Abstract
The southeastern margin of the Tibetan Plateau (SEMTP) is a particularly interesting region due to its topographic complexity and unique geologic history, but phylogeographic studies that focus on this region are rare. In this study, we investigated the phylogeography of the South China field mouse, Apodemus draco, in order to assess the role of geologic and climatic events on the Tibetan Plateau in shaping its genetic structure. We sequenced mitochondrial cytochrome b (cyt b) sequences in 103 individuals from 47 sampling sites. In addition, 23 cyt b sequences were collected from GenBank for analyses. Phylogenetic, demographic and landscape genetic methods were conducted. Seventy-six cyt b haplotypes were found and the genetic diversity was extremely high (π = 0.0368; h = 0.989). Five major evolutionary clades, based on geographic locations, were identified. Demographic analyses implied subclade 1A and subclade 1B experienced population expansions at about 0.052-0.013 Mya and 0.014-0.004 Mya, respectively. The divergence time analysis showed that the split between clade 1 and clade 2 occurred 0.26 Mya, which fell into the extensive glacial period (EGP, 0.5-0.17 Mya). The divergence times of other main clades (2.20-0.55 Mya) were congruent with the periods of the Qingzang Movement (3.6-1.7 Mya) and the Kun-Huang Movement (1.2-0.6 Mya), which were known as the most intense uplift events in the Tibetan Plateau. Our study supported the hypothesis that the SEMTP was a large late Pleistocene refugium, and further inferred that the Gongga Mountain Region and Hongya County were glacial refugia for A. draco in clade 1. We hypothesize that the evolutionary history of A. draco in the SEMTP primarily occurred in two stages. First, an initial divergence would have been shaped by uplift events of the Tibetan Plateau. Then, major glaciations in the Pleistocene added complexity to its demographic history and genetic structure.
Introduction
The Tibetan Plateau is a recognized biodiversity hotspot [1]. Recently, a number of phylogeographic or phylogenetic studies have focused on the Tibetan Plateau due to its complex geologic history [2]–[8]. Historically, it uplifted several times, and the most intense uplift events were known as the Qingzang Movement (3.6-1.7 million years ago, Mya) and the Kun-Huang Movement (1.2-0.6 Mya) [9]–[11]. These events caused dramatic environmental changes: grasslands replaced forests and glaciers and deserts developed as the climate gradually became drier, colder and windier [12]. Thus, as a result of the uplift events, the plateau began to diversify topographically by the continual developed mountains and watercourse [13]. Moreover, it also caused its elevation to increase by up to 3000 m, which consequently increased the exposure of the endemic fauna to glacial events [5], [9]. So, Quaternary glacial cycles have been regarded as an important factor in promoting the diversification of species in this region [2], [14]. Although there are different opinions about the range and extent of glaciers on the plateau [15]–[17], ice-sheets during the most extensive glaciation (0.5-0.17 Mya) were undoubtedly several times larger than those found today [9]. Taken together, the current distribution of endemic organisms and their patterns of genetic differentiation should be greatly influenced by these geologic and climatic events.
Several studies have demonstrated that species distributed on the Tibetan Plateau retreated to the east side of the plateau during cold periods [2], [7], [18], [19]. Qu et al. [19] reported that both the twite (Carduelis flavirostris) and the black redstart (Phoenicurus ochruros) inhabited the northeastern margin of the Tibetan Plateau and maintained stable population sizes through glacial cycles. On the contrary, three other avian species (Montifringilla adamsi, Pyrgilauda blanfordi and Eremophila alpestris), mostly distributed on the inner region of the plateau, experienced population expansions after glacial retreat [19]. These results indicate that the east side of the plateau probably constituted a large refugium during the major glaciations in the Pleistocene.
In this study, we focused on the southeastern margin of the Tibetan Plateau (SEMTP), including western Sichuan Province, northwestern Yunnan Province and eastern Xizang Autonomous Region (Tibet). The large rivers and mountains (e.g. Daxueshan Mountains, Shaluli Mountains, Gongga Mountains, Dadu River, Yalong River, and Jinsha River) in this region could be natural barriers for the postglacial colonization of small mammals (Fig. 1). The magnificent Gongga Mountain, with the highest peak of 7,556 m above the sea level, was an important glaciation centre and contemporary glaciers are still well developed [20]. Conversely, some previous studies indicated that Gonga Mountain region (GMR) was a suitable refuge during glacial cycles [21], [22]. Many ancient and endemic species inhabited in this region [21], [23]. But so far, none of the studies could provide detailed information.
