Abstract
Particulate methane monooxygenase (pMMO) is an integral membrane metalloenzyme that oxidizes methane to methanol in methanotrophic bacteria. Previous biochemical and structural studies of pMMO have focused on preparations from Methylococcus capsulatus (Bath) and Methylosinus trichosporium OB3b. A pMMO from a third organism, Methylocystis species strain M, has been isolated and characterized. Both membrane-bound and solubilized Methylocystis sp. strain M pMMO contain ~2 copper ions per 100 kDa protomer and exhibit copper-dependent propylene epoxidation activity. Spectroscopic data indicate that Methylocystis sp. strain M pMMO contains a mixture of CuI and CuII, of which the latter exhibits two distinct type 2 CuII electron paramagnetic resonance (EPR) signals. Extended X-ray absorption fine structure (EXAFS) data are best fit with a mixture of Cu–O/N and Cu–Cu ligand environments with a Cu–Cu interaction at 2.52–2.64 Å. The crystal structure of Methylocystis sp. strain M pMMO was determined to 2.68 Å resolution and is the best quality pMMO structure obtained to date. It provides a revised model for the pmoA and pmoC subunits and has led to an improved model of M. capsulatus (Bath) pMMO. In these new structures, the intramembrane zinc/copper binding site has a different coordination environment from that in previous models.
Methanotrophic bacteria are obligate C1 metabolizers that utilize methane as a carbon and energy source.1 The initial step in their metabolic pathway, the oxidation of methane to methanol, is catalyzed by two different methane mono-oxygenase (MMO) enzymes. Most methanotrophs produce a membrane-bound enzyme called particulate MMO (pMMO), and some can also produce a soluble, cytoplasmic enzyme (sMMO) under conditions of copper starvation.2 Although both MMOs can break the strong C–H bond in methane (104 kcal/mol), their overall structures are completely different. sMMO is an α2β2 γ2 dimer with a single metal binding site, a carboxylate-bridged diiron center responsible for methane oxidation.3 Much is known about O2 activation at the diiron center, O2 and substrate/product access to the active site, and activity modulation by other protein components.4,5 In contrast to sMMO, the chemistry of methane oxidation by pMMO is not well understood.
Crystal structures of Methylococcus capsulatus (Bath)6,7 and Methylosinus trichosporium OB3b8 pMMOs to 2.8 and 3.9 Å resolutions, respectively, reveal an α3β3 γ3 trimer composed of the pmoB, pmoA, and pmoC polypeptides and multiple metal binding sites. In both pMMO structures, the periplasmic regions of the pmoB subunit house a copper binding site modeled as a dicopper center on the basis of extended X-ray absorption fine structure (EXAFS) data9,10 and the M. capsulatus (Bath) pMMO crystallographic data.6 A second copper site, modeled as a monocopper center, is also present in the M. capsulatus (Bath) pmoB subunit. A metal binding site within the membrane-spanning pmoC and pmoA subunits is occupied by zinc in M. capsulatus (Bath) pMMO6 and copper in M. trichosporium OB3b pMMO.8 On the basis of activity data and characterization of recombinant pmoB fragments, we have proposed that the dicopper center in pmoB is the active site of pMMO.11
Other researchers have proposed that the pMMO active site is a diiron center located at the crystallographically observed transmembrane zinc/copper site12,13 or a tricopper center housed within a transmembrane hydrophilic patch of residues.14 The latter model includes the binding of 10 additional copper ions to the soluble C-terminus of pmoB.15 The differing views of the active site derive from variations in the metal content and spectroscopic properties of pMMO samples isolated by different laboratories. Most studies have focused on pMMO from M. capsulatus (Bath) and M. trichosporium OB3b.2,13,16 Purified M. capsulatus (Bath) pMMO has been reported to contain 2–20 Cu and 0–2 Fe per 100 kDa αβγ protomer,10,17–20 whereas purified M. trichosporium OB3b pMMO contains 2 Cu per protomer with only trace amounts of Fe.8,21 Electron paramagnetic resonance (EPR) spectra of both pMMOs indicate the presence of type 2 CuII,2,16 but the relationship between these EPR signals and the crystallographically detected copper centers remains unclear. Chan and co-workers also observe an isotropic signal at g ~ 2.1, which they interpret as a ferromagnetically coupled trinuclear CuII cluster.22 X-ray absorption spectroscopic (XAS) studies indicate that as-isolated purified pMMO contains a mixture of CuI and CuII and a Cu-containing cluster with a short Cu–Cu distance of ~2.5 Å,8–10 which is correlated with the presence of the crystallographic dicopper center and with methane oxidation activity.11
To obtain additional data regarding the structure and metal centers of pMMO, we have isolated and characterized pMMO from a third organism, the type II methanotroph Methylocystis species strain M. The pmoB, pmoA, and pmoC subunits from this organism are 82%, 87%, and 88% identical to the corresponding subunits from M. trichosporium OB3b, also a type II methanotroph, and 47%, 58%, and 62% identical to the corresponding subunits from M. capsulatus (Bath), a type X methanotroph. The metal content, activity, and spectroscopic properties of Methylocystis sp. strain M pMMO are consistent with what we have observed for the other two pMMOs. Importantly, Methylocystis sp. strain M pMMO crystallizes more reproducibly than M. capsulatus (Bath) pMMO, and the crystals diffract to higher resolution than those of both the M. capsulatus (Bath) and M. trichosporium OB3b pMMOs. The Methylocystis sp. strain M pMMO structure, determined to 2.68 Å resolution, is higher quality than the previously reported structures6,8 and provides a revised model for the pmoA and pmoC subunits. The new model has implications for the crystallographic zinc/copper site and has led to an improved structure of M. capsulatus (Bath) pMMO.
