The molecular mechanisms determining mitochondrial architecture are largely unclear. The C-terminal domain of Fcj1 and the TOB complex are shown to interact. Both are important for determining cristae morphology. The results explain how crista junctions are positioned at the outer membrane, assigning novel functions to both Fcj1 and the TOB complex.
Abstract
Crista junctions (CJs) are tubular invaginations of the inner membrane of mitochondria that connect the inner boundary with the cristae membrane. These architectural elements are critical for mitochondrial function. The yeast inner membrane protein Fcj1, called mitofilin in mammals, was reported to be preferentially located at CJs and crucial for their formation. Here we investigate the functional roles of individual domains of Fcj1. The most conserved part of Fcj1, the C-terminal domain, is essential for Fcj1 function. In its absence, formation of CJ is strongly impaired and irregular, and stacked cristae are present. This domain interacts with full-length Fcj1, suggesting a role in oligomer formation. It also interacts with Tob55 of the translocase of outer membrane β-barrel proteins (TOB)/sorting and assembly machinery (SAM) complex, which is required for the insertion of β-barrel proteins into the outer membrane. The association of the TOB/SAM complex with contact sites depends on the presence of Fcj1. The biogenesis of β-barrel proteins is not significantly affected in the absence of Fcj1. However, down-regulation of the TOB/SAM complex leads to altered cristae morphology and a moderate reduction in the number of CJs. We propose that the C-terminal domain of Fcj1 is critical for the interaction of Fcj1 with the TOB/SAM complex and thereby for stabilizing CJs in close proximity to the outer membrane. These results assign novel functions to both the C-terminal domain of Fcj1 and the TOB/SAM complex.
INTRODUCTION
Mitochondria have a number of essential functions in eukaryotic cells, including ATP synthesis, Fe–S cluster formation, and regulation of apoptotic pathways. They are delimited by an envelope, composed of the outer membrane and the inner boundary membrane. The latter membrane, together with the cristae membranes, which invaginate into the matrix space, forms the inner membrane. The inner boundary membrane is connected with the cristae membrane by crista junctions (CJs)—highly curved membrane structures with a tubular/slot-like morphology that are proposed to act as diffusion barriers for proteins, metabolites, and even protons (Mannella et al., 1994, 2001; Mannella, 2006; Frey and Mannella, 2000; Perkins et al., 1997, 2003; Nicastro et al., 2000; Renken et al., 2002; Zick et al., 2009). The ultrastructural appearance of cristae membranes is well known to exhibit high variability (Munn, 1974; Fawcett, 1981). This is true not only for different organisms and cell types; alterations have also been described to prevail in numerous pathological situations in humans (DiMauro et al., 1985; Wallace, 2005). Moreover, mitochondrial ultrastructure has been reported to be highly dynamic; it changes rapidly upon metabolic alterations or induction of apoptosis (Mannella et al., 2001; Scorrano et al., 2002).
We recently identified Fcj1 as a protein required for the formation of CJs in the yeast Saccharomyces cerevisiae (Rabl et al., 2009), a function also assigned to the mammalian orthologue mitofilin/IMMT (John et al., 2005). Fcj1 was the first protein to be localized to the CJ and was shown to modulate CJ formation in an antagonistic manner to the subunits e and g of the F1FO ATP synthase. Cristae structure depends on the oligomerization state of the F1FO ATP synthase (Paumard et al., 2002), which is influenced by Fcj1 and subunits e and g (Rabl et al., 2009). Protein interaction partners of Fcj1 and its orthologues have been suggested. Fcj1 was recently found to be part of a large multisubunit complex (MICOS/MINOS/MitOS complex) having a central role in the formation of CJs and in determining cristae morphology (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012). Fcj1 was reported to interact with outer membrane proteins such as Tom 40 (von der Malsburg et al., 2011), porin (Hoppins et al., 2011), or Ugo1 (Harner et al., 2011), thereby forming contact sites between the outer and the inner membrane. Mitofilin/IMMT in higher eukaryotes has been reported to physically interact with the outer membrane proteins SAMM50, metaxin-1 and -2, CHCHD3 and OPA1 (inner membrane/intermembrane space [IMS] proteins), Hsp70 (a matrix protein), and ChChd6 and DnaJC11 (two proteins of unknown location; Xie et al., 2007; Darshi et al., 2010). Genetic evidence suggested that IMMT-1, one of two mitofilin/IMMT homologues present in Caenorhabditis elegans, in concert with the outer membrane protein MOMA-1 and with ChChd3, acts in one pathway determining cristae morphology (Head et al., 2011). In addition, mitofilin was found to interact with DISC1, a mitochondrial protein involved in schizophrenia (Park et al., 2010). The functional roles of these interactions have been a matter of speculation.
A characteristic of CJs is that they are located almost exclusively at sites where the cristae meet the inner boundary membrane (IBM). Similar alterations of membrane curvature hardly occur internally at cristae membranes. The molecular mechanism underlying this positioning of CJ is largely unknown. One possible explanation is that CJ formation is controlled by unknown factors located in or at the outer membrane to which CJs are physically anchored. Here we investigate the role of individual domains of Fcj1 in the formation of CJ. We show that the C-terminal domain of Fcj1 mediates physical contact to the outer membrane via an interaction with the translocase of the outer membrane β-barrel proteins (TOB)/sorting and assembly machinery (SAM) complex. On the other hand, we show that the TOB/SAM complex affects cristae morphology and CJ formation. We propose that the interaction of the C-terminus of Fcj1 with the TOB/SAM complex provides a molecular basis for the positioning of CJ in the architecture of mitochondria.
RESULTS
Anchoring of Fcj1 to the inner membrane is important for Fcj1 function
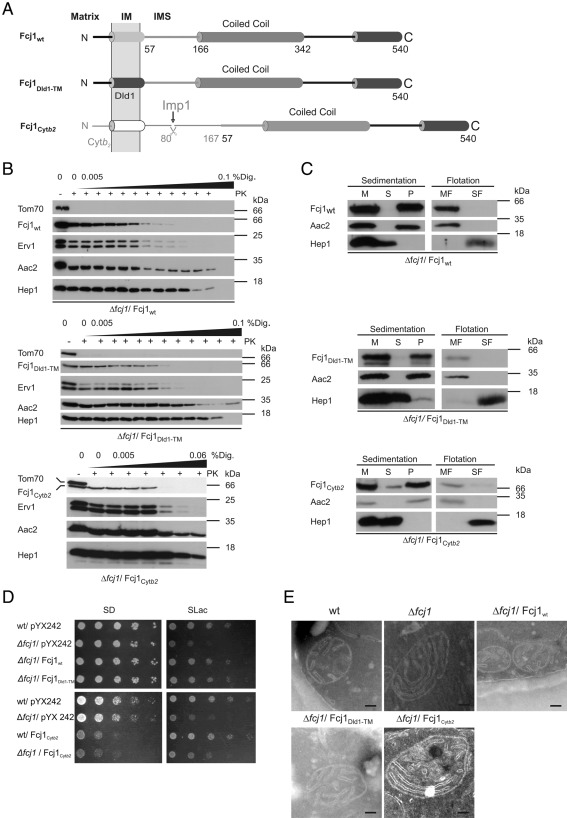
Several structural elements of Fcj1 can be distinguished. Fcj1 is a protein of ∼60 kDa molecular mass. It is anchored to the inner membrane by a hydrophobic segment close to the N-terminus, which faces the mitochondrial matrix (Figure 1A). The large hydrophilic part protruding into the IMS can be divided into at least two segments—the coiled-coil domain and the C-terminal domain. The latter domain is the only part of Fcj1 that is significantly conserved during evolution. To investigate the role of individual domains of Fcj1 in an initial step, we altered its transmembrane domain (Figure 1A and Supplemental Figure S1A). First, the putative dimerization motif AXXXG was disrupted by changing the amino acid sequence to AXXXL (Fcj1G52L). Second, the transmembrane segment of Fcj1 was replaced by that of Dld1 (Fcj1Dld1-TM), a protein with the same topology as Fcj1. Third, the N-terminus of Fcj1 was replaced by the N-terminus of cytochrome b2 (Cytb2) to yield Fcj1Cytb2. In contrast to the situation with regard to Fcj1, this part in Cytb2 is proteolytically cleaved in the IMS of mitochondria to release mature Cytb2 into the IMS. Therefore, the chimeric protein Fcj1Cytb2 after cleavage is expected to be present as a soluble protein in the IMS (Figure 1A).