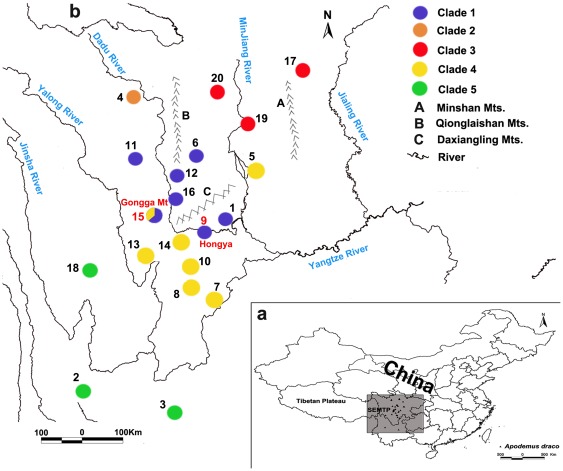
Figure 1. Sampling sites for the South China field mouse (Apodemus draco) in this study.
(a) Map of China shows the study area of this study. The area in the shadow represents the range of the southeastern margin of the Tibetan Plateau, which is our study area. (b) Sampling sites for the South China field mouse (Apodemus draco) on the Tibetan Plateau. Populations are numbered as in Table 1. Major rivers and mountain are also shown. The distribution of observed major evolutionary clades in phylogenetic analyses is shown in different colors. The two glacial refugia (Gongga Mountain and Hongya County) were labeled in red color.
In order to further assess the role of geologic and climatic events on the Tibetan Plateau in shaping the genetic structures of local fauna, we present a phylogeographic investigation of the South China field mouse (Apodemus draco). Small rodents are good candidates for inferring biotic history from contemporary patterns of genetic variation because their short generation time and relatively limited mobility [24]. This mouse is an endemic species to China, and is widely distributed from west to east, including in the Sichuan, Yunnan, Gansu, Qinghai, Shannxi, Hubei, Hunan, Fujian and Taiwan Provinces [23], [25]. However, the eastern part of the Tibetan Plateau was the centre of radiation of the genus Apodemus [18], [26]. One recent work sequenced the mitochondrial cytochrome b (cyt b) gene in six A. draco and 197 A. ilex with samples from Yunnan-Guizhou Plateau in China, they reported that A. ilex should be a valid species rather than synonym of A. draco [27]. They also indicated that the phylogenetic pattern of A. ilex was closely related to the complex geographical structures in southwest China [27].
The cyt b gene sequence has been extensively and successfully used in phylogeographic studies for rodents [27]–[33]. Therefore, the nucleotide sequences of the cyt b gene were applied to investigate the genetic variation and evolutionary history of A. draco. We examined: (1) the relationship between the genetic structure of A. draco and geologic or climatic events on the SEMTP, and (2) whether the SEMTP, especially the GMR, potentially functioned as a refuge during the Quaternary period. Details of the historical demography of A. draco revealed in this study could provide new insights into how this rodent and other small mammals in this region responded to the geologic or climatic events on the SEMTP.
Materials and Methods
Ethics Statement
We only used wild rodents in this study. All samples were obtained following the regulations for the implementation of China on the protection of terrestrial wild animals (State Council Decree [1992] No. 13). All our observational and field studies and lab work were approved by Wildlife Protection Office, Sichuan Provincial Forestry Departments (China) and by the Ethics Committee of Sichuan University, China.
Samples and DNA Extraction
One Hundred and three South China field mice were collected from 47 sampling sites in Sichuan Province (Table 1). In addition, 23 cyt b sequences from GenBank were retrieved (GenBank Accession Nos. AB096825, AB109397, AM945800-AM945819, AY389007), which were collected from an additional six sampling sites in the Sichuan and Yunnan Provinces (Table 1) [18], [34], [35]. We combined the very near sampling sites (which located at the same mountain or village) into one population. In total, we used 126 individuals from 20 populations (Table 1). Cyt b sequences from more individuals were in GenBank, but the length of the sequences was significantly shorter (319 bp or 402 bp) than our target sequences (1050 bp), thus we did not include them here. Detailed information on sampling localities is shown in Fig. 1 and Table 1. Voucher specimens were deposited in the Museum of the Sichuan Academy of Forestry. Total genomic DNA was extracted from muscle or liver tissues preserved in 95% ethanol using standard procedures [36].