MATERIALS AND METHODS
Bacterial Growth
Cultures of Methylocystis sp. strain M were grown at 30 °C using a nitrate mineral salts medium (0.85 g/L NaNO3, 0.17 g/L K2SO4, 0.04 g/L MgSO4·7H2O, 0.01 g/L CaCl2·2H2O) supplemented with a trace element solution (0.29 mg/L ZnSO4·7H2O, 0.17 mg/L MnCl2·6H2O, 0.06 mg/L H3BO3, 0.05 mg/L Na2MoO4·2H2O, 0.05 mg/L CoCl2·6H2O, 0.08 mg/L KI, 0.13 mg/L CuSO4·5H2O), 50 μM CuSO4·5H2O, and 40 μM FeSO4·7H2O. Fermentations (12 L) were initiated using 5–10 g of resuspended cell stocks directly or from single colonies grown on agar plates (supplemented with 10 μM CuSO4·5H2O) that were serially increased to 1 L (1 mL, 5 mL, 50 mL, etc.) before inoculation into 11 L of media. A 3:1 ratio of air to methane was maintained during growth with a constant agitation rate of 300 rpm. The pH was maintained between 7.0 and 7.3 using a 100 mM phosphate buffer with fine adjustments made during the growth using 2 M H2SO4 and 1 M NaOH. Cells were harvested at an OD600 of 5–6 and centrifuged at 8000g for 10 min. Pelleted cells were resuspended and washed in 25 mM PIPES, pH 7.0, centrifuged, flash frozen in liquid nitrogen, and stored at −80 °C.
Membrane Isolation and pMMO Solubilization
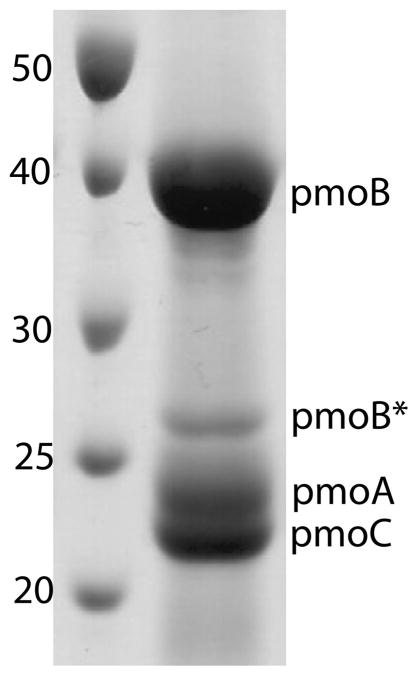
Frozen cells were resuspended in lysis buffer (25 mM PIPES, pH 7.3, 500 mM NaCl, 1 mM benzamidine) and either sonicated on ice for 10 min with 1 s on/off pulses or passed three times though a microfluidizer with the heat exchanging coil submerged in an ice–water bath at a constant pressure of 180 psi. Cell debris was removed by centrifugation at 22000g for 2 h. Intracytoplasmic membranes were pelleted using ultracentrifugation at 160000g for 2 h. These crude pMMO membrane isolations were washed by resuspending and homogenizing in fresh lysis buffer, followed by ultracentrifugation for an additional 2 h. Homogenized crude pMMO membranes diluted to 10–20 mg/mL were either used immediately or flash frozen in liquid nitrogen and stored at −80 °C. Membranes treated with CN− were prepared as described previously.11,23 pMMO was solubilized by adding 1.2 mg of n-dodecyl-β-D-maltopyranoside (DDM) per mg of protein as determined by the Bio-Rad DC protein assay. The solution was stirred at 4 °C for 15 min and ultracentrifuged for 30 min to remove insoluble material and excess lipids. This solubilization procedure results in approximately 90–95% pure pMMO with no further purification necessary (Figure 1). Attempts to further purify pMMO by size exclusion or anion exchange chromatography resulted in subunit degradation.
Figure 1.
SDS-PAGE gel of solubilized Methylocystis sp. strain M pMMO. Molecular mass standards are labeled in kDa. The band labeled pmoB* corresponds to a proteolytic fragment of pmoB with an N-terminus starting at residue Gln 260.
Metal Analysis and Activity Assays
A Varian Vista-MPX CCD Simultaneous ICP-OES instrument was used to determine the metal ion content of crude pMMO membranes and solubilized pMMO. Samples were digested in 5% trace metal grade nitric acid, and standard curves were generated using dilutions of copper, zinc, and iron atomic absorption standards (Sigma-Aldrich). All samples were measured in triplicate from at least two independent isolations. Activity was determined by the propylene epoxidation assay using duroquinol as a reductant as described11,23 with minor modifications. In brief, 200 μL of pMMO (10–20 mg/mL) was incubated with saturating amounts of freshly prepared duroquinol and 2 mL of propylene in a septum sealed vial and shaken at 200 rpm for 3 min in a 37 °C water bath. The amount of propylene oxide produced was detected by gas chromatography using either a Hewlett-Packard 5890A GC or an Agilent Technologies 7890A GC equipped with a Porapak Q column (Supelco) and quantitated using a standard curve generated with pure propylene oxide (Sigma-Aldrich). All samples were measured in duplicate from at least two independent isolations. For CN− treated samples, metal equivalents were added, mixed, and incubated at room temperature for 30 min prior to activity measurements.23
EPR Spectroscopy
Membrane-bound and solubilized pMMO samples were prepared as above with the addition of 10% glycerol (final concentration) to all the buffers. Reconstituted pMMO samples were incubated with copper equivalents for 30 min prior to glycerol addition. The reconstituted samples were prepared from the same protein batch as the activity assay samples in order to correlate the amount of EPR active copper with enzyme activity. All samples were measured on a modified Varian E-4 spectrometer at 77 K. Quantitation of EPR active CuII was performed by comparison with a standard curve generated from copper-EDTA solutions (CuSO4·5H2O in 1 mM EDTA, 0.5 M NaCl, 10% glycerol) of known concentrations. All samples were background and baseline corrected prior to quantitation by double integration. Spectra were simulated using a modified version of the program QPOWA.24,25
X-ray Absorption Spectroscopy
