cells. (B) Top, number of colonies measuring >50 μm; bottom, median size of colonies, with the bars representing the minimum and maximum values for each clone. (C) Western blot for the levels of PTEN and of phospho-ERK and phospho-AKT kinases using the total and phospho-specific antibodies listed in Materials and Methods. (D) In vivo tumor growth. Female BALB/c-nu/nu mice were subcutaneously injected with cell variants as indicated. Top, photographs of four tumors per cell variant at the day of dissection. Bottom, tumor volume and weight measured at 32 d postinjection; n = 4 mice/group; results are represented as mean ± SEM; the values of p are vs. pcDNA-injected animals.
The in vivo xenograft tumor formation ability of the cells was then examined by subcutaneous cell injection in the right flank of female Balb/c nu/nu mice (Figure 1D). It closely followed the pattern observed in soft agar. WT cell–induced tumors had both the lowest incidence (50%) and the smallest size, whereas pcDNA and PDZ1mut cells induced both 100% incidence and the biggest tumors. An intermediate condition was observed in mice injected with PDZ2mut cells, with a 75% tumor incidence and a slightly smaller tumor size.
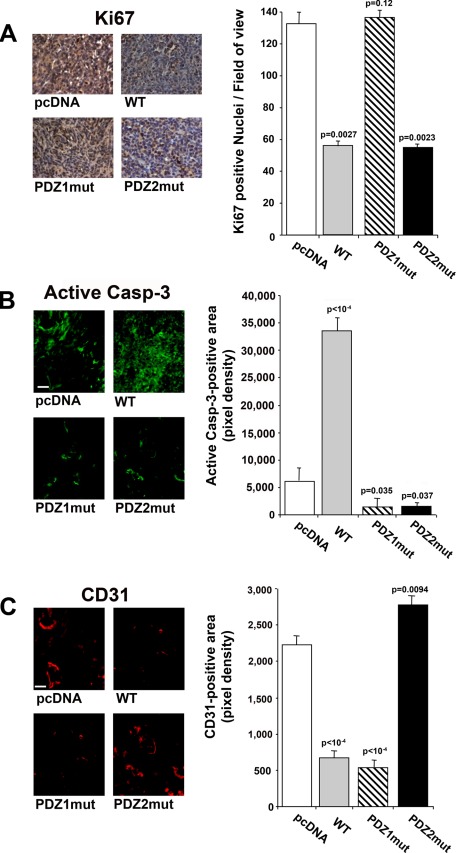
Analysis of the xenograft tumors revealed a significant decrease in Ki-67 expression in tumors derived from WT and HRF2 cells relative to tumors from parental or PDZ1mut cells (Figure 2A), confirming that NHERF1 overexpression inhibits tumor cell proliferation through the PDZ1 domain. Active caspase-3 expression (Figure 2B) revealed that tumors from WT cell–injected mice had a significantly increased apoptotic area, whereas those from PDZ1mut- and PDZ2mut-injected mice had a significantly reduced apoptotic area compared to that in the tumors of parental cell–injected mice. Tumor microvessel density (Figure 2C), evaluated by the endothelial marker CD31, significantly decreased in both WT- and PDZ1mut-derived tumors, whereas it increased in PDZ2mut-derived tumors compared with parental tumors. These data suggest that the inhibition of growth in the WT tumor xenografts results from a combination of decreased proliferation and tumor microvessel density with increased apoptosis.
FIGURE 2.
NHERF1 regulates tumor cell proliferation and apoptosis. Tumor sections from tumors obtained by subcutaneous injection of pcDNA-, WT-, PDZ1mut-, and PDZ2mut-transfected cells were subjected to (A) immunohistochemical analysis for the proliferative marker Ki-67, (B) immunofluorescence assay for the apoptosis marker active caspase-3, and (C) the endothelial marker CD31. Left, representative images at the original magnification (bar, 10 μm). Right, quantification of expression performed using ImageJ software, n = 4; results are represented as mean ± SEM; the values of p are compared with levels in pcDNA tumors.
NHERF1-induced invasion is mediated by an increase of extracellular matrix degradation via its PDZ2 domain
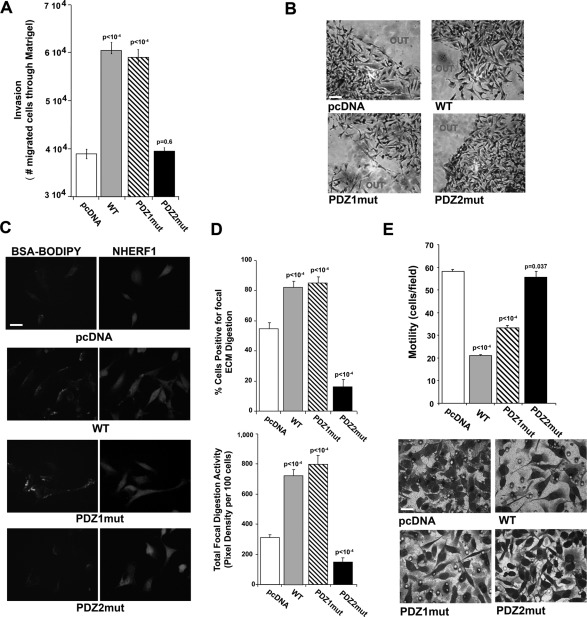
To determine the role of NHERF1 in cellular invasion and identify the PDZ domain involved, we analyzed the in vitro invasive ability of the cells. In Matrigel-based invasion assays overexpression of both WT and PDZ1mut significantly increased invasion, whereas PDZ2mut cells invaded through Matrigel at levels similar to the parental pcDNA cells (Figure 3A). When subjected to a three-dimensional (3D) invasion assay in which cells are entrapped in Matrigel, more WT and PDZ1mut cells exited from the drop than pcDNA cells, whereas in the same time frame, the PDZ2mut cells remained entrapped in the Matrigel (Figure 3B), indicating that, also in a 3D context, PDZ2-domain function confers an invasive advantage.
FIGURE 3:
The lack of functional PDZ2 domain decreases invasion of tumor cells. (A) The ability of the indicated transfected MDA-MB-231 cells to cross a Matrigel layer in 6 h was analyzed in Boyden chambers. Each cell variant was evaluated five times in triplicate, and results are represented as mean ± SEM. The values of p compared with the pcDNA-transfected cells. (B) Representative microphotographs of Matrigel evasion assays. Cells were included in growth factor–free Matrigel drops and incubated at 37°C for 6 d (bar, 30 μm). (C) NHERF1 regulation of ECM degradation measured by an in vitro fluorescence–Matrigel degradation assay. Cells were seeded on coverslips coated with Matrigel (4 mg/ml) and BSA-BODIPY (30 μg/ml) and incubated at 37°C for 24 h. Panels show representative pictures of proteolytic degradation in the Matrigel matrix in green (BSA-BODIPY) and immunofluorescence of NHERF1 protein expression in red (NHERF1; bar, 10 μm). (D) Quantification of invadopodia function (ECM degradation) analyzed as percentage of cells digesting the matrix (top) or total focal digestive activity of 100 cells (bottom); n = 5; mean ± SEM values of p test vs. pcDNA. (E) Migration assay of the indicated cell lines through collagen IV–coated filters in Boyden chambers. Results are expressed as numbers of migrated cells/field. Top, representative fields of migrated cells through filters. Bar, 10 μm. Mean ± SEM of three independent experiments performed in duplicate. The values of p are compared with the pcDNA-transfected cells.