FIGURE 1:
Anchoring of Fcj1 to the inner membrane is important for Fcj1 function. (A) Schematic representation of Fcj1 wild-type (Fcj1wt) and variants with altered N-terminal region: Fcj1DLD1-TM, replacement of Fcj1 transmembrane domain (TMD) by Dld1-TMD; Fcj1Cytb2, replacement of residues 1–57 of Fcj1 by residues 1–167 of cytochrome b2. (B) Submitochondrial localization of Fcj1 variants. Mitochondria isolated from Δfcj1 strains expressing the indicated Fcj1 variants were subjected to proteinase K treatment in the presence of increasing concentrations of digitonin (% Dig.). Further analysis by SDS–PAGE and immunoblotting using antibodies against Fcj1, Hep1 (matrix), Erv1 (intermembrane space), Aac2 (inner membrane), and Tom70 (outer membrane). (C) Membrane insertion of Fcj1 variants. Mitochondria from Δfcj1 strains expressing the indicated Fcj1 variants were subjected to carbonate extraction. Aliquots of untreated mitochondria (M), supernatant (S), and pellet (P) after sedimentation were analyzed by SDS–PAGE and immunoblotting using antibodies against Fcj1, Aac2, and Hep1. Membrane insertion was also tested by carbonate extraction, followed by flotation centrifugation. Equal amounts of membrane (MF) and soluble fraction (SF) were loaded and analyzed as described. (D) Expression of Fcj1wt and Fcj1Dld1-TM but not of Fcj1Cytb2 can rescue the growth phenotype of Δfcj1 on nonfermentable carbon source (SLac). Cells were grown to exponential phase on SLac at 30°C, consecutive fivefold dilution steps were made, and cells were spotted on SD-Leu and SLac-Leu plates. (E) Electron micrographs of mitochondria in chemically fixed whole yeast cells expressing the indicated Fcj1 variants. Cells were cultivated on selective SLac medium before fixation. Scale bars, 100 nm.
All variants were expressed in cells lacking Fcj1 (Δfcj1). The same levels of expression were observed, and neither variant affected the protein levels of other mitochondrial marker proteins (Supplemental Figure S2, A and B). All variants of Fcj1 were correctly located in the IMS, as revealed by proteolytic accessibility of digitonin-treated mitochondria isolated from Δfcj1 strains expressing the indicated variants (Figure 1B and Supplemental Figure S1B). On carbonate extraction followed by sedimentation or flotation of membranes, the Fcj1Dld1-TM and Fcj1G52L variants behaved like integral membrane proteins, similar to the Fcj1wt control (Figure 1C and Supplemental Figure S1C). Fcj1Cytb2 was efficiently, yet not completely, cleaved by the Imp1 peptidase (Supplemental Figure S3). Still, only a minor fraction was extracted with sodium carbonate, unlike Hep1, which was used as a control for soluble proteins. The major fraction behaved like the integral membrane protein ADP/ATP carrier (Aac2). This is most likely not due to the minor amount of unprocessed precursor protein present (see Supplemental Figure S3), but rather due to some other type of firm association of Fcj1Cytb2 with the membrane.
Neither inactivating the putative dimerization motif (Fcj1G52L) nor exchanging the transmembrane segment (Fcj1Dld1-TM) led to a growth defect on fermentable carbon source (SD; Figure 1D and Supplemental Figure S1D, left). Similarly, on a nonfermentable carbon source (SLac) a strain deficient in Fcj1 (Δfcj1) was fully rescued by these variants (Figure 1D and Supplemental Figure S1D, right). In contrast, expression of Fcj1Cytb2 did not rescue the growth defect of the Δfcj1 strain (Figure 1D, right) nor did it reduce the generation of respiratory deficient cells typical for the Δfcj1 strain (Table 1), suggesting the functional importance of anchoring Fcj1 to the inner membrane. Moreover, expression of Fcj1Cytb2 caused growth impairment on a fermentable carbon source in the Δfcj1 strain. This was also observed when Fcj1Cytb2 was expressed in a wild-type background, demonstrating a dominant-negative effect of this construct (Figure 1D, left). Taken together, these data indicate the functional importance of anchoring the entire pool of Fcj1 to the inner membrane by a transmembrane segment.
TABLE 1:
Formation of respiratory-deficient cells in S. cerevisiae expressing various Fcj1 variants.
| Strain | Respiratory-deficient cells (%) |
|---|---|
| Wild type | 1 |
| Δfcj1 | 17 |
| Δfcj1/ Fcj1wt | 2 |
| Δfcj1/ Fcj1DLD1-TM | 6 |
| Δfcj1/ Fcj1G52L | 5 |
| Δfcj1/Fcj1Cytb2 | 15 |
| Δfcj1/ Fcj1His | 3 |
| Δfcj1/Fcj1Δ166-342His | 8 |
| Δfcj1/ Fcj11-472 | 16 |
The percentages of respiratory-deficient cells as indicated by petite colony morphology (rho−/rho0 cells) on complete medium containing lactate supplemented with 0.1% glucose were determined (ntotal cells > 100) for the indicated strains.
The ultrastructure of mitochondria in cells expressing the Fcj1 variants in a Δfcj1 background was studied by electron microscopy. With Fcj1G52L and Fcj1Dld1-TM no alterations were observed compared with wild type (Figure 1E and Supplemental Figure S1E), and the number of CJs per mitochondrial section was not grossly affected (Table 2). Expression of Fcj1Cytb2 resulted in a reduction of the number of CJs per mitochondrial section to ∼50% compared with expression of Fcj1wt (Table 2). We attribute the remaining ability to form CJs to unprocessed molecules that are anchored to the inner membrane by the transmembrane segment of the Cytb2 presequence (Supplemental Figure S3). We also tested the effect of expressing these variants on mitochondrial morphology by fluorescence microscopy. Fcj1G52L and Fcj1Dld1-TM fully reverted the aberrant mitochondrial morphology, in contrast to Fcj1Cytb2, which did so only partially (Supplemental Figure S4). Considering these results together, we propose that the presence of a transmembrane segment in Fcj1 is important for full functionality but that a specific primary amino acid sequence of this part of Fcj1 is not critical.
TABLE 2:
Frequency of CJs in mitochondrial sections of whole cells of S. cerevisiae expressing various Fcj1 variants.
| Strain | Relative number of CJs per mitochondrial section (%) |
|---|---|
| Δfcj1/Fcj1DLD1-TM | 68 |
| Δfcj1/Fcj1G52L | 124 |
| Δfcj1/Fcj1Cytb2 | 46 |
| Δfcj1/Fcj1Δ166-342His | 4 |
| Δfcj1/Fcj11-472 | 9 |
Indicated strains were grown on nonfermentable carbon source, chemically fixed, and cryosectioned, and electron micrographs were analyzed. The strain Δfcj1/Fcj1wt was used as a control, and the frequency of CJs per mitochondrial section was set to 100%.