Samples of as-isolated, reduced, copper-reconstituted, and oxidized pMMO were prepared in 50 mM HEPES, pH 7.5, containing 0.03% n-undecyl-β-D-maltopyranoside (UDM) and contained 0.5–1.0 mM copper. All samples except for the copper-reconstituted sample were solubilized as described above. The copper-reconstituted sample was membrane-bound and was prepared by CN− treatment as described previously.11,23 XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline 9-3 using a Si(220) double crystal monochromator equipped with a harmonic rejection mirror. Samples were maintained at 10 K using an Oxford Instruments continuous-flow liquid helium cryostat, and protein fluorescence excitation spectra were collected using a 30-element Ge solid-state array detector. A 0.6 μm wide nickel filter was placed between the cryostat and detector to filter scattering fluorescence not associated with protein Cu signals. XAS spectra were measured as detailed previously.11 All data represent the average of 5–6 scans, and spectra were collected on at least two independent and reproducible samples. XAS data were processed with the Macintosh OS X version of the EXAFSPAK program suite26 integrated with the Feff v7 software27 for theoretical model generation. Data reduction and fitting analysis were performed according to published protocols (refs 9 and 11, Supporting Information) using single scattering Feff v7 theoretical models, calculated for carbon, oxygen, sulfur, and copper coordination to simulate copper-ligand environments, with values for the scale factors (Sc) and E0 calibrated as described elsewhere.9
Crystallization and Structure Determination
Solubilized pMMO was concentrated to ~250 μL using a 15 mL Amicon Ultra 50,000 MWCO filter and exchanged into crystallization buffer (50 mM HEPES, pH 7.5, 0.03% n-undecyl-β-D-maltopyranoside (UDM)) by diluting to ~15 mL and concentrating back to 250 μL. The concentrated sample was then diluted to 10 mg/mL with crystallization buffer. Initial screening using the Qiagen Classics with 96-well sitting drop trays (50 μL well solution, 1 μL protein, and 1 μL well solution in drops) produced microcrystals in condition #79 (0.1 M MES, pH 6.5, 25% PEG 550 MME, 0.01 M ZnSO4) at 22 °C. This condition was reproducible in 24-well sitting drop trays (750 μL well solution, 2 μL protein, and 2 μL well solution in drops) and formed small crystals that diffracted to ~7 Å resolution. Screening different molecular weight PEG solutions together with the Hampton additive screen yielded crystals that diffracted to better than 5 Å resolution. Subsequent optimization resulted in a condition (0.1 M MES, pH 6.5, 18% PEG 750 MME, 0.03 M glycyl glycyl glycine, 0.01 M ZnSO4) that reproducibly gave crystals diffracting to ~3 Å resolution. The best of these crystals diffracted to better than 2.8 Å resolution and were produced in 1–2 weeks by mixing 2 μL of protein with 2 μL of crystallization buffer in sitting drops at 22 °C.
Crystallographic data collection was performed at the Advanced Photon Source (Argonne National Laboratory) using LS-CAT beamline 21-ID-D and GM/CA-CAT beamline 23-ID-D. The data were integrated and scaled using HKL2000.28 The crystals belong to the space group P212121 with unit cell dimensions of a = 107.7, b = 178.3, and c = 183.1 Å. A molecular replacement solution was obtained with PHASER29 using the coordinates of M. capsulatus (Bath) pMMO (PDB code 1YEW) as a starting model. Multiple rounds of model building and refinement with COOT30 and REFMAC531 were needed to fit the Methylocystis sp. strain M pMMO subunit sequences to the electron density. Tight noncrystallographic symmetry (NCS) was used throughout the refinement for pmoB residues 30–414, pmoA residues 35–223, and pmoC residues 26–179. The final model contains pmoB residues 29–414, pmoA residues 11–252, pmoC residues 16–197 and 225–252, two polyalanine helices comprising 16 and 19 residues, respectively, four copper ions, 9 zinc ions, and three water molecules. A Ramachandran plot calculated with MolProbity32 indicates that 90.3% of residues are in the favored regions and 96.7% of residues are in the allowed regions. The final Methylocystis sp. strain M pMMO coordinates were then used as a molecular replacement model to solve and refine the M. capsulatus (Bath) pMMO structure using the original data.6 The new refined model contains pmoB residues 33–414, pmoA residues 7–191 and 213–245, pmoC residues 45–224 and 254–286, 9 copper ions, and 9 zinc ions. A Ramachandran plot calculated with MolProbity32 indicates that 89.5% of residues are in the favored regions and 97.7% of residues are in the allowed regions.
RESULTS AND DISCUSSION
Metal Content and Activity
Membrane-bound and solubilized pMMO from Methylocystis sp. strain M contain ~2 Cu per 100 kDa protomer (Table 1). This is significantly less copper than that measured in membrane-bound preparations of M. capsulatus (Bath) pMMO (4–59 Cu/100 kDa pMMO)2,16 but similar to the amounts reported for M. trichosporium OB3b pMMO (4–5 Cu per 100 kDa membrane-bound pMMO and 1–2 Cu per 100 kDa purified pMMO).8,21 Recent studies indicate that only 2–3 copper ions are required for M. capsulatus (Bath) pMMO activity, suggesting that some copper is bound adventitiously and/or associated with the copper chelator methanobactin.11 Unlike pMMO from M. trichosporium OB3b,8,21 the Methylocystis sp. strain M samples do contain measurable iron (0.59 ± 0.09 Fe/100 kDa pMMO for membrane-bound and 0.43 ± 0.02 Fe/100 kDa pMMO for solubilized) (Table 1). The optical spectrum of solubilized Methylocystis sp. strain M pMMO exhibits a peak at ~412 nm, suggestive of heme contamination, which is commonly observed for M. capsulatus (Bath) pMMO9,10 and may account for at least some of the measured iron. Negligible amounts of zinc are detected.
Table 1.
Metal Content and Activity of Methylocystis sp. strain M pMMOa
| sample | Cu per 100 kDa pMMO | Fe per 100 kDa pMMO | Zn per 100 kDa pMMO | activity (nmol propylene oxide/(min mg)) |
|---|---|---|---|---|
| membrane-bound | 2.29 ± 0.22 | 0.59 ± 0.09 | 0.10 ± 0.02 | 5.3 ± 1.4 |
| CN− treated | 0.05 ± 0.01 | 0.29 ± 0.11 | ND | 0 |
| CN− treated + 2Cu | 1.34 ± 0.06 | |||
| solubilized | 2.11 ± 0.46 | 0.43 ± 0.02 | 0.06 ± 0.02 | 1.24 ± 0.21 |
All samples were measured in triplicate and are the average of at least two independent isolations.