Proteolytic degradation of the extracellular matrix (ECM) and high level of directed motility are two of the key steps of invasion in primary cancer lesions (Basbaum and Werb, 1996). Therefore we evaluated living cell in situ proteolytic activity (Busco et al. 2010; Figure 3C), analyzing either the percentage of cells positive for proteolysis per low-power microscopic field (Figure 3D, top) or the total focal digestive activity per 100 cells (Figure 3D, bottom). These data demonstrate an increased ability of overexpressed WT and PDZ1mut to degrade the ECM compared with parental cells, whereas PDZ2mut cells had a reduced ECM degradative capacity.
Finally, migration assays in Boyden chambers (Figure 3E) and in scratch wound assays (Supplemental Figure S3) demonstrated that WT and PDZ1mut cells had significantly reduced motility with respect to pcDNA cells, whereas no changes were observed in PDZ2mut cells. Moreover, in these assays WT and PDZ1mut cells displayed an elongated morphology (Figure 3E, bottom) and an increase in stress fiber formation (Supplemental Figure S4A) typical of epithelial-to-mesenchymal transition (EMT)–like phenotype. This was further confirmed by the enhanced expression of mesenchymal markers (vimentin and smooth muscle actin) together with a diminished expression of epithelial markers (E-cadherin and occludin; Supplemental Figure S4B), whereas PDZ2mut cells had low levels of stress fiber formation and a higher expression of epithelial-toward-mesenchymal markers, suggesting that the PDZ2 domain is required for EMT.
NHERF1 PDZ1 and PDZ2 domains differently regulate invadopodia and podosome dynamics
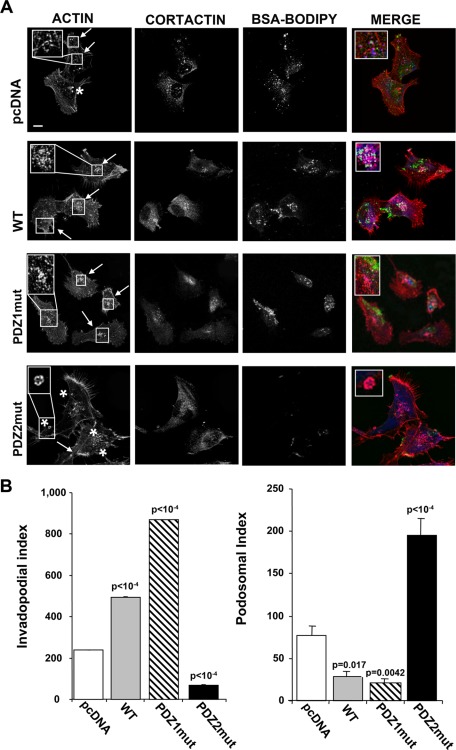
The general consensus is that the highly dynamic, cortactin/actin–rich, ring-like plasma membrane structures involved in adhesion and migration are called podosomes (Yoshio et al., 2007), whereas invadopodia are more stable, actin-based, cortactin-positive, filament-like protrusions emanating from the surface of invasive tumor cells and displaying high focalized proteolytic activity inside the ECM (Gimona et al., 2008; Buccione et al., 2009, Stylli et al., 2009). We therefore examined the distribution of invadopodia, podosome rings, and ECM focal digestion (Busco et al., 2010) by confocal fluorescence microscopy of cells cultured on Matrigel containing quenched DQ-green–labeled bovine serum albumin (BSA)–BODIPY (Figure 4A). pcDNA cells formed both podosome rings (asterisks) at the ventral cell surface that were rarely associated with focal proteolysis and punctate actin/cortactin–rich invadopodia structures that entered the ECM and were sites of active focal ECM proteolysis (arrows). Three-dimensional reconstructions of typical invadopodia and podosomes with their respective RGB analyses are shown in Supplemental Figure 6, A and B.
FIGURE 4:
The PDZ2 domain regulates invadopodia and the PDZ1 domain regulates podosome dynamics in NHERF1-overexpressing cells. Invadopodia and podosome rings in the cells cultured on glass coverslips coated with Matrigel containing BSA-BODIPY for 24 h to visualize ECM digestion (green) and stained for F-actin (red) and cortactin (blue). (A) Confocal immunofluorescence localization shows invadopodia (arrows) visible as F-actin dots colocalizing with cortactin and overlapping with areas of ECM degradation. Podosomal ring structures are marked with cortactin and surround F-actin rich cores in pcDNA and PDZ2mut cells (asterisk). Bar, 10 μm. (B) Quantification of the Invadopodia Index (left) and Podosomal Index (right) for each cell variant calculated as described in Materials and Methods. Data are representative or the mean ± SEM of cells from three independent experiments; the values of p are compared with the pcDNA-transfected cells.
The relative expression of invadopodia and podosomes were quantified by scoring the cells for the Invadopodial Index and the Podosomal Index (Figure 4B) as defined in Materials and Methods and in Supplemental Figure S5. Indeed, WT and PDZ1mut cells had an increased Invadopodial Index compared with the parental cells, whereas the PDZ2mut cells had a very low Invadopodial Index (Figure 4B, left). In contrast, WT and PDZ1mut cells had a reduced Podosomal Index compared with the pcDNA cells, whereas PDZ2mut cells showed a potent increase of this parameter (Figure 4B, right). Taken together, these results demonstrate that NHERF1-induced ECM digestive invadopodia formation is stimulated by the PDZ2 domain and slightly inhibited by the PDZ1 domain, whereas the opposite occurs in NHERF1-induced podosome formation.