The coiled-coil domain of Fcj1 is crucial for formation of crista junctions
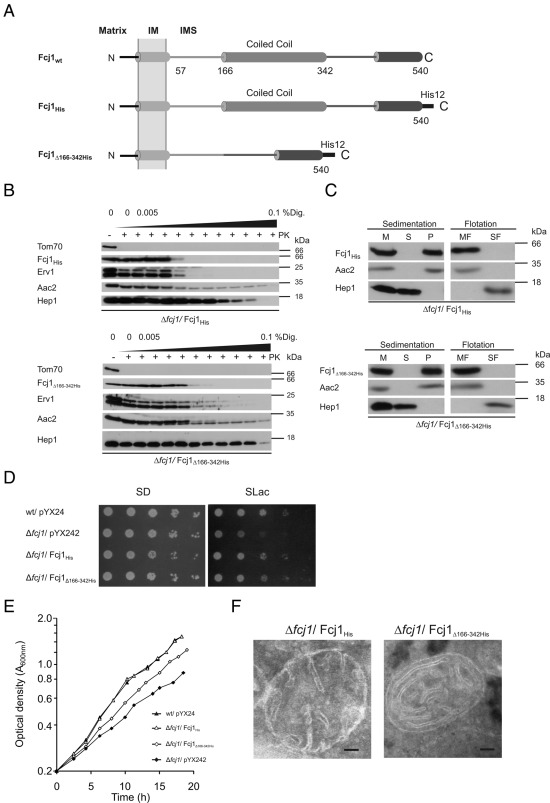
To assess the functional role of the coiled-coil domain of Fcj1, we expressed in the Δfcj1 strain a variant lacking the coiled-coil domain (Fcj1Δ166-342His, Figure 2A). Because the deleted region contained the epitope recognized by our antibody, we added a C-terminal dodecahistidine (His12) tag to this truncated construct and expressed a His12-tagged, full-length variant of Fcj1 (Fcj1His) as a control. All variants were localized to the IMS and integrated into or strongly associated with mitochondrial membranes (Figure 2, B and C), consistent with the presence of a transmembrane segment. Unlike the wild-type control (Fcj1His), Fcj1Δ166-342His rescued only partially the growth defect of the Δfcj1 strain on a nonfermentable (SLac) carbon source, as demonstrated by drop dilution growth assays (Figure 2D) and by monitoring the increase in optical density during exponential growth in liquid media (Figure 2E). This incomplete growth suppression could be caused by an increased loss of mitochondrial DNA. Indeed, the percentage of respiratory-deficient cells in the Fcj1Δ166-342His strain was higher than in the Fcj1His strain but lower than in the Δfcj1 strain (Table 1). Mitochondrial morphology was restored by Fcj1His but not by Fcj1Δ166-342His (Supplemental Figure S4). Cristae morphology in Fcj1Δ166-342His cells was very similar of that in Δfcj1 cells, characterized by concentric cristae stacks with no or very few CJs (Figure 2F). The number of CJs per mitochondrial section was strongly reduced to ∼4% of control (Table 2). Taken together, these results demonstrate the importance of the coiled-coil domain of Fcj1 for the formation of CJs. How the architecture of mitochondria is connected to functionality like growth is unclear. Similar observations were reported in a recent study in which a component of the MICOS/MINOS/MitOS complex, Aim13/Mcs19, was deleted, resulting in a reduced number of CJs but not in a reduced cellular growth rate of yeast cells (Harner et al., 2011).
FIGURE 2:
The coiled-coil domain of Fcj1 is essential for crista junction formation. (A) Schematic representation of Fcj1 wild-type (Fcj1wt) and the following variants: Fcj1His, C-terminally His12-tagged version of wild-type Fcj1; Fcj1Δ166-342His, C-terminal His12-tagged version of Fcj1Δ166-342His (deletion of coiled-coil domain). (B, C) Submitochondrial localization and membrane insertion of Fcj1 variants were performed as in Figure 1, B and C. (D, E) Expression of Fcj1Δ166-342His can partially rescue the growth phenotype of Δfcj1 on nonfermentable carbon source. (D) Drop dilution tests were performed as in Figure 1D. (E) Growth rates of indicated strains were monitored during exponential growth phase by determining the optical density at A600nm. The y-axis is represented on a logarithmic scale. (F) Electron micrographs of mitochondria, as in Figure 1E.
The conserved C-terminal domain of Fcj1 is essential for Fcj1 function
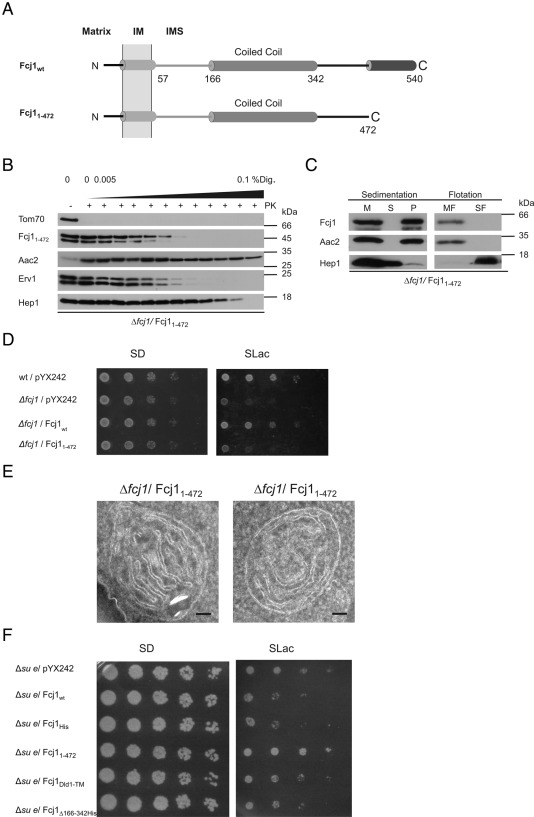
The role of the most-conserved region within Fcj1, its C-terminal domain, was addressed by expressing Fcj11-472 that lacks the C-terminal 68 amino acid residues. This variant was present in mitochondria at a similar level as Fcj1wt (Supplemental Figure S2A) and was correctly localized and integrated into the membrane (Figure 3, B and C). Fcj11-472 did not rescue the growth defect of the Δfcj1 strain on SLac (Figure 3D). In addition, the high level of respiratory-deficient cells formed during exponential growth (Table 1) and mitochondrial morphology (Supplemental Figure S4) were not rescued. Concentric cristae stacks with no or very few CJs were abundant (Figures 1E and 3E), as in Δfcj1 cells. The number of CJs per mitochondrial section was strongly reduced to ∼9% of control (Table 2). Overexpression of wild-type Fcj1 in a strain lacking subunit e of the F1FO ATP synthase (Δsu e) exerted a dominant-negative effect on growth on SLac (Figure 3F), consistent with the reported genetic interaction between Fcj1 and Su e (Rabl et al. 2009). This effect was observed upon expression of the variants described previously but not of Fcj11-472 (Figure 3F). Thus the C-terminal domain of Fcj1 appears to be required for the genetic interaction of Fcj1 with subunit e of the F1FO ATP synthase, confirming the necessity of this domain for Fcj1 function. In conclusion, we propose that the conserved C-terminal domain of Fcj1 is crucial for formation of stable CJ. This domain is unique compared with the other regions in Fcj1 investigated, as it is the only one required for the genetic interaction of Fcj1 with the F1FO ATP synthase.