The specific activity of membrane-bound Methylocystis sp. strain M pMMO is 5.3 ± 1.4 nmol propylene oxide/(min mg) (average of 10 samples) (Table 1), which is comparable to the specific activity of 2–5 nmol propylene oxide/(min mg) reported for pMMO from M. trichosporium OB3b.8,21,33 The addition of 1 equiv of CuSO4 per 100 kDa pMMO prior to activity measurements typically increases the membrane-bound pMMO specific activity by ~5–20%, suggesting that the copper active site is not fully loaded (vide infra). Further copper additions decrease activity. Solubilized Methylocystis sp. strain M pMMO has a specific activity of 1.24 ± 0.21 nmol propylene oxide/(min mg) (average of two samples), which is in the range of that reported for solubilized M. trichosporium OB3b pMMO.8 Since membrane-bound and solubilized M. trichosporium OB3b pMMO contain less than 0.01 Fe/100 kDa pMMO,8 it is unlikely that the iron in Methylocystis sp. strain M pMMO (Table 1) accounts for the observed enzyme activity. However, these data should be interpreted with the caveat that the activity levels for all isolated pMMO samples are typically 1–10% of that measured for whole cells.13
Cyanide treatment of Methylocystis sp. strain M membrane-bound pMMO removes ~98% of the copper and ~49% of the iron (Table 1) and abolishes all activity. Addition of 2 equiv of copper restores ~25% of the as-isolated activity (1.34 ± 0.06 nmol propylene oxide/(min mg), average of two samples). Similar results were obtained upon treatment of M. capsulatus (Bath) pMMO with cyanide, with addition of 2–3 equiv of copper restoring ~70% of activity.11,23 Typically, adding more than 2 equiv of copper results in inhibition, which is attributed to hydrogen peroxide formation.11,33 The difference in the level of activity restored by addition of copper to cyanide treated pMMO from Methylocystis sp. strain M and M. capsulatus (Bath) is consistent with the copper in Methylocystis sp. strain M pMMO being more labile. This increased lability may be related to the absence of the monocopper center in pmoB (ref 8; vide infra). While apparently not catalytically active, the presence of the monocopper site likely stabilizes the residues involved in copper binding at the dicopper site.11
EPR Spectroscopy
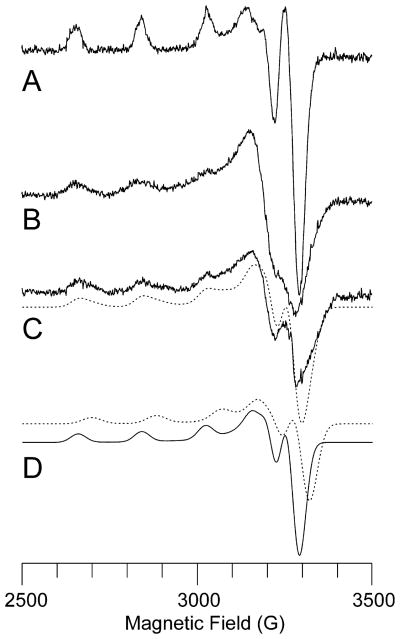
The X-band EPR spectra of membrane-bound and solubilized Methylocystis sp. strain M pMMO exhibit a typical type 2 CuII signal (Figure 2B,C). According to spin quantitation using a CuII standard, ~69% of the copper in the membrane-bound sample and ~43% of the copper in the solubilized sample are EPR active. A type 2 CuII signal has been observed in whole cell, membrane-bound, and purified pMMO samples from several different organisms.8,10,17,18,34–36 There is no evidence for the signal attributed to a trinuclear copper cluster.22,37,38 A simulation of the solubilized pMMO EPR spectrum indicates the presence of two type 2 CuII signals (Figure 2D). The major component, with parameters g⊥ = 2.052, g|| = 2.247, A (63Cu)|| = 570 MHz, and A (63Cu)⊥ = 60 MHz, is identical to the major component that constitutes at least 80% of the signal in the EPR spectrum of purified M. trichosporium OB3b pMMO.8 The M. trichosporium OB3b pMMO minor component has g values g⊥ = 2.060 and g|| = 2.225 and resolved hyperfine coupling of about 40 MHz to two equivalent 14N nuclei.8 By contrast, the second component of the Methylocystis sp. strain M pMMO EPR spectrum has parameters g⊥ = 2.04 and g|| = 2.215, exhibits no observable 14N hyperfine coupling, and is at least 70% of the intensity of the major component (an approximation due to uncertainty in the perpendicular region at this microwave frequency). For both M. trichosporium OB3b and Methylocystis sp. strain M pMMO, the A(63Cu)|| value for the major and minor components is indistinguishable.
Figure 2.
X-band EPR spectra of Methylocystis sp. strain M pMMO. (A) Cu-EDTA standard (250 μM). (B) Membrane-bound pMMO. (C) Solubilized pMMO with simulation (dashed line) generated by summing two contributing type 2 CuII centers. (D) Individual simulated spectra of the two components in (C). The experimental spectra are offset for clarity, but their intensities are meaningful since the spectra were recorded under identical conditions. Experimental parameters: microwave frequency, 9.21 GHz; microwave power, 15 dB; modulation amplitude, 5.000 G; receiver gain, 1.25 × 102; time constant, 0.3 s; temperature, 77 K; 2 min scan. Simulation parameters for major component (solid line in part D): relative integrated intensity, 1.0; g⊥ = 2.052, g|| = 2.247, A(63Cu)|| = 570 MHz, A (63Cu)⊥ = 60 MHz. Simulation parameters for minor component (dashed line in part D): relative integrated intensity, 0.7; g⊥ = 2.04, g|| = 2.215, A(63Cu)|| = 570 MHz, A(63Cu)⊥ = 60 MHz.
The X-band EPR spectrum of cyanide-treated membrane-bound pMMO following addition of 2 equiv of copper is similar to that of as-isolated pMMO (Figure S1). Interestingly, quantitative comparisons to a CuII standard indicate that only ~50% of the total copper in these reconstituted samples is EPR-active. Assuming that these copper ions occupy the dicopper center, these data could be consistent with a previously suggested trapped valence CuICuII site.39 Upon addition of a third equivalent of copper, ~67% of the total copper is EPR-active. Thus, the third copper is CuII, but it is not clear if or where it is binding. One possibility is that this third equivalent binds in the transmembrane site occupied by zinc in the M. capsulatus (Bath)6 and Methylocystis sp. strain M (vide infra) pMMO crystal structures.