The PDZ2 domain regulates NHERF1-mediated neoangiogenesis and vasculogenic mimicry–like ability
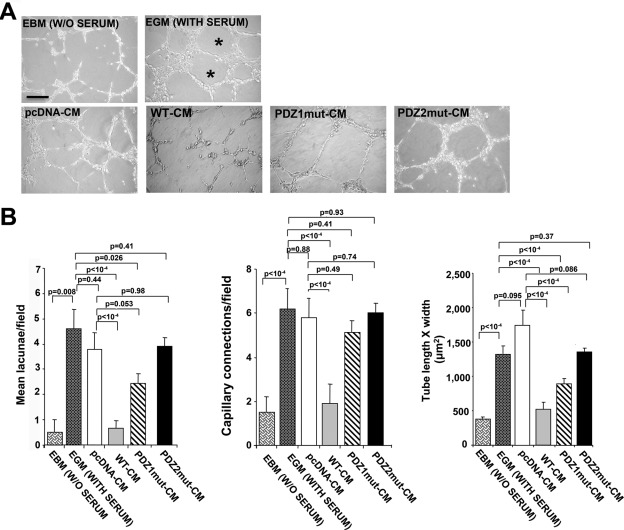
To determine the angiogenic role of NHERF1, we first measured the ability of tumor conditioned medium (CM) from each cell variant to affect the ability of human umbilical vein endothelial cells (HUVECs) to form capillary-like structures. Figure 5A shows that CM from pcDNA, PDZ1mut, and PDZ2mut cells induced networks of capillary-like structures. In contrast, capillary-like outgrowth was totally blocked in HUVECs treated with WT-CM. Of interest, networks formed in the presence of pcDNA-CM and PDZ2mut-CM displayed typical irregular capillary-like networks anastomosing to multicentric nodes to delimit completely closed areas (lacunae, asterisk), whereas cells treated with PDZ1mut-CM had large, regular, orthogonal networks with long, thin ramifications formed by a few cells. Histograms quantifying these morphological data (Figure 5B) for mean number of lacunae (left), number of capillary-like connections (middle), and capillary tube morphology (right) verified these angiogenic patterns. These data suggest that especially the PDZ2 domain is critical for efficient NHERF1 overexpression-mediated inhibition of endothelial cell vascular network.
FIGURE 5:
NHERF1 inhibits neoangiogenesis via the PDZ2 domain. (A) Representative microphotographs of capillary network formation of HUVECs seeded on Matrigel and incubated with EBM (medium without serum), EGM (medium with serum), or CM of each cell variant for 24 h. Asterisks indicate empty areas (lacunae) bordered by the capillary-like network. (B) Graphs showing quantification of mean number of lacunae/field (left) and of capillary-like connections/field (center), and tube morphology (tube length × width; right) in HUVECs in the presence of EBM, EGM, or CM from each cell variant. Mean ± SEM; the values of p compared with the EBM-, EGM-, or pcDNA-transfected cells as indicated.
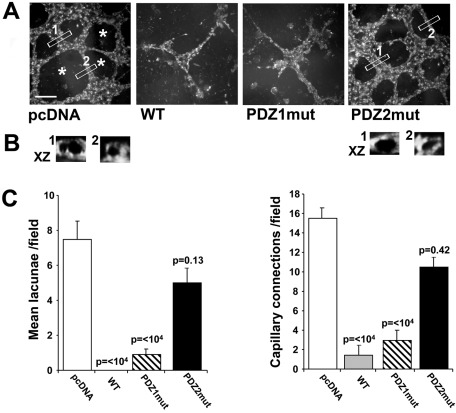
We next analyzed the cells for their innate vasculogenic-like ability when grown embedded in growth factor–reduced Matrigel without added serum (Figure 6A). Both the PDZ2mut cells and the pcDNA cells generated a complex 3D vascular-like tubular network emerging from multicentric cellular nodes. Vertical sections of the 3D-reconstructed tubes from the pcDNA and PDZ2mut fields (Figure 6B) revealed that these are open lumen-like structures surrounded by or delineated by fluorescent cells. In contrast, PDZ1mut cells generated short, thin, cord-like structures issuing out of the nodes but not forming lacunae, whereas WT cells did not form cords at all. The data quantified in Figure 6C verified these vasculogenic-like patterns.
FIGURE 6:
NHERF1 inhibits vasculogenic mimicry–like ability via the PDZ2 domain. (A) Representative microphotographs of capillary-like tubule formations obtained with the cells when seeded inside Matrigel for 5 d in their growth medium. Cells were stained with Oregon Green 488 DHPE and acquired by confocal microscopy. Asterisks indicate empty areas (lacunae) bordered by the capillary-like network. Bar, 10 μm. (B) Bottom, XZ-zoomed vertical cross-section views of 3D reconstruction of Z-stacked VM tubes of the regions of interest, indicated by 1 or 2, in each field from pcDNA cells and PDZ2mut cells. Vertical sections of the 3D-reconstructed tubes show that they are open lumen-like structures surrounded by fluorescent cells. (C) The mean number of lacunae/field (left) and the capillary connections number (right) were quantified. Results are expressed as mean ± SEM; p value vs. pcDNA. All assays were performed in quadruplicate.
NHERF1 orchestrates in vivo organotropism of metastases
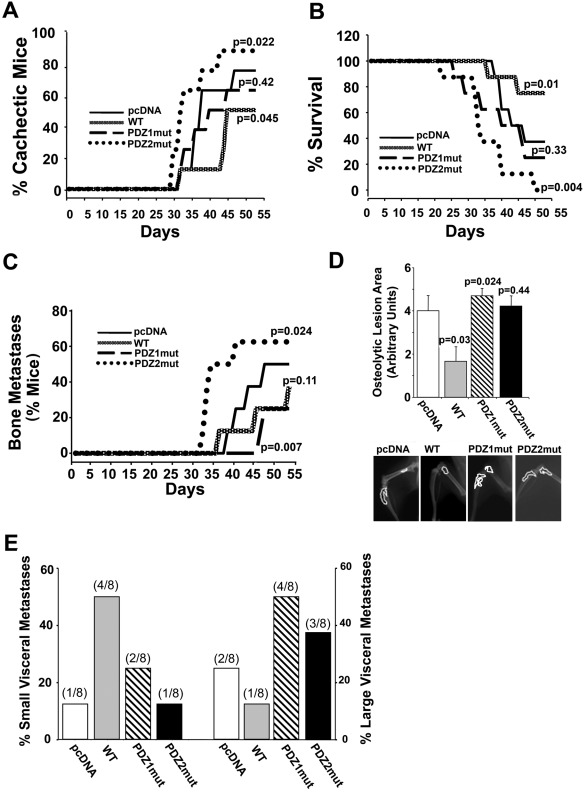
Because of this strong in vitro tumor phenotype selection through the two NHERF1 PDZ domains, we tested whether NHERF1 modulation could affect the ability of these cells to develop in vivo experimental metastases by intracardiac injection with our stable transfected variants (Rucci et al., 2006). As reported by many groups, animals injected with the parental pcDNA cells formed both bone (Figure 7, C and D) and visceral metastases (Figure 7E) and had medium levels of both cachexia (Figure 7A) and mortality (Figure 7B). Remarkably, mice injected with cells having differential PDZ-domain function displayed a very strong stratification in metastasis formation and organotropism. Mice injected with cells overexpressing WT NHERF1 displayed a strong reduction in both bone osteolytic lesions and large visceral metastases and was the only variant to form a large number of micrometastases (<30 mm3). These characteristics translated into reduced cachexia and increased survival. Of importance, mice injected with PDZ2mut cells displayed a much stronger osteolytic bone tropism than the parental, pcDNA-injected mice (higher incidence and shorter time to first incidence) and formed large visceral metastases, whereas the PDZ1mut cell–injected mice formed only visceral metastases. Consistently, cachexia and mortality appeared earlier and had the highest incidence in the mice injected with PDZ2mut cells, whereas mice injected with parental, pcDNA, or PDZ1mut cells had intermediate rates of both cachexia and mortality.