FIGURE 3:
The conserved C-terminal domain of Fcj1 is essential for respiratory growth and crista junction formation. (A) Schematic representation of Fcj1wt and Fcj11-472, truncation of C-terminal amino acid residues 473–540. (B, C) Submitochondrial localization and membrane insertion of Fcj11-472 variant was performed as in Figure 1, B and C, respectively. (D) Expression of Fcj11-472 does not rescue the growth phenotype of Δfcj1 on nonfermentable carbon source. Drop dilution tests were performed as in Figure 1D. (E) Electron micrographs of Fcj11-472 mitochondria, as in Figure 1E. F, Overexpression of Fcj1 variants in cells deficient in subunit e of the F1F0 ATP synthase (Δsu e). Drop dilution tests were performed as in Figure 1D.
Physical interaction of the C terminus of Fcj1 with the TOB55 complex
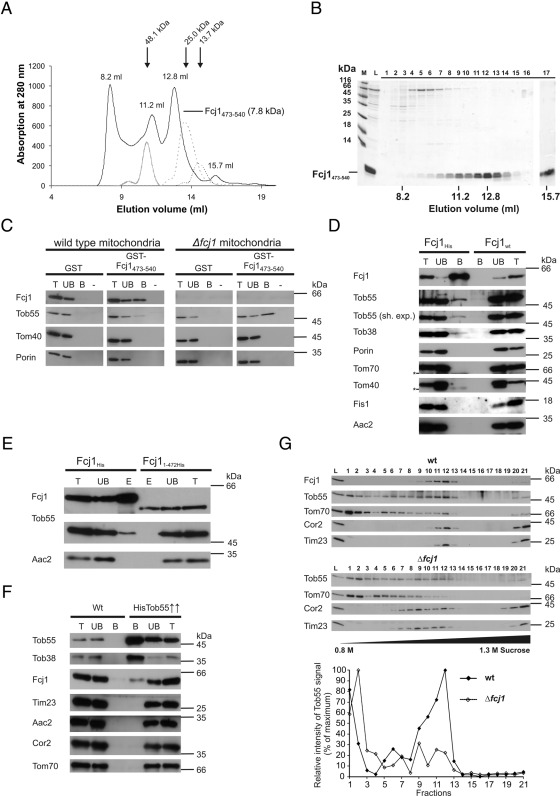
To study the role of the C-terminal domain of Fcj1, we expressed Fcj1473-540 as a fusion protein with glutathione S-transferase (GST) in bacteria. The protein was purified by binding to glutathione Sepharose beads and, after cleavage from the GST moiety, analyzed by size exclusion chromatography (Figure 4, A and B). Most of the protein was recovered as oligomeric complexes corresponding to tetramers to hexamers. This is consistent with the homotypic mode of protein–protein interaction of Fcj1 reported earlier and the observation of dimeric, trimeric, or tetrameric Fcj1 complexes upon solubilization of mitochondria in Triton X-100 (Rabl et al. 2009). In accordance with these findings, purified Fcj1473-540 efficiently bound full-length Fcj1 from a mitochondrial lysate (Figure 4C, left). Thus the C-terminal domain appears to play an important role in the formation of homo-oligomers of Fcj1.
FIGURE 4:
The C-terminal domain of Fcj1 interacts with the TOB complex. (A, B) The C-terminal domain of Fcj1 forms oligomers. (A) Purified recombinant C-terminal domain of Fcj1 (amino acid residues 473–540) was subjected to size exclusion chromatography and the eluate monitored by recording the absorption at 280 nm (continuous line). Protein markers for sizing, ovalbumin (48.1 kDa), chymotrypsinogen (25.0 kDa), and ribonuclease A (13.7 kDa; broken lines) are indicated by arrows. (B) Fractions (1–17) of the eluate were subjected to SDS–PAGE and staining with Coomassie blue. L, load (purified C-terminal domain); M, markers. Fraction 17 corresponding to 15.7 ml was trichloroacetic acid precipitated, representing ∼25-fold higher amounts compared with the other fractions. (C) The C-terminal domain of Fcj1 interacts with full-length Fcj1 and Tob55. Purified N-terminally GST-tagged C-terminal domain of Fcj1 (GST-Fcj1473-540) and recombinant GST (control) were bound to glutathione Sepharose beads. Mitochondria from wild-type or Δfcj1 cells were lysed with detergent and subjected to affinity chromatography. Fractions corresponding to total (T, 5%), unbound (UB, 5%), and bound (B, 100%) material were analyzed by SDS–PAGE and immunoblotting with antibodies raised against the indicated proteins. M, mock treatment, no lysate applied. (D) Fcj1 interacts with Tob55. Mitochondria from cells expressing Fcj1His and Fcj1wt (control) were lysed with detergent and subjected to Ni-NTA affinity chromatography. Further analysis as in C using indicated antibodies. For Tob55, long and short (sh.exp.) exposures are shown. Asterisks indicate unspecific cross-reactions. (E) Fcj1 lacking the C-terminal domain shows no interaction with Tob55. Mitochondria from cells expressing Fcj11-472His and Fcj1His (control) were lysed with detergent and subjected to Ni-NTA affinity chromatography. Further analysis as in C using indicated antibodies. (F) Tob55 interacts with Fcj1. Mitochondria from cells overexpressing His-tagged Tob55 (Tob55↑↑) and from wild-type (control) were lysed with detergent and subjected to Ni-NTA affinity chromatography. Further analysis as in C. (G) A subpopulation of the TOB complex comigrates with Fcj1. Submitochondrial vesicles from wild-type (top) and Δfcj1 (middle) mitochondria were subjected to sucrose density gradient centrifugation as described in a recent study (Harner et al., 2011). Equal volumes of each fraction were analyzed by SDS–PAGE and immunoblotting with antibodies raised against the indicated proteins. The relative intensities for Tob55 were quantified and plotted (bottom).
We then asked whether this domain would specifically interact with other proteins present in mitochondrial detergent lysates. Indeed, we found that Tob55 specifically binds to the C-terminal domain of Fcj1 (Figure 4C). Of interest, binding of Tob55 was enhanced when lysates were used from mitochondria lacking Fcj1 (Figure 4C, right). Apparently, binding of Tob55 to the C-terminus of Fcj1 does not depend on the presence of full-length Fcj1. This suggests that the three interaction partners—the C-terminal domain of Fcj1, full-length Fcj1, and the TOB complex—compete for binding to each other in such a way that possible binary interactions are promoted, whereas formation of a trimeric complex is less favored.
For further analysis of the interaction between Fcj1 and the TOB complex we performed nickel-nitriloacetic acid (Ni-NTA) affinity purification from isolated mitochondria harboring a His-tagged version of Fcj1. Tob55 and Tob38, both subunits of the TOB complex, were copurified, whereas control outer and inner membrane proteins were not (Figure 4D). Performance of these experiments with mitochondria isolated from a strain expressing nontagged Fcj1 did not show an interaction of Fcj1 with the TOB complex. When mitochondria expressing a His-tagged variant of Fcj11-472 were used, the interaction to Tob55 was also lost, corroborating our earlier results (Figure 4E). We cannot, however, fully exclude the possibility that the observed lack of Tob55 is partly caused by a somewhat reduced solubility of the Fcj11-472His variant. Conversely, when using isolated mitochondria from a strain overexpressing His-tagged Tob55 for Ni-NTA affinity purification, we observed a specific copurification of Fcj1 (Figure 4F). In addition, we subjected detergent-solubilized, isolated, wild-type mitochondria to glycerol gradient density centrifugation. Subunit Tob55 of the TOB/SAM complex was observed to comigrate with a major subpopulation of a high–molecular weight Fcj1-containing complex (Supplemental Figure S5A). We also prepared submitochondrial vesicles from wild-type mitochondria, fractionated them by sucrose density gradient ultracentrifugation, and analyzed the obtained fractions by SDS–PAGE and immunodecoration following a procedure recently described in Harner et al. (2011). We observed that the TOB/SAM complex was present not only in the outer membrane fractions (1–3), but also in fractions containing Fcj1 in the middle of the gradient (Figure 4G, top and bottom), consistent with a recent study (Harner et al., 2011). These last-named fractions were shown to contain the MICOS complex (Harner et al., 2011). When we performed the same analysis but with submitochondrial vesicles from mitochondria lacking Fcj1 we noted that the level of the last-named subpopulation of the TOB/SAM complex was markedly reduced (Figure 4G, middle and bottom), demonstrating that in the absence of Fcj1 the association of the TOB/SAM complex with the inner membrane is strongly weakened or lost. In summary, several lines of evidence demonstrate a physical interaction between Fcj1 and the TOB complex.