XAS Spectroscopy
The XANES spectrum of dithionite-reduced, solubilized Methylocystis sp. strain M pMMO (Figure 3A) closely resembles spectra reported for three to four-coordinate CuI compounds, based on the intensity of the 1s → 4p transition, the overall edge structure, and the lack of any discernible 1s → 3d feature.40 The XANES spectrum for as-isolated, solubilized pMMO (Figure 3B) has a reduced, but observable, 1s → 4p transition and a weak 1s → 3d feature, consistent with copper in this sample being a mixture of CuI and CuII or a predominantly square or tetragonal CuII complex. The XANES spectrum of Cu-reconstituted, membrane-bound pMMO (Figure 3C) is nearly identical to that of as-isolated solubilized pMMO (Figure 3B). The oxidized pMMO sample treated with hydrogen peroxide has a XANES spectrum consistent with a square or tetragonal CuII coordination environment (Figure 3D).
Figure 3.
Copper XANES spectra of Methylocystis sp. strain M pMMO. The spectra for (A) reduced, (B) as-isolated, (C) Cu-reconstituted, membrane-bound, and (D) oxidized samples are shown. The reduced, as-isolated, and oxidized samples were solubilized. The dashed vertical line at ca. 8984 eV identifies spectral features corresponding to the CuI 1s → 4p transition. Spectra are offset for clarity.
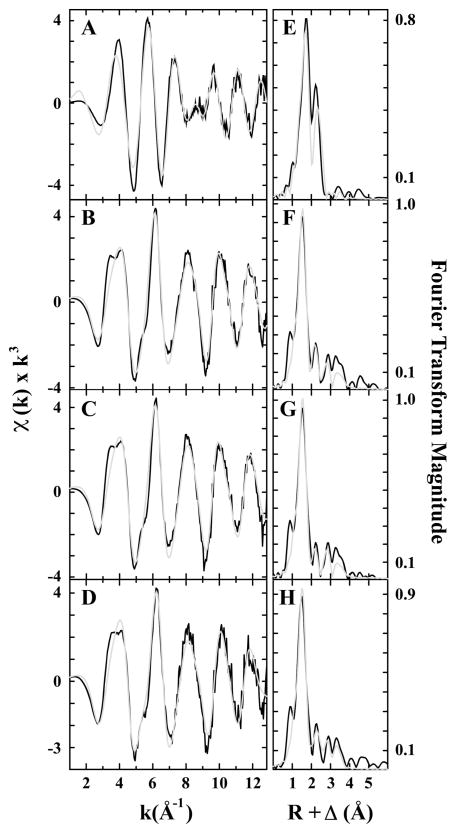
Fourier transforms of the Cu EXAFS data for as-isolated, reduced, Cu-reconstituted, and oxidized pMMO all show two scattering interactions corresponding to nearest-neighbor ligands at approximately 2 and 2.5 Å (Figure 4 and Table 2). In addition, long-range ligand interactions (>3 Å) are evident in all the spectra. EXAFS simulations show a mixture of Cu–O/N and Cu–Cu ligand environments in each of the samples, and an additional Cu–O/N environment was justifiably included for the reduced sample. Best-fit simulations to the average nearest-neighbor ligand environment included only Cu–O/N ligands with bond distances gradually decreasing from 2.02 Å (reduced pMMO) to 1.97 Å (as-isolated and Cu-reconstituted pMMO) to 1.96 Å (oxidized pMMO) (Tables 2, Tables S1–S4). The additional nearest-neighbor Cu–O/N environment in the reduced pMMO sample is at 2.15 Å. There is no evidence for Cu–S scattering. There was a significant improvement in all fits when a Cu–Cu scattering environment at >2.5 Å was included (Tables S1–S4). The Cu–Cu bond length of 2.64 Å for reduced pMMO is substantially longer than the 2.53 and 2.52 Å bond lengths observed for as-isolated or oxidized pMMO and Cu-reconstituted pMMO, respectively. The 2.64 Å Cu–Cu interaction in the reduced sample is different from the 2.58 Å interatomic distance expected for Cu metal, suggesting that the integrity of the dinuclear metal center is maintained upon reduction. The occupancy for the Cu–Cu vector in the reduced sample is dramatically larger than the other three samples (CN of 0.75 vs 0.25, 0.25, and 0.25), suggesting that the Cu–Cu environment in all samples except the reduced may be heterogeneous with slightly different Cu–Cu vectors. In all samples, we were able to fit the data with long-range C scattering using single-scattering models. We were not able to fit long-range (>3 Å) scattering with a Cu–Cu interaction, also eliminating the possibility that our sample contains any appreciable Cu metal. Although fingerprints of Cu–imidazole scattering are apparent in the EXAFS of all samples (camelback beat pattern between k = 3–4 Å−1 and the shoulder at k = 5.5 Å−1), attempts to fit any of the Cu EXAFS with a multiple scattering Cu–imidazole model were unsuccessful. Long-range Cu···C/N scattering, certainly resulting from the coordinated histidine residues, could, however, be adequately fit to the as-isolated, Cu-reconstituted, and oxidized samples EXAFS with single scattering models at bond lengths between 2.85 and 3.0 Å, at ca. 3.3 and 3.9 Å, bond lengths consistent with imidazoles. Overall, these results are consistent with data obtained for pMMO samples from M. capsulatus (Bath)9 and M. trichosporium OB3b.8
Figure 4.
Copper EXAFS fitting analysis for Methylocystis sp. strain M pMMO. Raw EXAFS data for (A) reduced, (B) as-isolated, (C) Cu-reconstituted, membrane-bound, and (D) oxidized pMMO are shown in black. The reduced, as-isolated, and oxidized samples were solubilized. The phase shifted Fourier transforms of the copper data for (E) reduced, (F) as-isolated, (G) Cu-reconstituted, membrane-bound, and (H) oxidized are shown in black. Best-fit simulations are shown in gray.
Table 2.