FIGURE 7:
NHERF1 PDZ domain function dictates organotropism. Four-week-old female BALB/c-nu/nu mice were inoculated in the left ventricle with a suspension of each cell variant. n = 8 mice/group. Mice were monitored daily for (A) cachexia (decrease of body weight) and (B) survival (p values vs. pcDNA-injected mice are presented) and (C) monitored weekly by x-ray analysis to determine onset and incidence of bone metastasis. p value vs. pcDNA-injected mice. (D) Evaluation of osteolytic area by densitometric analysis of hindlimb x-ray. Bottom, representative radiographic images of one hindlimb that developed an osteolytic lesion for each group. Data are presented as mean ± SEM; p value vs. pcDNA. (E) At the end of the experiment animals were killed and subjected to anatomical dissection to evaluate the incidence of visceral metastases, which were scored, according to the size, as percentage of small (left x-axis) or large (right x-axis) metastases.
Consistent with this pattern of in vivo induction of osteolytic bone metastases, treatment with pcDNA-CM and PDZ2mut-CM stimulated in vitro osteoclast formation that was very strongly inhibited in the presence of WT-CM and partially reduced by PDZ1mut-CM (Supplemental Figure S7). These data also indicate that, although PDZ1mut cells induced the lowest incidence of bone metastases, once they reached the bone microenvironment, their ability to induce osteolysis was similar to that of pcDNA and PDZ2mut cells.
DISCUSSION
The complex set of processes required to produce metastatic lesions is based on tumor cell phenotypic plasticity, which enables, while still within the primary tumor, the acquisition of special prometastatic phenotypes that function as cell-fate determinants, in that they specify cell destiny by governing organotropism (Kang et al., 2003; Minn et al., 2005, 2007). This phenotypic plasticity relies on specific signal transduction systems, which coordinate defined cellular programs (Firestone and Chen, 2010). Signaling modules are orchestrated by scaffolding proteins, and here we show that one such scaffolding protein, NHERF1, finely regulates the development of metastatic lesions by selecting and eventually integrating the various metastatic behaviors that the cells are capable of expressing. Indeed, the present study identified a novel role for NHERF1 in cellular plasticity, in that by turning off one or the other of its PDZ domains, we could control the shift from one to another phenotypic outcome, thus both conferring a specific metastatic competence to the cell and determining organotropic metastatic choice. We observed that this can occur even in cells considered to be “endpoint” in their malignant evolution.
Although clinical expression studies have found NHERF1 overexpression to be associated with aggressive tumor characteristics and poor prognosis (Stemmer-Rachamimov et al., 2001; Cardone et al., 2007; Song et al., 2007; Bellizzi et al., 2010; Hayashi et al., 2010; Tabrizi et al., 2010; Karn et al., 2011; Molina et al., 2012), in vitro studies have observed a negative role of WT NHERF1 in tumor growth (Pan et al., 2006; Kreimann et al., 2007; Zheng et al., 2010; Wheeler et al., 2011; Molina et al., 2012), suggesting that NHERF1 functions instead as a tumor suppressor. Furthermore, this tumor suppressor role of WT-NHERF1 in cell lines contrasts with reports that WT-NHERF1 overexpression enhances invasion in tumor cell lines (Cardone et al., 2007). In the present study, these apparently divergent results are reconciled, as we confirmed that overexpression of WT-NHERF1 in highly metastatic human breast cancer cells greatly reduces both anchorage-independent and xenograft tumor growth while increasing invasion and ECM digestion via stimulation of invadopodia formation.
Various signaling mechanisms have been put forward for the NHERF1-growth suppression function: 1) NHERF1-PDZ1–mediated PTEN membrane recruitment and subsequent PI3K/AKT signaling pathway inactivation (Molina et al., 2012); 2) a PTEN-independent Akt-modulating effect by formation of a complex with PHLPP1/PHLPP2 Akt phosphatases (Molina et al., 2012); 3) a PDZ2-dependent down-regulation of the Wnt/β-catenin axis (Wheeler et al., 2011), and 4) an increased apoptosis due to WT NHERF1 overexpression mediated by the suppression of ERK (Zheng et al., 2010). The data of the present study demonstrate that the PDZ1 domain is the main player in growth, as it completely reversed the WT growth repression, supporting previous reports in which NHERF1 regulates proliferation primarily through the PDZ1-PTEN complex via the PI3K/AKT signaling pathway rather than via the PDZ2-dependent down-regulation of the Wnt/β-catenin axis. This conclusion was confirmed with WB measurements (Figure 1C) showing that 1) the p-AKT survival pathway was attenuated in the WT transfected cells while being increased especially in the PDZ1-mutated (PDZ1mut) cells and 2) the p-ERK–induced proliferation pathway was inhibited in the WT cells via the PDZ1 domain. Moreover, the increased apoptosis observed in the WT xenograft supports the previously observed suppression of tumor expansion by WT NHERF1 overexpression.
In recent years, we have better understood that motility/adhesion and invasion occur through the intervention of specific membrane organelles—podosomes (Yoshio et al., 2007) and invadopodia (Gimona et al., 2008; Buccione et al., 2009; Stylli et al., 2009), respectively. Some cell types can simultaneously form both podosomes and invadopodia, whereas other types primarily form only podosomes or only invadopodia. Very recent work shows that they are indeed quite different structures in both their dynamics and regulation (Artym et al., 2011; Oser et al., 2011). An important question is whether these two structures are under separate regulatory control. We find that overexpression of either the WT or PDZ1mut constructs increased invadopodia formation and ECM digestion with a reduction in podosome formation, whereas in cells expressing PDZ2mut, invadopodia disappeared, and this was accompanied by an increase in podosome number and motility. These results demonstrate that there are important dynamic differences between podosomes and invadopodia in the regulation of their formation, in that there is a mechanistic “switch” regulated by the binding of NHERF1 to specific, and yet-to-be-determined, protein partners. In this regard, it is possible that the presence of an excess of one active domain sequesters that domains' binding proteins from wild-type signaling complexes. A recent article by Mamonova et al. (2012) showed that PDZ-partner interactions involve competition between the two NHERF1 PDZ domains and their target proteins. Therefore it is reasonable to assume that the mutation of one of the domains, while abolishing the ability of the mutated domain to bind its network of partner proteins, also alters the binding capacity and specificity of the remaining PDZ domain, which reinforces and creates new signal networks.