Fcj1 plays a minor role in the biogenesis of β-barrel proteins
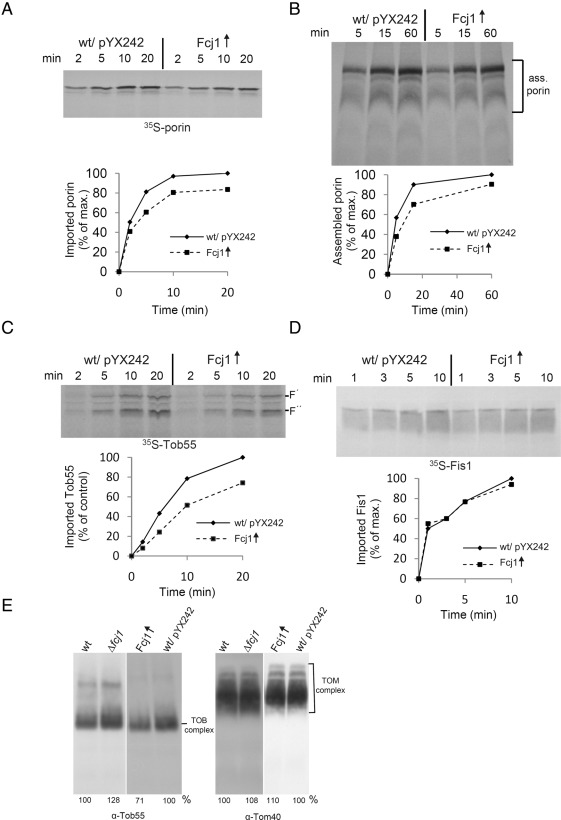
Prompted by the observed interaction of Fcj1 with Tob55, we tested whether Fcj1 has a role in the biogenesis of β-barrel proteins. We determined whether down-regulation of Tob38 or overexpression of Tob55 affected the protein levels of Fcj1. This was not the case (Supplemental Figure S5BC). In addition, deletion of Fcj1 and overexpression of Fcj1 or variants thereof did not affect the levels of Tob55 (Supplemental Figure S2A). In further experiments we analyzed the import of β-barrel proteins into isolated mitochondria. Deletion of Fcj1 affected neither the protein import of porin, Tob55, or Tom40 (Supplemental Figure S5, D F, and G) nor the assembly kinetics of porin or Tom40 (Supplemental Figure S5, E and G). Overexpression of Fcj1 led to a modest reduction in the import and assembly of porin and of import of Tom40 and Tob55 but not of Fis1 (Figure 5, A–D, and Supplemental Figure S5H). Furthermore, assembly of the TOB complex was only slightly affected in both mitochondria deficient in Fcj1 and mitochondria overexpressing Fcj1; the amount of assembled TOM complex was unaffected (Figure 5E). We conclude that overexpression of Fcj1 moderately impairs the function of the TOB complex but that Fcj1 does not appear to play a prominent role in the biogenesis of β-barrel membrane proteins.
FIGURE 5:
Overexpression of Fcj1 has a minor effect on the biogenesis of β-barrel proteins. (A, B) Overexpression of Fcj1 mildly impairs porin biogenesis. Mitochondria isolated from wild-type strain (Wt/pYX242) and or from the Δfcj1/pYX242-Fcj1wt strain overexpressing Fcj1 (Fcj1↑) were incubated with radiolabeled precursor of porin for the indicated time periods. (A) Samples were treated with proteinase K, and mitochondria were analyzed by SDS–PAGE and autoradiography (top), and imported porin was quantified (bottom). The amount of imported porin into wild-type mitochondria after the longest incubation period was set to 100%. (B) After import as in A, samples were solubilized with 1% digitonin and analyzed by BN-PAGE and autoradiography (top). Assembled porin complexes as indicated were quantified (bottom). (C) Overexpression of Fcj1 mildly impairs Tob55 biogenesis. Mitochondria as in A were incubated with radiolabeled precursor of Tob55 for the indicated time periods. Further analysis as in A. Two characteristic proteolytic fragments of imported Tob55, with molecular masses of ∼30 and 25 kDa, are indicated as F′ and F″, respectively (Habib et al., 2005). The bands corresponding to F′ were quantified (bottom). (D) Overexpression of Fcj1 does not impair biogenesis of Fis1. Mitochondria as in A were incubated with radiolabeled precursor of Fis1. The levels of Fis1 present in carbonate pellets were analyzed by SDS–PAGE and autoradiography (top), and imported Fis1 was quantified (bottom). (E) Overexpression of Fcj1 mildly impairs assembly of the TOB complex. Mitochondria isolated from the indicated strains were solubilized in digitonin and analyzed by BN-PAGE. TOB complex (left) and TOM complex (right) were visualized by immunoblotting with antibodies against Tob55 or Tom40, respectively. The relative amounts relative to total mitochondrial protein are indicated at the bottom (wild-type controls were set to 100%).
The TOB/SAM complex is involved in determining cristae morphology and CJ formation
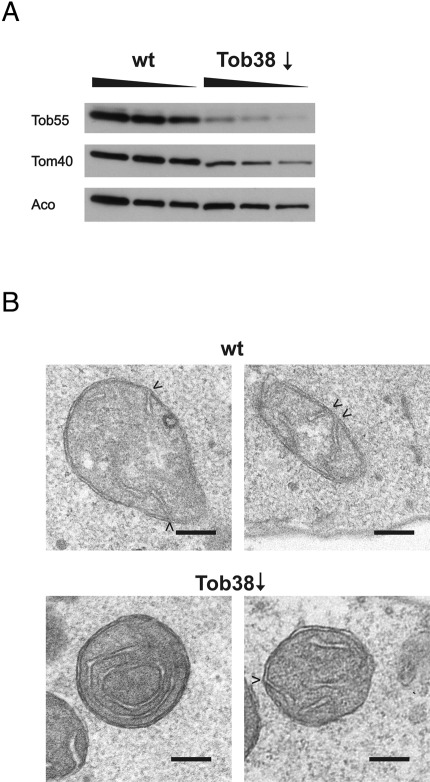
To address whether the TOB/SAM complex influences cristae morphology, we down-regulated Tob38, an essential component of the TOB/SAM complex, and determined mitochondrial ultrastructure in whole cells by electron microscopy. The levels of Tob55 in these cells were markedly reduced, whereas the levels of the matrix protein aconitase were hardly affected (Figure 6A). The levels of Tom40 were reduced, consistent with the fact that Tom40 is a β-barrel protein. On down-regulation of Tob38 mitochondrial ultrastructure appeared altered, as in some instances irregularly shaped, concentric cristae membranes resembling mitochondria lacking Fcj1 were observed (Figure 6B, bottom). Such alterations were not observed when the wild-type strain was cultured under the same conditions (Figure 6B, top). To further evaluate the effect of Tob38 down-regulation, we determined the ratio of inner membrane to outer membrane in mitochondrial sections, as it was reported that this ratio is increase about twofold in cells lacking Fcj1 (Rabl et al., 2009). In Tob38 down-regulated cells this ratio was significantly increased as compared with the wild-type control (2.0 ± 0.4 [Tob38↓; n = 12] vs. 1.6 ± 0.2 [wild type; n = 10]; *p = 0.012 according to Student's t test). Next we determined the number of CJs per mitochondrial section. Down-regulation of Tob38 led to a moderate reduction of this value from 1.6 (wild type; 45 CJs in 29 mitochondrial sections) to 1.2 (Tob38↓; 54 CJs in 45 mitochondrial sections). In summary, down-regulation of the TOB complex leads to altered cristae morphology and a slight reduction of the number of CJs.