Table 1. Sampling information of Apodemus draco.
| Populations | Map label | Sample size | Sample names | Haplotypes | Latitude (oN)/Longitude (oE) | Altitude |
| Emeishan, Sichuan | 1 (EMS) | 1 | EMS_1 | EMS1 | – | – |
| Dali, Yunnan | 2 (DL) | 1 | DL_1 | DL1 | – | – |
| Kunming, Yunnan | 3 (KM) | 1 | KM_1 | KM1 | – | – |
| Maerkang, Sichuan | 4 (MEK) | 13 | Mar_1 | MEK1 | – | – |
| Mar_2 | MEK1 | – | – | |||
| Mar_3 | MEK1 | – | – | |||
| Mar_4 | MEK1 | – | – | |||
| Mar_5 | MEK1 | – | – | |||
| Mar_6 | MEK1 | – | – | |||
| Mar_7 | MEK1 | – | – | |||
| Mar_8 | MEK2 | – | – | |||
| Mar_9 | MEK1 | – | – | |||
| Mar_10 | MEK3 | – | – | |||
| Mar_11 | MEK3 | – | – | |||
| Mar_12 | MEK3 | – | – | |||
| Mar_13 | MEK1 | – | – | |||
| Zhongguo, Sichuan | 5 (ZG) | 7 | Zhong_1 | ZG1 | – | – |
| Zhong_2 | ZG2 | – | – | |||
| Zhong_3 | ZG3 | – | – | |||
| Zhong_4 | ZG1 | – | – | |||
| Zhong_5 | ZG4 | – | – | |||
| Zhong_6 | ZG1 | – | – | |||
| Zhong_7 | YX2 | – | – | |||
| Jiajin Mountain, Sichuan | 6 (JM) | 16 | Jiajin_1 | JM4 | 30.84/102.752 | 2720 |
| Jiajin_2 | JM5 | 30.843/102.689 | 1800 | |||
| Jiajin_3 | JM6 | 30.843/102.689 | 3440 | |||
| Jiajin_4 | JM7 | 30.84/102.752 | 2720 | |||
| Jiajin_5 | JM8 | 30.886/102.655 | 3700 | |||
| Jiajin_6 | JM4 | 30.909/102.648 | 1800 | |||
| Jiajin_7 | JM9 | 30.84/102.752 | 2720 | |||
| Jiajin_8 | JM10 | 30.84/102.752 | 2720 | |||
| Jiajin_9 | JM11 | 30.909/102.648 | 1800 | |||
| Jiajin_10 | JM12 | 30.886/102.655 | 3700 | |||
| Jiajin_11 | JM2 | 30.601/102.753 | 3210 | |||
| Jiajin_12 | JM2 | 30.601/102.753 | 2780 | |||
| Jiajin_13 | JM3 | 30.614/102.798 | 2780 | |||
| Jiajin_14 | JM2 | 30.629/102.855 | 3700 | |||
| Jiajin_15 | JM2 | 30.629/102.855 | 3700 | |||
| Jiajin_16 | JM1 | 30.629/102.855 | 1800 | |||
| Jinyang, Sichuan | 7 (JY) | 3 | Jinyang_1 | JY2 | 27.808/103.187 | 3090 |
| Jinyang_2 | ZG3 | 27.808/103.187 | 3090 | |||
| Jinyang_3 | JY1 | 27.808/103.187 | 3090 | |||
| Zhaojue, Sichuan | 8 (ZJ) | 1 | Zhaojue_1 | ZJ1 | 27.985/102.786 | 2140 |
| Hongya, Sichuan | 9 (HY) | 9 | Hongya_1 | HY4 | 29.373/103.034 | 1880 |
| Hongya_2 | HY1 | 29.373/103.034 | 1880 | |||
| Hongya_3 | HY1 | 29.374/103.039 | 1820 | |||
| Hongya_4 | HY2 | 29.373/103.034 | 1880 | |||
| Hongya_5 | HY3 | 29.373/103.034 | 1880 | |||
| Hongya_6 | HY1 | 29.373/103.034 | 1880 | |||
| Hongya_7 | HY6 | 29.374/103.039 | 1820 | |||
| Hongya_8 | HY5 | 29.374/103.039 | 1820 | |||
| Hongya_9 | HY1 | 29.374/103.039 | 1820 | |||
| Yuexi, Sichuan | 10 (YX) | 5 | Yuexi_1 | YX1 | 28.489/102.594 | 2130 |
| Yuexi_2 | YX2 | 28.489/102.594 | 2130 | |||
| Yuexi_3 | YX4 | 28.409/102.745 | 2720 | |||
| Yuexi_4 | GM1 | 28.409/102.745 | 2720 | |||
| Yuexi_5 | YX3 | 28.489/102.594 | 2130 | |||