Summary of Cu EXAFS Fitting Analysis for Methylocystis sp. strain M pMMOa
| sample | nearest-neighbor ligand environmentb |
nearest-neighbor ligand environmentb |
F′g | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| atomc | R (Å)d | CNe | σ2f | atomc | R (Å)d | CNe | σ2f | atomc | R (Å)d | CNe | σ2f | ||
| reduced | O/N | 2.02 | 1 | 0.8 | O/N | 2.15 | 3 | 1.1 | C | 4.01 | 0.25 | 4.1 | 0.24 |
| Cu | 2.64 | 0.75 | 4.3 | ||||||||||
| as-isolated | O/N | 1.97 | 3 | 4.6 | Cu | 2.53 | 0.25 | 5.0 | C | 2.99 | 0.75 | 3.4 | 0.21 |
| C | 3.30 | 1 | 5.9 | ||||||||||
| C | 3.90 | 2 | 4.9 | ||||||||||
| Cu-reconstituted | O/N | 1.97 | 3 | 4.3 | Cu | 2.52 | 0.25 | 6.0 | C | 2.97 | 0.75 | 5.5 | 0.20 |
| C | 3.30 | 1 | 3.6 | ||||||||||
| C | 3.89 | 2 | 5.4 | ||||||||||
| oxidized | O/N | 1.96 | 3 | 5.1 | Cu | 2.53 | 0.25 | 3.7 | C | 2.85 | 0.75 | 5.6 | 0.25 |
| C | 3.29 | 1 | 5.4 | ||||||||||
| C | 3.86 | 2 | 5.0 | ||||||||||
Data were fit over a k range of 1–12.85 Å−1.
Independent metal–ligand scattering environment.
Scattering atoms: O (oxygen), N (nitrogen), and C (carbon), and Cu (copper).
Average metal–ligand bond length from three independent samples.
Average metal–ligand coordination number from three independent samples.
Average Debye–Waller factor in Å2 × 103 from three independent samples.
Number of degrees of freedom weighted mean-square deviation between data and fit.
Crystal Structure
The crystal structure of pMMO from Methylocystis sp. strain M was solved to 2.68 Å resolution (Table 3). Like pMMO from M. capsulatus (Bath) and M. trichosporium OB3b, Methylocystis sp. strain M pMMO is a trimer. Notably, strong density corresponding to a helix was observed adjacent to two of the three pmoC subunits and was modeled as polyalanine (Figure 5A). An unidentified helix is present at the same location in the 3.9 Å resolution M. trichosporium OB3b pMMO structure.8 Although some side-chain electron density is apparent, it was not possible to determine the sequence of this helix.
Table 3.
Data Collection and Refinement Statistics
| native (Cu anomalous) | Zn anomalous | |
|---|---|---|
| data collection | ||
| wavelength (Å) | 1.37765 | 1.27816 |
| resolution (Å)a | 50–2.68 (2.75–2.68) | 50–3.6 (3.66–3.60) |
| Rsym | 0.146 (0.549) | 0.201 (0.571) |
| I/σI | 19.2 (1.6) | 20.1 (5.3) |
| completeness (%) | 88.9 (54.4) | 99.2 (94.3) |
| redundancy | 12.6 (6.8) | 12.3 (9.4) |
| refinement | ||
| no. of reflections | 83 726 | |
| Rwork/Rfreeb | 0.249/0.281 | |
| B-factor (Å2) | 49.7 | |
| rms deviations | ||
| bond lengths (Å) | 0.016 | |
| bond angles (deg) | 1.735 | |
Values in parentheses refer to the highest-resolution shell.
For calculation of Rfree, 5% of the reflections were reserved.
Figure 5.
Structures of Methylocystis sp. strain M pMMO subunits. (A) One of the three protomers in the trimer. The pmoB, pmoA, and pmoC subunits are shown in gray, blue, and purple, respectively. Copper ions are shown in cyan, and a zinc ion is shown in gray. The additional helix is shown in yellow. (B) Superposition of the pmoA subunits from the Methylocystis sp. strain M (blue) and the original M. capsulatus (Bath) (gray) structures. (C) Superposition of the pmoC subunits from the Methylocystis sp. strain M (purple) and the original M. capsulatus (Bath) (light pink) structures. Residues discussed in the text are labeled.
The Methylocystis sp. strain M pmoB subunit is similar to the M. capsulatus (Bath) pmoB subunit with a root-mean-square deviation of 0.972 Å. The most pronounced differences are at the C-terminus, starting with an insertion in the Methylocystis sp. strain M pmoB sequence spanning residues 272–283. This region forms an extra loop structure, and as a result, the β strands encompassing residues 283–291, 302–308, and 358–364 are offset from the corresponding strands in the M. capsulatus (Bath) pmoB structure. The electron density for the pmoA subunit revealed a discrepancy with the model for M. capsulatus (Bath) pmoA. At residue Thr 196 (Thr 191 in M. capsulatus (Bath) pmoB), the electron density clearly extends along the periplasmic side of the membrane interface rather than leading through the membrane as in M. capsulatus (Bath) pmoA (Figure 5B). This extended loop region connects to a transmembrane helix spanning residues 217–244 and terminating at residue 252 on the cytoplasmic side of membrane. In M. capsulatus (Bath) pmoA, the region starting at Thr 191 is modeled as spanning the membrane to the cytoplasmic side and the helical region corresponding to Methylocystis sp. strain M residues 217–244 is modeled in the opposite direction (Figure 5B).
After tracing pmoA according to this new model, it became clear that the residues modeled as pmoA 195–215 in M. capsulatus (Bath) pMMO (Figure 5B) are actually part of pmoC and were therefore modeled as residues 178–197 of Methylocystis sp. strain M pmoC (Figure 5C). Finally, the C-terminal region of pmoC was traced with residues 225–252 extending from the periplasmic to cytoplasmic side, as opposed to residues 231–259 extending from the cytoplasmic to periplasmic side in the M. capsulatus (Bath) pmoC model. The orientation of this helix was also reversed in the M. trichosporium OB3b pMMO structure, but in that case, the low resolution precluded assigning any side chains.8 The residues between 197 and 225 (Figure 5C) are not visible in the electron density map. The improved model for pmoC clearly indicates that it is not possible to connect the extra helix to this subunit and suggests that this helix derives from another yet to be identified polypeptide. Interestingly, this extra helix is not observed in the M. capsulatus (Bath) pMMO structure, suggesting its presence may be specific to pMMO from type II methanotrophs.