FIGURE 6:
Downregulation of the TOB complex alters cristae morphology and impairs CJ formation. Wild-type cells and cells expressing Tob38 under the control of the GAL10 promoter were grown on lactate containing 0.1% glucose for down-regulation of the TOB complex. (A) Total cell extracts of wild-type and GAL10-Tob38 strains were subjected to SDS–PAGE and analyzed by immunoblotting with antibodies against the indicated proteins. For each sample increasing amounts were loaded as indicated. (B) Downregulation of Tob38 led to altered cristae morphology. Electron micrographs of indicated strains, as in Figure 1E, after growth in lactate medium supplemented with 0.1% glucose.
DISCUSSION
The mutational analysis of Fcj1 reported here revealed a number of insights into the function of this protein, which is known for its role in determining the architecture of mitochondria and in maintaining cristae and CJ structure. Three modules of Fcj1 were analyzed: 1) the membrane anchor, 2) the coiled-coil domain, making up the major part of Fcj1, and 3) the C-terminal domain, which is the only part conserved in amino acid sequence among all mitofilins. The results of this part of the study show the following. 1) The transmembrane module is important to put and keep the protein in place, but a major specific function beyond this was not found. 2) The coiled-coil domain is not important for the genetic interaction of Fcj1 with the F1FO ATP synthase but is required for CJ formation. In addition, in the absence of this domain Fcj1 is still partially functional regarding its ability to complement the growth phenotype of Δfcj1 on a nonfermentable carbon source. 3) Several lines of evidence assign an important function to the most-conserved part of Fcj1, the C-terminal domain. The presence of this domain is required for the formation of CJs. It is also required for the ability of Fcj1 to exert a dominant-negative growth effect on cells lacking subunit e (Su e) of the F1FO ATP synthase. Apparently, the reported antagonism between Fcj1 and Su e (Rabl et al., 2009) depends on the C-terminal domain of Fcj1. In addition, this domain is sufficient to allow formation of homo-oligomers, consistent with the reported ability of full-length Fcj1 to engage in homotypic interactions (Rabl et al., 2009). The important role of the C-terminal domain may thus be linked to its ability to form homo-oligomers and to confer this ability on Fcj1. It is possible that the higher-order homo-oligomeric structure of Fcj1 mediated by its C-terminal domain is the structural basis for the construction of the narrow tubules that form the CJs.
The function of the C-terminal domain is particularly interesting since it is also involved in the interaction with the TOB complex. This protein–protein interaction is supported by several lines of evidence. The isolated C-terminal domain fused to GST was able to bind to Fcj1 as well as to the TOB complex present in detergent-solubilized, wild-type mitochondria. The amount of TOB complex bound was increased when mitochondrial lysates of the Δfcj1 strain were analyzed. This suggests that the isolated C-terminal domain of Fcj1 binds to other Fcj1 molecules and to the TOB complex in a competitive manner. Affinity chromatography using tagged Fcj1 or Tob55 confirmed their physical interaction. Moreover, an Fcj1-containing protein complex was detected to partially comigrate with the TOB complex in a linear glycerol density gradient separating protein complexes of detergent-solubilized, wild-type mitochondria. The interaction of Fcj1 with the TOB complex was further corroborated by cofractionation of both components in submitochondrial vesicles on a sucrose density gradient as described previously (Harner et al., 2011). In addition, upon deletion of Fcj1, this subpopulation of the TOB complex lost its association with inner membrane vesicles. The observation that Fcj1 interacts with the TOB complex is in accordance with reports on a physical interaction of the mammalian Fcj1 orthologue mitofilin/IMMT with subunits of the mammalian SAMM50 complex (Xie et al., 2007; Darshi et al., 2010).
Our study shows that the C-terminal domain of Fcj1 serves as the domain interacting with the TOB complex. What could be the functional role of this interaction? One possibility was that Fcj1 has a so-far-unknown role in the import of β-barrel proteins. However, we did not find an indication for a major or direct role of Fcj1 in the biogenesis or function of the TOB complex. Another rationale for explaining the physical interaction between Fcj1 and the TOB complex relates to the observation that CJs are present at sites where the cristae meet the outer membrane. Internal branches of cristae that have similarities to CJs with respect to the general appearance of negative membrane curvatures are very rarely observed in wild-type cells. On the other hand, overexpression of Fcj1 was reported to lead to the formation of such internal branches (Rabl et al., 2009). Three recent studies reported that Fcj1 in S. cerevisiae is part of a large protein complex—MICOS (Harner et al., 2011), MINOS (von der Malsburg et al., 2011), and MitOS (Hoppins et al., 2011)—making contact sites to the outer membrane by interaction with Ugo1, the TOB complex, and with Tom40 (Harner et al., 2011; von der Malsburg et al., 2011). In context with the data presented here as well as with these recently published studies we suggest that the interaction between Fcj1 and proteins in the outer membrane serves to stabilize CJs at this particular location. Thereby they create contact sites that are visible by electron microscopy in isolated mitochondria subjected to hyperosmotic treatment, leading to retraction of the IBM from the outer membrane, revealing sites of adhesion (Hackenbrock, 1968). Taken together, the results suggest that Fcj1 is part of a large protein complex spanning from the inner membrane to the outer membrane. How all these components are arranged with respect to each other is an open question. It is not unexpected that modulating the level of a number of these components affects cristae morphology or impairs CJ formation. However, suspending the Fcj1–TOB interaction by deletion of the C-terminal domain of Fcj1 is sufficient to impair CJ formation and cristae morphology, pointing to a functional role of TOB/SAM and the C-terminus of Fcj1 in determining the architecture of cristae. On the other hand, depletion of the TOB complex leads to a reduced number of CJs and altered cristae morphology. We cannot exclude that the concomitant depletion of Tom40 contributes to this effect. However, this seems unlikely, as Tom40 levels were not reduced to an extent that might affect protein import into various mitochondrial compartments (Figure 6A and Supplemental Figure S5B). Furthermore, the view that the TOB/SAM complex plays a role in determining cristae structure is in agreement with a recent study in mammalian cells demonstrating that SAM mutants but not Tom40 and VDAC/porin mutants adversely affected cristae morphology (Ott et al., 2012). Still, a role of certain outer membrane proteins in crista morphogenesis seems quite possible, as, for example, Fcj1 interacts also with Ugo1, a protein involved in mitochondrial fusion (Harner et al., 2011). The biochemical mechanism by which the TOB/SAM complex and Ugo1 determine the morphology of the crista membrane remains to be elucidated.