Much of solid tumor growth is dependent on the ready supply of nutrients and oxygen from a local blood supply and, as a consequence, it can stimulate angiogenesis, or, due to malignant cell plasticity, can form its own tumor vascular channel–like structures without the participation of endothelial cells (vasculogenic mimicry; Dome et al., 2007). As has been reported (Buchanan et al., 2011; Cuddy et al., 2012), we found that MDA-MB-231 cells secrete proteins that are able to influence the network-forming ability of endothelial cells. Our data reveal that NHERF1 regulates endothelial activation and vascular channel formation by breast cancer cells, and this could potentially influence the organotropic choice. We find that parental MDA-MB-231 cell CM positively influences angiogenesis, whereas WT-CM inhibits HUVEC network formation. These data also fit well with the xenograft results, in which a strong reduction of WT tumor size resulted from increased apoptosis, moderate decrease in proliferation, and also strong reduction in vessel density. This inability to recruit blood vessels is likely caused by the lack of secretion of proangiogenic factors and/or high secretion of antiangiogenic factors (Ossowski and Aguirre-Ghiso, 2010). In preliminary measurements analyzing WT-CM with an angiogenesis antibody array, the expression of the antiangiogenic molecule angiostatin was up-regulated, whereas the expression of proangiogenic molecules VEGF, UPA, and TF3 were strongly decreased with respect to the CM of all the other cell variants.
Of importance, we found that NHERF1 PDZ-domain function also differentially regulates vasculogenic mimicry (VM)–like channel formation, in that the ability of pcDNA cells to differentiate into extracellular matrix–rich, vascular-like channels in 3D Matrigel cultures was impaired in the highly invasive WT- and PDZ1-mutated clones, whereas the PDZ2-mutated clone (PDZ2mut) had full VM formation ability. These results suggest that NHERF1 can orchestrate a bidirectional transition between in vitro mesenchymal/invasive and vasculogenic programs that defines a novel cancer cell transition, the mesenchymal–vasculogenic transition (Figure 8). The regulation of this transdifferentiation by NHERF1 is in line with the inhibitory effect of cAMP on VM (Fan and Sun, 2010) and angiogenesis (Bianco et al., 1997) and its stimulation of invasion (Cardone et al., 2007), as we previously observed that cAMP levels increase upon overexpression of both WT and PDZ1mut but not PDZ2mut (Cardone et al., 2007). Because VM was reported to be resistant to angiogenesis inhibitors in tumor therapy (van der Schaft et al., 2004), elucidation of molecular mechanisms by which dysregulated NHERF1 signaling promotes both angiogenesis and VM could open new perspectives for developing therapeutic strategies with maximized efficacy against tumor microcirculation.
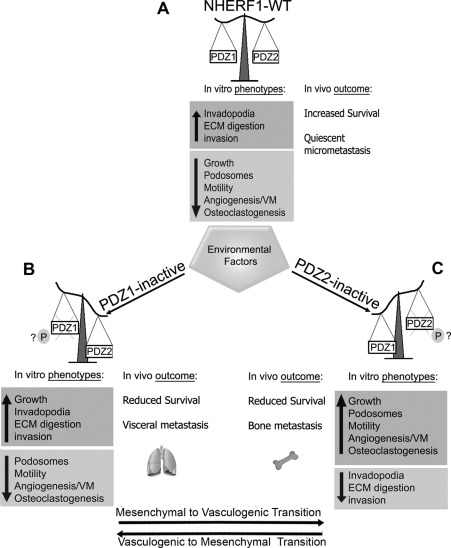
FIGURE 8:
Proposed model for NHERF1 PDZ domain-orchestrated switches regulating in vitro tumor phenotypes and in vivo metastatic organotropism. On the basis of its expression, NHERF1 may behave as (A) an oncosuppressor (NHERF1-WT) or (B, C) a metastasis-organotropic–specific protein (NHERF1 PDZ1 or PDZ2inactive). Although it is not yet clear which cellular signal mechanisms are responsible for this functional switch, exposure to an aberrant tumor microenvironment (hypoxic stress, nutrient stress, low extracellular pHe, ECM components) may play a role in affecting NHERF1 posttranslational modifications and/or shuttling from one cellular compartment to another, probably via altered phosphorylation and activity of one of its PDZ domains, thus attenuating its oncosuppressor role and activating specific oncogenic pathways. Moreover, the specific loss of function of one of the two PDZ domains, through the selection of a wide and flexible spectrum of possible hallmark behaviors, regulates the shift from one set of metastatic phenotypes to another and the spread to one specific organ to another (metastatic switch). (B) Specifically, when NHERF1 is overexpressed and PDZ1 is blocked (PDZ1mut cells) a growth program is activated, and visceral organs are preferred for spreading because invadopodia-dependent invasion and growth are up-regulated, whereas podosome/motility/angiogenic/osteoclastogenic programs are inhibited. (C) On the other hand, when PDZ2 is blocked (PDZ2mut cells) bone metastases are promoted because podosome/motility, neoangiogenesis, vasculogenic mimicry–like ability, and osteoclastogenesis are stimulated, whereas the invasive program is turned off. The shifting from one program to the other could be controlled by the well-documented phosphorylation-dependent alteration in PDZ domain function of NHERF1. If the switch between these phenotypic programs is bidirectional due to reversible phosphorylation-dependent alterations of the activity of one or the other PDZ domains, then this could define the phenotypic transition termed the mesenchymal–vasculogenic transition.
In agreement with the master role of NHERF1 to orchestrate the expression of in vitro phenotypic programs, we find here that the two PDZ domains of NHERF1 also control in vivo metastatic fate by actuating a switch in organotropic metastatic choice. Remarkably, although the parental pcDNA cells have the generalized capacity to form metastases in both bone and visceral tissues, the PDZ2mut cells were highly osteotropic, forming bone metastases much earlier and more efficiently than even the parental cells, whereas the PDZ1mut cells could no longer form bone metastases and showed enhanced metastatic growth in viscera. These data permit us to associate in vitro metastatic hallmark behaviors with in vivo organotropism (Figure 8), in that PDZ1mut cells in vitro exhibited a preeminent mesenchymal/invasive phenotype with restricted vasculogenic capacity that favors visceral metastases, whereas the PDZ2mut cells had increased in vitro capability to 1) form podosomes, which is highly associated with bone metastasis (Blouw et al., 2008; Sturge et al., 2011); 2) induce a vasculogenic program that conferred an additional advantage in the bone microenvironmental, as bone is one of the most physiologically hypoxic areas of the body (Dunn et al., 2009); and 3) induce osteoclastogenesis (Supplemental Figure S7), which acts synergistically to increase bone metastasis.
In mice injected with WT-NHERF1–overexpressing cells we found a striking decrease in both visceral and bone metastasis, with a general increase in survival. The high number of visceral micrometastases and the low number of visceral outgrowth metastases in WT-injected mice suggest that whereas in WT cells the activation of the invasive program permits the cells to infiltrate the visceral parenchyma of the secondary tissue, the lack of sustaining growth signals restrains metastasis outgrowth. These results provide functional evidence for a metastasis suppressor role of NHERF1 in response to its stringent blockade of the neovascularization/growth programs. A further protective role of overexpression of WT-NHERF1 in bone could be due to its reduced osteoclastogenic ability (Supplemental Figure S7).