Examination of the electron density map for the original M. capsulatus (Bath) pMMO structure reveals poor density in the pmoA and pmoC regions that differ from those in the Methylocystis sp. strain M pMMO structure. Therefore, the M. capsulatus (Bath) pMMO structure was solved again by molecular replacement using the Methylocystis sp. strain M pMMO structure as a starting model. Following several refinement cycles, the final values of Rwork and Rfree were 0.269 and 0.296, respectively, as compared to 0.270 and 0.302 for the original structure.6 Analysis with MolProbity41 indicates that the new model is much better quality than the original model, with a Molprobity score of 3.20, corresponding to the 49th percentile of 4482 structures in the 2.80 ± 0.25 Å resolution range. The original model has a score of 3.85, corresponding to the 12th percentile. For comparison, the Methylocystis sp. strain M pMMO structure has a score of 2.93, corresponding to the 60th percentile of 5385 structures at 2.68 ± 0.25 Å resolution.
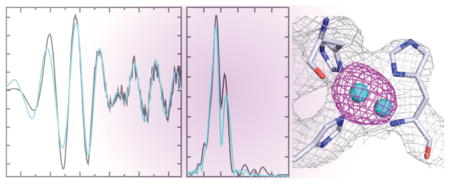
Anomalous Fourier maps of Methylocystis sp. strain M pMMO calculated using data collected at the Cu absorption edge (Table 3) exhibit strong (>15σ) peaks at the proposed dicopper active site. There are no peaks elsewhere in the structure, including within the C-terminal domain of pmoB, which is proposed by Chan and co-workers to bind 10 copper ions.15 A dicopper site could be modeled in one of three pmoB subunits whereas the other two were best modeled with a single copper ion (Figure 6A,B). It is possible that the buffer exchange procedure involving multiple concentration and dilution steps removes some copper, which is consistent with the increased lability of copper binding to both Methylocystis sp. strain M and M. trichosporium OB3b pMMO.8 The EXAFS data (Figure 4 and Table 2) indicate the presence of a short Cu–Cu interaction, supporting the modeling of a dicopper center in one of the pmoB subunits. A monocopper center corresponding to that in the M. capsulatus (Bath) pMMO structure6 is not present, consistent with the substitution of an asparagine (Asn 44) for one of the histidine ligands. This monocopper site is not observed in the M. trichosporium OB3b pMMO structure either.8
Figure 6.
Metal centers in Methylocystis sp. strain M pMMO. The copper sites in (A) and (B) were modeled as dinuclear and mononuclear, respectively. The Cu anomalous map (magenta, 5σ) is superposed on the 2Fo – Fc map (1.4σ). (C) The zinc site is solvent exposed. The Zn anomalous map (cyan, 4.5σ) is superposed on the 2Fo – Fc map (1.4σ). (D) The zinc site as modeled in the original M. capsulatus (Bath) pMMO structure. The pmoC and pmoA subunits are colored light pink and gray as in Figure 5.
Like M. capsulatus (Bath) pMMO, Methylocystis sp. strain M pMMO could be crystallized only in the presence of zinc. In addition to zinc ions mediating crystal contacts, the intra-membrane site occupied by zinc in the M. capsulatus (Bath) pMMO structure is also occupied by zinc (Figure 6C,D). In the original M. capsulatus (Bath) pMMO structure, the zinc ion is coordinated by Asp 156, His 160, and His 173 from pmoC and Glu 195 from pmoA (Figure 6D). The new model for the pmoA and pmoC subunits drastically changes the environment at this site. While Methylocystis sp. strain M pmoC residues Asp 129, His 133, and His 146 are coordinated, Glu 200 from pmoA, which corresponds to Glu 195 in M. capsulatus (Bath) pMMO, points toward the N-terminal cupredoxin domain of pmoB (Figure 5B) and forms hydrogen bonds with the amide nitrogen atoms of pmoB residues Ser 109 and Gly 263. The fourth ligand to the zinc ion is instead modeled as a water molecule, which is within 4 Å of the carbonyl oxygen of Asn 196 (Figure 6C). The electron density is poor around Asn 196, and residues 197–225 could not be modeled. Therefore, it is also possible that the fourth coordination site is partially occupied by a disordered side chain from this region. As a result of the new model, the zinc site is exposed in the center of the pMMO trimer. The accessibility of this site is consistent with its occupation by zinc in the Methylocystis sp. strain M and M. capsulatus (Bath) pMMO structures and by copper in the M. trichosporium OB3b structure.8 However, it seems less likely that a diiron active site could be accommodated in this position, as proposed by DiSpirito and co-workers.12 Moreover, there is no evidence from anomalous data collected at the Fe absorption edge for iron binding sites in the Methylocystis sp. strain M pMMO structure. Finally, the hydrophilic patch of residues proposed to house a tricopper center14 is devoid of metal ions in Methylocystis sp. strain M pMMO as it is in the M. capsulatus (Bath) and M. trichosporium OB3b structures.
In sum, pMMO from Methylocystis sp. strain M binds ~2 copper ions per 100 kDa protomer and exhibits specific activity similar to that reported for M. trichosporium OB3b pMMO. As in the M. trichosporium OB3b and M. capsulatus (Bath) pMMOs, Methylocystis sp. strain M pMMO contains a mixture of CuI and CuII and exhibits type 2 CuII EPR signals. XAS data are also consistent with the presence of CuI and CuII. EXAFS data indicate the presence of a short Cu–Cu interaction, also observed in the other two pMMOs, and consistent with the dicopper active site model. The Methylocystis sp. strain M crystal structure, which is the best resolution and quality pMMO structure to date, provides revised models for the pmoA and pmoC subunits. These models led to an improved M. capsulatus (Bath) pMMO structure and a revised coordination environment for the crystallographic zinc/copper site. Taken together, these data are consistent with the dicopper active site model for pMMO.11 The improvement of the pMMO structure is promising and suggests that a higher resolution view of the metal centers may be within grasp.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH grants GM70473 (A.C.R.), DK068139 (T.L.S.), and HL13531 (B.M.H).
We thank Professor Hiroo Uchiyama for providing a culture of Methylocystis sp. strain M. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). XAS data were collected at SSRL, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program. We thank Sarah Sirajuddin for performing key crystallization screens and Dr. Rama Balasubramanian for valuable discussions and assistance with figure preparation.
ABBREVIATIONS
- MMO
methane monooxygenase
- sMMO
soluble MMO
- pMMO
particulate MMO
- EXAFS
extended X-ray absorption fine structure
- EPR
electron paramagnetic resonance
- XAS
X-ray absorption spectroscopy
Footnotes
Accession Codes
The coordinates of Methylocystis species strain M pMMO and of Methylococcus capsulatus (Bath) pMMO have been deposited in the Protein Data Bank with accession codes 3RFR and 3RGB, respectively.