The TOB complex has apparently evolved a second function in addition to its established one—the import of β-barrel proteins into the outer membrane. It is unclear when during evolution the TOB complex was selected for linking CJs to the outer membrane. Possibly, this is related to the fact that, in evolutionary terms, the insertion of β-barrel proteins is an old invention. Not only is the central component of the TOB complex, Tob55, well conserved among eukaryotes, but obviously it is derived from the Omp85 proteins of bacteria, which have the corresponding function of insertion of β-barrel proteins into the outer membrane (Paschen et al., 2005; Dolezal et al., 2006). Cristae-like membrane infoldings are observed in some bacteria and mitochondria of eukaryotes and, of importance, were proposed to be present early during the endosymbiotic origin of eukaryotes (Cavalier-Smith, 2007). Thus it seems possible that anchoring of ancestral CJ-like structures to the outer membrane occurred early in evolution. Future studies will have to characterize the complete machinery determining cristae morphology and contact sites and how these complexes are regulated.
MATERIALS AND METHODS
Yeast strains, plasmids, media, and growth conditions
Yeast strains, plasmids, and primers used in this study are summarized in the Supplemental Information. Culturing of yeast strains was performed using standard methods (Sherman et al., 1986) at 30°C on complete liquid media containing 2% (vol/vol) lactate. Strains containing plasmids (Supplemental Tables S1 and S2) were grown on selective liquid media containing 2% (vol/vol) lactate supplemented with 0.1% (wt/vol) glucose. Drop dilution assays were performed to determine the growth phenotype. Cells were grown on liquid selective lactate medium at 30°C to exponential phase adjusted to 0.4 A578nm/ml and subjected to consecutive fivefold dilution steps. We spotted 3-μl aliquots of each dilution on glucose- and lactate-selective plates, and plates were incubated for 2 d (glucose) and 3 d (lactate) at 24°C. In addition, we monitored the increase in optical density at A600nm during exponential growth in liquid media. For determination of respiratory growth competence, indicated strains were grown on nonfermentable carbon source; during the exponential growth phase, cells were plated on agar plates with complete media containing 2% (vol/vol) lactate supplemented with 0.1% (wt/vol) glucose. The percentage of respiratory-deficient cells as indicated by petite colony morphology (e.g., rho−/rho0 cells) was calculated (ntotal cells > 100).
Construction of Fcj1 variants
Details for generating Fcj1 variants are given in the Supplemental Information. Oligonucleotide sequences are given in Supplemental Table S3.
Submitochondrial localization of Fcj1 variants
Yeast total cell extracts were prepared by alkaline lysis as previously described (Herlan et al., 2003). Mitochondria were isolated according to standard procedures (Herrmann et al., 1994) and diluted to a final concentration of 1 mg/ml in HS buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 0.6 M sorbitol). Mitochondria (1.9 mg) were mixed with increasing amounts of digitonin (0–1.0% [wt/vol]) and proteinase K (100 μg/ml) and incubated for 30 min on ice. Digestion of proteins was stopped by adding 1 mM phenylmethylsulfonyl fluoride (PMSF) for 5 min on ice, and cells were reisolated by centrifugation (13,000 × g, 10 min, 4°C) and washed once with 1 ml of HS buffer containing 1 mM PMSF. Proteins were precipitated with trichloroacetic acid, washed once with ice-cold acetone, and dissolved in Laemmli gel loading buffer. All samples were analyzed by SDS–PAGE and immunoblotting.
Analysis of membrane insertion
To extract peripherally bound membrane proteins, we diluted mitochondria to a final concentration of 1 mg/ml in HS buffer. Equal volume of freshly prepared 0.2 M sodium carbonate was added, and the sample was incubated for 30 min on ice. Membrane and soluble fractions were then separated by centrifugation (90,000 × g, 30 min, 4°C) and analyzed by SDS–PAGE and immunoblotting. For membrane floating, carbonate extraction was performed as described. After 30 min of incubation the mitochondria/carbonate solution was mixed with 20 mM HEPES buffer containing 2.4 M sucrose to a final concentration of 1.7 M sucrose and overlaid with a sucrose step gradient containing 0.25, 1.4, and 1.6 M sucrose and centrifuged as described. Bottom (soluble) and top (membrane) fractions were analyzed by SDS–PAGE and immunoblotting.
Electron microscopy
Transmission electron microscopy (TEM) on chemically fixed, cryosectioned yeast cells using the Tokuyashu method (Figures 1–3 and Supplemental Figure S1) was performed as previously described (Vogel et al., 2006; Rabl et al., 2009). TEM for Figure 6 was performed as described in Harner et al. (2011).
Fluorescence microscopy
Strains were transformed with pVTU100-mtGFP expressing mitochondrial targeted green fluorescent protein (Westermann and Neupert, 2000). Cells were grown in selective medium (lactate) containing 0.2% glucose to exponential phase and analyzed by standard fluorescence microscopy. From 50 to 150 cells were classified for each strain.
Protein expression and purification of Fcj1473-540
An overnight culture of BL21 (DE3) pLysS Escherichia coli cells (Stratagene, Santa Clara, CA), transformed with pGEX-Fcj1473-540, was diluted at 1:40 ratio in 3 l of LB-Medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (33 μg/ml) and grown to OD595 of 0.3 at 37°C. Cells were shifted to 24°C and grown to OD595 of 0.6, and expression was induced by addition of 0.1 mM isopropyl-β-d-thiogalactoside. After 3 h of expression, cells were harvested by centrifugation, resuspended in 40 ml of buffer A (300 mM NaCl, 1 mM PMSF, 1 mM EDTA, 20 mM Tris/HCl, pH 7.0) and disrupted by sonication. After centrifugation the supernatant was incubated for 3 h with glutathione (GSH) Sepharose beads (GE Healthcare, Piscataway, NJ) at 4°C for protein binding. After removal of unbound material, beads were washed with 120 ml of buffer A and resuspended in 30 ml of the same buffer. Recombinant protein was eluted from glutathione beads by cleavage with PreScission Protease at 4°C overnight.
GST-Fcj1473-540 affinity chromatography
For affinity chromatography GSH-GST/Fcj1473-540 beads were prepared. A 50-ml amount of cultured BL21 (DE3) pLysS E. coli cells expressing GST-Fcj1473-540 was sonicated in 1 ml of buffer GST-A (30 mM HEPES, pH 7.4, 150 mM KAc, 2 mM ε-amino-N-caproic acid,1 mM PMSF, 0.5 mM EDTA). After centrifugation (16,000 × g, 15 min, 4°C) the supernatant was incubated for 30 min at 4°C with 50 μl of GSH Sepharose beads (GE Healthcare) equilibrated in buffer GST-A containing 0.05% Triton X-100. Beads were washed five times in 1 ml of buffer GST-A containing 0.05% Triton X-100. Isolated mitochondria from wild-type or Δfcj1 strains were solubilized at a Triton X-100/protein (wt/wt) ratio of 1 in solubilization buffer (30 mM HEPES, pH 7.4150 mM KAc, 2 mM ε-amino-N-caproic acid, 1 mM PMSF, 0.5 mM EDTA, protease complete inhibitor cocktail, 1% Triton X-100). After a clarifying spin (57,000 × g, 20 min, 4°C) equilibrated GSH-GST/Fcj1473-540 beads were incubated with the supernatant and an equal volume of buffer GST-A for 30 min at 4°C and washed three times in 1 ml buffer of GST-A containing 0.05% Triton X-100. Proteins were eluted with Laemmli gel loading buffer preheated to 95°C.