This putative metastasis-suppressor role of WT-NHERF1 seems, paradoxically, not in line with clinical studies in which overexpression of WT-NHERF1 is strongly associated with highly aggressive tumors. This suggests that in vivo the activities of one or the other PDZ domains must be altered to convert NHERF1 from a metastasis repressor to a metastasis activator and raises the question as to how these activities of NHERF1 could be regulated. Whereas published data suggests that NHERF1 mutation is of minor importance (Dai et al., 2004), changes in phosphorylation are known to be among the major mechanisms for regulating NHERF1 activity and function, including cellular localization, protein conformation, and protein–protein interaction and complex formation (Garbett et al., 2010). NHERF1 is phosphorylated both constitutively and by physiological stimuli, and phosphorylation modifies both 1) its localization and the binding ability/specificity of the PDZ domains through serines 77 (Voltz et al., 2007; Weinman et al., 2007) and 162 (Raghuram et al., 2003; Li et al., 2007) and 2) formation of homodimers via the phosphorylation of serine 289 (Hall et al., 1999) and/or the stimulus-driven phosphorylation of serines 339 and 340 (Fouassier et al., 2005; Voltz et al., 2007). During mitosis in HeLa cells NHERF1 is phosphorylated by cdc2 on serines 279/301, and this acts as a “switch,” in that decreases NHERF1 oligomerization while increasing its association with the cell-cycle regulator Pin1 (He et al., 2001). Moreover, it was recently observed that HPV-16 E6/E7-induced neoplastic transformation increases cdc2-dependent NHERF1 phosphorylation at serines 279/301, and this activity was linked to increased PI3K signaling and Akt activity (Accardi et al., 2011). Taken together, these studies provided strong evidence that many critical events involved in cellular responses are mediated by NHERF1 phosphorylation We hypothesize that metastatic cells respond to different microenvironments inherent to different secondary organs, thus altering their phenotypic outcome, through the modulation of NHERF1 phosphorylation. In support of this hypothesis, we observe in preliminary experiments that breast tumors express hyperphosphorylated NHERF1 compared with the local nontumor tissue (Supplemental Figure S8). The understanding of the specific phosphorylations in different tumors and the mechanisms regulating changes in phosphorylation will be an important line of future research.
Our findings identified NHERF1 as a key protein in the regulation of metastatic phenotype expression and organotropism. This regulatory system will permit the deciphering of the molecular mechanisms largely responsible for cell metastatic fate choices and suggests that there is a NHERF1 form as a novel signaling nexus that could be exploited for antimetastasis therapy (Mayasundari et al., 2008).
MATERIALS AND METHODS
Cells and expression vectors containing NHERF1 mutants
The human breast cancer cell line MDA-MB-231 was grown as described (Cardone et al., 2007). Cells were transfected with FuGene6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN) and 3 μg of DNA construct according to the manufacturer's protocol. Expression vectors for NHERF1 WT and NHERF1 mutated in the PDZ1 or PDZ2 domains (PDZ1mut and PDZ2mut, respectively) were developed as described (Weinman et al., 2003) and used for the transfection of MDA-MB-231 cells (Supplemental Figure S1). The transfected cells were selected and maintained in complete medium containing 500 μg/ml hygromycin B (Calbiochem, Schwalbach, Germany). Multiple stable NHERF1-transfected clones (WT) and PDZ1/PDZ2-defective, mutant, stably transfected clones (PDZ1mut and PDZ2mut) expressing approximately three -times the NHERF1 levels, together with a pcDNA empty vector–expressing clone in which NHERF1 was expressed at endogenous levels, were obtained and screened as described in Supplemental Figure S2. Adherent HUVECs were propagated in Endothelial Basal Medium (EBM) plus 2% fetal bovine serum, bovine brain extract, hydrocortisone, human endothelial growth factor, and gentamicin/amphotericin B (EGM) in 5% CO2 at 37°C.
ECM digestion by in situ zymography
For in vitro dequenching assay, experiments were conducted in Matrigel (diluted to a final concentration of 4 mg/ml) containing quenched BODIPYs linked to BSA (DQ-green BSA, DQ-red BSA) as described (Busco et al., 2010). Focal proteolysis produces fluorescence in a black background. which is used both to quantify proteolytic activity levels and in colocalization analysis.
Colony formation in soft agar, in vitro invasion, three-dimensional evasion, migration, and wound-healing assays
Details on these measurements are supplied in the Supplemental Materials and Methods.
Invadopodia and podosomes
The visualization and analysis of invadopodia and podosomes formed in Matrigel were performed as described (Busco et al., 2010).
The Invadopodia Index was calculated as the product of the percentage of invadopodia-positive cells for the mean number of invadopodia per cell, and the Podosomal Index was calculated as the product of the percentage of F-actin and cortactin ring–positive cells for the mean number of rings per cell. The individual histograms displaying the data for these parameters are shown in Supplemental Figure S5.
In vitro angiogenesis and three-dimensional vasculogenic mimicry network formation assays
Details on the measurement and analysis of angiogenic and vasculogenic mimicry capacity are supplied in the Supplemental Materials and Methods.
In vivo experimental tumors and analysis
Four-week-old female immunocompromised BALB/c nu/nu mice (Charles River, Milan, Italy) were maintained under sterile conditions and used for all in vivo experiments. Procedures involving animals and their care were conducted in conformity with national and international laws and policies and were approved by our Institutional Review Board. Intracardiac and subcutaneously injections were performed as described (Arguello et al., 1988; Yoneda et al., 1997; Rucci et al., 2006). Further details can be found in the Supplemental Materials and Methods.
Evaluation of osteolytic lesions
Radiographs were scanned using the Bio-Rad scanning densitometer (Hercules, CA), model GS800, and quantification of the area of interest was done using the Bio-Rad Quantity One image analysis software.
Statistical analysis
Data are presented as means ± SEM. One-way ANOVA Kruskal–Wallis test was used to compare more than two groups, and Student's t test (two-tailed) was used to compare two groups. A value of p < 0.05 was considered significant, assuming equal variances on all experimental data sets. The incidence of cachexia, survival, and bone and visceral metastases was analyzed by a chi-squared contingency table analysis (Fisher's exact test), and a value of p < 0.05 was considered significant. All analyses were performed with InStat (GraphPad Software, La Jolla, CA).
Supplementary Material
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro Grant 5167 and PIO Grant RFPS-2006-3-340260 to S.J.R. and by European Commission Grant METABRE (#LSHM-CT-2003-503049) and a grant from the Associazione Italiana per la Ricerca sul Cancro to A.T. The Reshkin laboratory is part of the Italian network Istituto Nazionale Biostrutture e Biosistemi, the Centro di Eccellenza di Genomica in Campo Biomedico ed Agrario of the University of Bari, and project BioBoP of the Region Puglia.