Methods of EXAFS data analysis, tables summarizing progress of EXAFS simulations for all four samples, and EPR spectra of cyanide treated membrane-bound pMMO upon addition of copper. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 3.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 4.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew Chem, Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Tinberg CE, Lippard SJ. Dioxygen activation in soluble methane monooxygenase. Acc Chem Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman RL, Rosenzweig AC. The quest for the particulate methane monooxygenase active site. Dalton Trans. 2005;21:3390–3396. doi: 10.1039/b506651d. [DOI] [PubMed] [Google Scholar]
- 8.Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry. 2008;47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC, Stemmler TL. Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg Chem. 2006;45:8372–8381. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman RL, Shrestha DB, Doan PE, Hoffman BM, Stemmler TL, Rosenzweig AC. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc Natl Acad Sci U S A. 2003;100:3820–3825. doi: 10.1073/pnas.0536703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian R, Smith SM, Rawat S, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J Am Chem Soc. 2007;129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Lett. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 14.Chan SI, Wang VCC, Lai JCH, Yu SSF, Chen PPY, Chen KHC, Chen CL, Chan MK. Redox potentiometry studies of particulate methane monooxygenase: support for a trinuclear copper cluster active site. Angew Chem, Int Ed. 2007;46:1992–1994. doi: 10.1002/anie.200604647. [DOI] [PubMed] [Google Scholar]
- 15.Yu SSF, Ji CZ, Wu YP, Lee TL, Lai CH, Lin SC, Yang ZL, Wang VCC, Chen KHC, Chan SI. The C-terminal aqueous-exposed domain of the 45 kDa subunit of the particulate methane monooxygenase in Methylococcus capsulatus (Bath) is a Cu(I) sponge. Biochemistry. 2007;46:13762–13774. doi: 10.1021/bi700883g. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman RL, Rosenzweig AC. Biological methane oxidation: regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit Rev Biochem Mol Biol. 2004;39:147–164. doi: 10.1080/10409230490475507. [DOI] [PubMed] [Google Scholar]
- 17.Basu P, Katterle B, Andersson KK, Dalton H. The membrane-associated form of methane monooxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem J. 2003;369:417–427. doi: 10.1042/BJ20020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM, DiSpirito AA. The membrane-associated methane mono-oxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen HH, Elliott SJ, Yip JH, Chan SI. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. Isolation and characterization. J Biol Chem. 1998;273:7957–7966. doi: 10.1074/jbc.273.14.7957. [DOI] [PubMed] [Google Scholar]
- 20.Yu SS-F, Chen KH-C, Tseng MY-H, Wang Y-S, Tseng C-F, Chen Y-J, Huang DS, Chan SI. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J Bacteriol. 2003;185:5915–5924. doi: 10.1128/JB.185.20.5915-5924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyaji A, Kamachi T, Okura I. Improvement of the purification method for retaining the activity of the particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biotechnol Lett. 2002;24:1883–1887. [Google Scholar]
- 22.Chan SI, Yu SSF. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc Chem Res. 2008;41:969–979. doi: 10.1021/ar700277n. [DOI] [PubMed] [Google Scholar]
- 23.Smith SM, Balasubramanian R, Rosenzweig AC. Metal reconstitution of particulate methane monooxygenase and heterologous expression of the pmoB subunit. Methods Enzymol. 2011;495:195–210. doi: 10.1016/B978-0-12-386905-0.00013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belford RL, Belford GG. Eigenfield expansion technique for efficient computation of field-swept fixed-frequency spectra from relaxation master equations. J Chem Phys. 1973;59:853–854. [Google Scholar]
- 25.Belford RL, Nilges MJ. Computer simulations of powder spectra, in EPR Symposium. 21st Rocky Mountain Conference; Denver, CO. 1979. [Google Scholar]
- 26.George GN, George SJ, Pickering IJ. EXAFSPAK. 2001 http://www-ssrl.slac.stanford.edu/~george/exafspak/exafs.htm.
- 27.Ankudinov AL, Rehr JJ. Relativistic calculations of spin-dependent X-ray absorption spectra. Phys Rev B. 1997;56:R1712–R1715. [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyaji A, Suzuki M, Baba T, Kamachi T, Okura I. Hydrogen peroxide as an effecter on the inactivation of particulate methane monooxygenase under aerobic conditions. J Mol Catal B. 2009;57:211–215. [Google Scholar]
- 34.Katterle B, Gvozdev RI, Abudu N, Ljones T, Andersson KK. A continuous-wave electron-nuclear double resonance (X-band) study of the Cu2+ sites of particulate methane mono-oxygenase of Methylococcus capsulatus (strain M) in membrane and pure dopamine beta-mono-oxygenase of the adrenal medulla. Biochem J. 2002;363:677–686. doi: 10.1042/0264-6021:3630677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemos SS, Collins MLP, Eaton SS, Eaton GR, Antholine WE. Comparison of EPR-visible Cu2+ sites in pMMO from Methylococcus capsulatus (Bath) and Methlyomicrobium album BG8. Biophys J. 2000;79:1085–1094. doi: 10.1016/s0006-3495(00)76362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen H-HT, Nakagawa KH, Hedman B, Elliott SJ, Lidstrom ME, Hodgson KO, Chan SI. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu(I) ions and their implications. J Am Chem Soc. 1996;118:12766–12776. [Google Scholar]
- 37.Chen KH-C, Chen C-L, Tseng C-F, Yu SS-F, Ke S-C, Lee J-F, Nguyen HT, Elliott SJ, Alben JO, Chan SI. The copper clusters in the particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) J Chin Chem Soc. 2004;51:1081–1098. [Google Scholar]
- 38.Hung S-C, Chen C-L, Chen KH-C, Yu SS-F, Chan SI. The catalytic copper clusters of the particulate methane monooxygenase from methanotropic bacteria: electron paramagnetic resonance spectral simulations. J Chin Chem Soc. 2004;51:1229–1244. [Google Scholar]
- 39.Balasubramanian R, Rosenzweig AC. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 40.Kau L-S, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. X-ray absorption edge determination of the oxidation state and coordination number of copper. Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 41.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.