Protein purification by Ni-NTA affinity chromatography
For Ni-NTA purification, isolated mitochondria were solubilized in NTA-solubilization buffer containing 30 mM HEPES, pH 7.4, 150 mM KAc, 20 mM imidazole, 1 mM PMSF, protease complete inhibitor cocktail, and 1% Triton X-100 on ice for 30 min. After a clarifying spin the supernatant was incubated for 30 min on ice with 50 μl of washed Ni-NTA beads slurry. After four washing steps in buffer containing 30 mM HEPES, pH 7.4, 150 mM KAc, 30 mM imidazole,1 mM PMSF, and 0.05% Triton X-100, protein was eluted with Laemmli gel loading buffer containing 300 mM imidazole.
Blue native gel electrophoresis and import of proteins
Mitochondria were resuspended in import buffer (250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 3% (wt/vol) fatty acid–free bovine serum albumin, 10 mM 3-(N-morpholino)propanesulfonic acid, 2 mM ATP, 2 mM NADH, pH 7.2) to a protein concentration of 0.5 mg/ml. Samples, which were analyzed further by blue native (BN)-PAGE, were additionally supplemented with 5 mM creatine phosphate and 0.1 mg/ml creatine kinase. Import of precursor proteins was initiated by addition of reticulocyte lysate (5–10% [vol/vol)]) containing 35S-labeled protein, and reaction mixtures were incubated for various time periods at 25°C. Import reaction was stopped by adding proteinase K (50 μg/ml) or cold SEM buffer containing 80 mM KCl and 2 mM PMSF. The protease was inhibited after 15 min of incubation on ice by addition of 2 mM PMSF and additional incubation for 15 min on ice. Mitochondria were finally harvested by centrifugation (36,000 × g, 15 min, 4°C) and resuspended in either 30 μl of 2× Laemmli gel loading buffer (for further SDS–PAGE analysis) or 30 μl of BN-PAGE solubilization buffer (0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, 1 mM PMSF, 20 mM Tris, 1% [wt/vol] digitonin, pH 7.4) for further analysis by BN-PAGE according to Schägger (2001). Isolated mitochondria (40–100 μg) were resuspended in 30 μl of BN-PAGE solubilization buffer on ice for 15 min. After a clarifying spin (37,000 × g, 15 min at 2°C) solubilized material was transferred into a new tube and mixed with 3 μl of 10× loading dye (5% [wt/vol] Coomassie blue G, 500 mM ε-amino-N-caproic acid, 100 mM bis-Tris, pH 7.0). Samples were loaded on the precooled 6–13% native gradient gel and run in a cold room (4°C).
Size exclusion chromatography
Fcj1473-540 purified as described was subjected on Superdex 75 size exclusion column (GE Healthcare) using 20 mM Tris/HCl, pH 7.0, and 150 mM NaCl as running buffer. Protein markers (200 μg) were dissolved in running buffer and analyzed under the same conditions. Fractions were analyzed by SDS–PAGE and Coomassie staining. Markers were detected by absorption spectroscopy at 280 nm.
Density gradient centrifugation
Isolated wild-type mitochondria (1 mg) were solubilized for 30 min on ice at a digitonin/protein ratio of 2:1 in solubilization buffer 1 (30 mM HEPES, pH 7.4, 150 mM KAc, 10% [vol/vol] glycerol, 2 mM ε-amino-N-caproic acid, 1 mM PMSF, 1 mM EDTA). After a clarifying spin (20 min, 57,000 × g, 4°C) the supernatant was loaded on a linear 10–50% (vol/vol) glycerol gradient in a buffer containing 20 mM HEPES, pH 7.4, and 150 mM KAc. Protein complexes were separated by ultracentrifugation (4 h, 250,000 × g, 4°C), and 500-μl fractions were collected and analyzed by SDS–PAGE. Subfractionation of mitochondrial vesicles by sucrose gradient ultracentrifugation was performed as previously described by Harner et al. (2011).
Supplementary Material
Acknowledgments
We are grateful to Werner Kühlbrandt and Achilleas Frangakis for access to electron microscopy equipment, Michael Zick and Kai Hell for inspiring scientific discussions, and Gabi Ludwig, Simone Grau, Christiane Kotthoff, Christian Bach, and Friederike Joos for excellent technical assistance. This work was supported by the Cluster of Excellence Macromolecular Complexes at the Goethe University Frankfurt DFG Project EXC 115 (M.B., K.E., A.R.), SFB 594 (C.K., R.R., W.N., A.R.), and DFG Project RA 1028/2-2 (J.D., D.R.).
Abbreviations used:
- BN
blue native
- CJ
crista junction
- IBM
inner boundary membrane
- IMS
intermembrane space
- SAM
sorting and assembly machinery
- TOB
translocase of outer membrane β-barrel proteins
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0831) on April 11, 2012.
The authors declare no conflict of interest.
REFERENCES
- Alkhaja AK, et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The chimaeric origin of mitochondria: photosynthetic cell enslavement, gene-transfer pressure, and compartmentation efficiency. In: Martin WF, Müller M, editors. Origin of Mitochondria and Hydrogenosomes. Berlin: Springer-Verlag; pp. 161–200. [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2010;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Bonilla E, Zeviani M, Nakagawa M, DeVivo DC. Mitochondrial myopathies. Ann Neurol. 1985;17:521–538. doi: 10.1002/ana.410170602. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. The Cell. Philadelphia: WB Saunders; Mitochondria; pp. 410–468. [Google Scholar]
- Frey TG, Mannella CA. The internal structure of mitochondria. Trends Biochem Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. J Biol Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. [DOI] [PubMed] [Google Scholar]
- Hackenbrock CR. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci USA. 1968;61:598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head BP, Zulaika M, Ryazantsev S, van der Bliek AM. A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol Biol Cell. 2011;22:831–841. doi: 10.1091/mbc.E10-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Vogel F, Bornhövd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella CA. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta. 2006;1762:140–147. doi: 10.1016/j.bbadis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Mannella CA, Marko M, Penczek P, Barnard D, Frank J. The internal compartmentation of rat-liver mitochondria: tomographic study using the high-voltage transmission electron microscope. Microsc Res Tech. 1994;27:278–283. doi: 10.1002/jemt.1070270403. [DOI] [PubMed] [Google Scholar]
- Mannella CA, Pfeiffer DR, Bradshaw PC, Moraru II, Slepchenko B, Loew LM, Hsieh CE, Buttle K, Marko M. Topology of the mitochondrial inner membrane: dynamics and bioenergetic implications. IUBMB Life. 2001;52:93–100. doi: 10.1080/15216540152845885. [DOI] [PubMed] [Google Scholar]
- Munn EA. The Structure of Mitochondria. New York: Academic Press; 1974. [Google Scholar]
- Nicastro D, Frangakis AS, Typke D, Baumeister W. Cryo-electron tomography of Neurospora mitochondria. J Struct Biol. 2000;129:48–56. doi: 10.1006/jsbi.1999.4204. [DOI] [PubMed] [Google Scholar]
- Ott C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, Nguyen MD, Han SS, Suh PG, Park SK. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with mitofilin. Proc Natl Acad Sci USA. 2010;107:17785–17790. doi: 10.1073/pnas.1004361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Neupert W, Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey T. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Ellisman MH, Fox DA. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol Vis. 2003;9:60–73. [PubMed] [Google Scholar]
- Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renken C, Siragusa G, Perkins G, Washington L, Nulton J, Salamon P, Frey TG. A thermodynamic model describing the nature of the crista junction: a structural motif in the mitochondrion. J Struct Biol. 2002;138:137–144. doi: 10.1016/s1047-8477(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Schagger H. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in Yeast Genetics: A Laboratory Course. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Vogel F, Bornhövd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.