Abbreviations used:
- CM
conditioned medium
- EBM
endothelial basal medium
- ECM
extracellular matrix
- EGM
endothelial growth medium
- PDZ1mut cells
cells expressing NHERF1 mutated in PDZ1 domain
- PDZ2mut cells
cells expressing NHERF1 mutated in PDZ2 domain
- VM
vasculogenic mimicry
- WB
Western blotting
- WT cells
cells expressing wild-type NHERF1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0911) on April 11, 2012.
There are no conflicts of interest.
REFERENCES
- Accardi R, et al. E6 and E7 from human papillomavirus type 16 cooperate to target the PDZ protein Na/H exchange regulatory factor 1. J Virol. 2011;85:8208–8216. doi: 10.1128/JVI.00114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello F, Baggs RB, Frantz CN. A murine model of experimental metastasis to bone and bone marrow. Cancer Res. 1988;48:6876–6881. [PubMed] [Google Scholar]
- Artym VV, Matsumoto K, Mueller SC, Yamada KM. Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. Eur J Cell Biol. 2011;90:172–180. doi: 10.1016/j.ejcb.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum CB, Werb Z. Focalized proteolysis: spatial and temporal regulation of extracellular matrix degradation at the cell surface. Curr Opin Cell Biol. 1996;8:731–738. doi: 10.1016/s0955-0674(96)80116-5. [DOI] [PubMed] [Google Scholar]
- Bellizzi A, Malfettone A, Cardone RA, Mangia A. NHERF1/EBP50 in breast cancer: clinical perspectives. Breast Care (Basel) 2010;5:86–90. doi: 10.1159/000298962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Tortora G, Baldassarre G, Caputo R, Fontanini G, Chine S, Bianco AR, Ciardiello F. 8-Chloro-cyclic AMP inhibits autocrine and angiogenic growth factor production in human colorectal and breast cancer. Clin Cancer Res. 1997;3:439–448. [PubMed] [Google Scholar]
- Blouw B, Seals DF, Pass I, Diaz B, Courtneidge SA. A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. Eur J Cell Biol. 2008;87:555–567. doi: 10.1016/j.ejcb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccione R, Caldieri G, Ayala I. Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metastasis Rev. 2009;28:137–149. doi: 10.1007/s10555-008-9176-1. [DOI] [PubMed] [Google Scholar]
- Buchanan CF, Szot CS, Wilson TD, Akman S, Metheny-Barlow LJ, Robertson JL, Freeman JW, Rylander MN. Cross-talk between endothelial and breast cancer cells regulates reciprocal expression of angiogenic factors in vitro. J Cell Biochem. 2011;113:1142–1151. doi: 10.1002/jcb.23447. [DOI] [PubMed] [Google Scholar]
- Busco G, et al. NHE1 promotes invadopodial ECM proteolysis through acidification of the peri-invadopodial space. FASEB J. 2010;24:3903–3915. doi: 10.1096/fj.09-149518. [DOI] [PubMed] [Google Scholar]
- Cardone RA, Bellizzi A, Busco G, Weinman EJ, Dell'Aquila ME, Casavola V, Azzariti A, Mangia A, Paradiso A, Reshkin SJ. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18:1768–1780. doi: 10.1091/mbc.E06-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Cuddy AC, Qiu C, Gerecht S. Breast cancer cell-derived matrix supports vascular morphogenesis. Am J Physiol Cell Physiol. 2012;302:C1243–C1256. doi: 10.1152/ajpcell.00011.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JL, Wang L, Sahin AA, Broemeling LD, Schutte M, Pan Y. NHERF (Na+/H+ exchanger regulatory factor) gene mutations in human breast cancer. Oncogene. 2004;23:8681–8687. doi: 10.1038/sj.onc.1207962. [DOI] [PubMed] [Google Scholar]
- Dome B, Hendrix MJ, Paku S, Tovari J, Timar J. Alternative vascularization mechanisms in cancer: pathology and therapeutic implications. Am J Pathol. 2007;170:1–15. doi: 10.2353/ajpath.2007.060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LK, Mohammad KS, Fournier PG, McKenna CR, Davis HW, Niewolna M, Peng XH, Chirgwin JM, Guise TA. Hypoxia and TGF-beta drive breast cancer bone metastases through parallel signaling pathways in tumor cells and the bone microenvironment. PLoS One. 2009;4:e6896. doi: 10.1371/journal.pone.0006896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YZ, Sun W. Molecular regulation of vasculogenic mimicry in tumors and potential tumor-target therapy. World J Gastrointest Surg. 2010;2:117–127. doi: 10.4240/wjgs.v2.i4.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig HH, Maier A, Burger AM. Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur J Cancer. 2004;40:802–820. doi: 10.1016/j.ejca.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Chen JK. Controlling destiny through chemistry: small-molecule regulators of cell fate. ACS Chem Biol. 2010;5:15–34. doi: 10.1021/cb900249y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouassier L, Nichols MT, Gidey E, McWilliams RR, Robin H, Finnigan C, Howell KE, Housset C, Doctor RB. Protein kinase C regulates the phosphorylation and oligomerization of ERM binding phosphoprotein 50. Exp Cell Res. 2005;306:264–273. doi: 10.1016/j.yexcr.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Garbett D, LaLonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Georgescu MM, Morales FC, Molina JR, Hayashi Y. Roles of NHERF1/EBP50 in cancer. Curr Mol Med. 2008;8:459–468. doi: 10.2174/156652408785748031. [DOI] [PubMed] [Google Scholar]
- Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Curr Opin Cell Biol. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Hall RA, Spurney RF, Premont RT, Rahman N, Blitzer JT, Pitcher JA, Lefkowitz RJ. G protein-coupled receptor kinase 6A phosphorylates the Na(+)/H(+) exchanger regulatory factor via a PDZ domain-mediated interaction. J Biol Chem. 1999;274:24328–24334. doi: 10.1074/jbc.274.34.24328. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Molina JR, Hamilton SR, Georgescu MM. NHERF1/EBP50 is a new marker in colorectal cancer. Neoplasia. 2010;12:1013–1022. doi: 10.1593/neo.10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Lau AG, Yaffe MB, Hall RA. Phosphorylation and cell cycle-dependent regulation of Na+/H+ exchanger regulatory factor-1 by Cdc2 kinase. J Biol Chem. 2001;276:41559–41565. doi: 10.1074/jbc.M106859200. [DOI] [PubMed] [Google Scholar]
- Hunter KW, Alsarraj J. Gene expression profiles and breast cancer metastasis: a genetic perspective. Clin Exp Metastasis. 2009;26:497–503. doi: 10.1007/s10585-009-9249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Xie X, Chen C, Park JG, Stark C, James DA, Olhovsky M, Linding R, Mao Y, Pawson T. Eukaryotic protein domains as functional units of cellular evolution. Sci Signal. 2009;2:ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Karn T, et al. Gene expression profiling of luminal B breast cancers reveals NHERF1 as a new marker of endocrine resistance. Breast Cancer Res Treat. 2011;130:409–420. doi: 10.1007/s10549-010-1333-x. [DOI] [PubMed] [Google Scholar]
- Kislin KL, McDonough WS, Eschbacher JM, Armstrong BA, Berens ME. NHERF-1: modulator of glioblastoma cell migration and invasion. Neoplasia. 2009;11:377–387. doi: 10.1593/neo.81572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimann EL, Morales FC, de Orbeta-Cruz J, Takahashi Y, Adams H, Liu TJ, McCrea PD, Georgescu MM. Cortical stabilization of beta-catenin contributes to NHERF1/EBP50 tumor suppressor function. Oncogene. 2007;26:5290–5299. doi: 10.1038/sj.onc.1210336. [DOI] [PubMed] [Google Scholar]
- Li J, Poulikakos PI, Dai Z, Testa JR, Callaway D. Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J Biol Chem. 2007;282:27086–27099. doi: 10.1074/jbc.M702019200. [DOI] [PubMed] [Google Scholar]
- Mamonova T, Kurnikova M, Friedman PA. Structural basis for NHERF1 PDZ domain binding. Biochemistry. 2012;51:3110–3120. doi: 10.1021/bi201213w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayasundari A, Ferreira AM, He L, Mahindroo N, Bashford D, Fujii N. Rational design of the first small-molecule antagonists of NHERF1/EBP50 PDZ domains. Bioorg Med Chem Lett. 2008;18:942–945. doi: 10.1016/j.bmcl.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Minn AJ, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, Georgescu MM. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31:1264–1274. doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Dovas A, Cox D, Condeelis J. Nck1 and Grb2 localization patterns can distinguish invadopodia from podosomes. Eur J Cell Biol. 2011;90:181–188. doi: 10.1016/j.ejcb.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski L, Aguirre-Ghiso JA. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res. 2010;23:41–56. doi: 10.1111/j.1755-148X.2009.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Wang L, Dai JL. Suppression of breast cancer cell growth by Na+/H+ exchanger regulatory factor 1 (NHERF1) Breast Cancer Res. 2006;8:R63. doi: 10.1186/bcr1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram V, Hormuth H, Foskett JK. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc Natl Acad Sci USA. 2003;100:9620–9625. doi: 10.1073/pnas.1633250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucci N, Recchia I, Angelucci A, Alamanou M, Del Fattore A, Fortunati D, Susa M, Fabbro D, Bologna M, Teti A. Inhibition of protein kinase c-Src reduces the incidence of breast cancer metastases and increases survival in mice: implications for therapy. J Pharmacol Exp Ther. 2006;318:161–172. doi: 10.1124/jpet.106.102004. [DOI] [PubMed] [Google Scholar]
- Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology (Bethesda) 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- Song J, Bai J, Yang W, Gabrielson EW, Chan DW, Zhang Z. Expression and clinicopathological significance of oestrogen-responsive ezrin-radixin-moesin-binding phosphoprotein 50 in breast cancer. Histopathology. 2007;51:40–53. doi: 10.1111/j.1365-2559.2007.02730.x. [DOI] [PubMed] [Google Scholar]
- Stemmer-Rachamimov AO, Wiederhold T, Nielsen GP, James M, Pinney-Michalowski D, Roy JE, Cohen WA, Ramesh V, Louis DN. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas. Am J Pathol. 2001;158:57–62. doi: 10.1016/S0002-9440(10)63944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol. 2011;8:357–368. doi: 10.1038/nrclinonc.2011.67. [DOI] [PubMed] [Google Scholar]
- Stylli SS, Stacey TT, Verhagen AM, Xu SS, Pass I, Courtneidge SA, Lock P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J Cell Sci. 2009;122:2727–2740. doi: 10.1242/jcs.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi AD, Kalloger SE, Kobel M, Cipollone J, Roskelley CD, Mehl E, Gilks CB. Primary ovarian mucinous carcinoma of intestinal type: significance of pattern of invasion and immunohistochemical expression profile in a series of 31 cases. Int J Gynecol Pathol. 2010;29:99–107. doi: 10.1097/PGP.0b013e3181bbbcc1. [DOI] [PubMed] [Google Scholar]
- van der Schaft DW, Seftor RE, Seftor EA, Hess AR, Gruman LM, Kirschmann DA, Yokoyama Y, Griffioen AW, Hendrix MJ. Effects of angiogenesis inhibitors on vascular network formation by human endothelial and melanoma cells. J Natl Cancer Inst. 2004;96:1473–1477. doi: 10.1093/jnci/djh267. [DOI] [PubMed] [Google Scholar]
- Voltz JW, Brush M, Sikes S, Steplock D, Weinman EJ, Shenolikar S. Phosphorylation of PDZ1 domain attenuates NHERF-1 binding to cellular targets. J Biol Chem. 2007;282:33879–33887. doi: 10.1074/jbc.M703481200. [DOI] [PubMed] [Google Scholar]
- Weinman EJ, Biswas RS, Peng Q, Shen L, Turner CL, Xiaofei E, Steplock D, Shenolikar S, Cunningham R. Parathyroid hormone inhibits renal phosphate transport by phosphorylation of serine 77 of sodium-hydrogen exchanger regulatory factor-1. J Clin Invest. 2007;117:3412–3420. doi: 10.1172/JCI32738. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Weinman EJ, Wang Y, Wang F, Greer C, Steplock D, Shenolikar S. A C-terminal PDZ motif in NHE3 binds NHERF-1 and enhances cAMP inhibition of sodium-hydrogen exchange. Biochemistry. 2003;42:12662–12668. doi: 10.1021/bi035244l. [DOI] [PubMed] [Google Scholar]
- Wheeler DS, Barrick SR, Grubisha MJ, Brufsky AM, Friedman PA, Romero G. Direct interaction between NHERF1 and Frizzled regulates beta-catenin signaling. Oncogene. 2011;30:32–42. doi: 10.1038/onc.2010.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Sasaki A, Dunstan C, Williams PJ, Bauss F, De Clerck YA, Mundy GR. Inhibition of osteolytic bone metastasis of breast cancer by combined treatment with the bisphosphonate ibandronate and tissue inhibitor of the matrix metalloproteinase-2. J Clin Invest. 1997;99:2509–2517. doi: 10.1172/JCI119435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshio T, Morita T, Kimura Y, Tsujii M, Hayashi N, Sobue K. Caldesmon suppresses cancer cell invasion by regulating podosome/invadopodium formation. FEBS Lett. 2007;581:3777–3782. doi: 10.1016/j.febslet.2007.06.073. [DOI] [PubMed] [Google Scholar]
- Zheng JF, Sun LC, Liu H, Huang Y, Li Y, He J. EBP50 exerts tumor suppressor activity by promoting cell apoptosis and retarding extracellular signal-regulated kinase activity. Amino Acids. 2010;38:1261–1268. doi: 10.1007/s00726-009-0437-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.