| Danba, Sichuan | 11 (DB) | 1 | Danba_1 | DB1 | 30.614/101.736 | 3610 |
| Kangding, Sichuan | 12 (KD) | 1 | Kangding_1 | KD1 | 30.499/102.297 | 2400 |
| Jiulong, Sichuan | 13 (JL) | 3 | Jiulong_1 | ZG3 | 29.222/102.958 | 2570 |
| Jiulong_2 | JL1 | 28.675/101.610 | 2238 | |||
| Jiulong_3 | JL2 | 28.675/101.610 | 2238 | |||
| Shimian, Sichuan | 14 (SM) | 1 | Shimian_1 | SM1 | 29.025/102.391 | 2080 |
| Gongga Mountain, Sichuan | 15 (GM) | 25 | Gongga_1 | GM1 | 29.351/101.873 | 2912 |
| Gongga_2 | GM3 | 29.410/101.928 | 2602 | |||
| Gongga_3 | GM2 | 29.396/101.971 | 2388 | |||
| Gongga_4 | GM4 | 29.410/101.928 | 2602 | |||
| Gongga_5 | GM9 | 29.702/102.017 | 2360 | |||
| Gongga_6 | GM11 | 29.702/102.017 | 2360 | |||
| Gongga_7 | GM7 | 29.574/101.999 | 3010 | |||
| Gongga_8 | GM9 | 29.574/101.999 | 3010 | |||
| Gongga_9 | GM9 | 29.574/101.999 | 3010 | |||
| Gongga_10 | GM5 | 29.574/101.999 | 3010 | |||
| Gongga_11 | GM9 | 29.576/102.002 | 3000 | |||
| Gongga_12 | GM5 | 29.576/102.002 | 3000 | |||
| Gongga_13 | GM9 | 29.698/102.002 | 3150 | |||
| Gongga_14 | GM6 | 29.576/102.002 | 3000 | |||
| Gongga_15 | GM8 | 29.574/101.999 | 3010 | |||
| Gongga_16 | GM5 | 29.574/101.999 | 3010 | |||
| Gongga_17 | GM9 | 29.574/101.999 | 3010 | |||
| Gongga_18 | JM3 | 29.698/102.002 | 3150 | |||
| Gongga_19 | GM10 | 29.698/102.002 | 3150 | |||
| Gongga_20 | JM3 | 29.685/101.929 | 3460 | |||
| Gongga_21 | JM3 | 29.574/101.999 | 3010 | |||
| Gongga_22 | GM9 | 29.698/102.002 | 3150 | |||
| Gongga_23 | JM3 | 29.698/102.002 | 3150 | |||
| Gongga_24 | GM9 | 29.685/101.929 | 3460 | |||
| Gongga_25 | GM5 | 29.574/101.999 | 3010 | |||
| Erlang Mountain, Sichuan | 16 (EM) | 13 | Erlang_1 | EM2 | 29.852/102.263 | 2780 |
| Erlang_2 | EM5 | 29.852/102.263 | 2780 | |||
| Erlang_3 | EM5 | 29.852/102.263 | 2780 | |||
| Erlang_4 | EM5 | 29.856/102.262 | 2780 | |||
| Erlang_5 | EM6 | 29.856/102.261 | 2750 | |||
| Erlang_6 | EM5 | 29.843/102.251 | 2450 | |||
| Erlang_7 | HY1 | 29.865/102.278 | 3130 | |||
| Erlang_8 | EM3 | 29.872/102.312 | 2250 | |||
| Erlang_9 | EM4 | 29.873/102.309 | 2220 | |||
| Erlang_10 | HY1 | 29.873/102.309 | 2220 | |||
| Erlang_11 | EM5 | 29.879/102.345 | 1920 | |||
| Erlang_12 | EM1 | 29.885/102.299 | 2450 | |||
| Erlang_13 | HY1 | 29.885/102.299 | 2450 | |||
| Qichuan, Sichuan | 17 (QC) | 10 | Qingchuan_1 | QC1 | 32.568/104.764 | 1450 |
| Qingchuan_2 | QC4 | 32.581/104.755 | 1450 | |||
| Qingchuan_3 | QC5 | 32.578/104.742 | 1500 | |||
| Qingchuan_4 | QC2 | 32.568/104.764 | 1450 | |||
| Qingchuan_5 | QC6 | 32.581/104.755 | 1450 | |||
| Qingchuan_6 | QC7 | 32.581/104.755 | 1450 | |||
| Qingchuan_7 | QC3 | 32.579/104.759 | 1450 | |||
| Qingchuan_8 | QC8 | 32.579/104.759 | 1450 | |||
| Qingchuan_9 | QC3 | 32.568/104.764 | 1450 | |||
| Qingchuan_10 | QC9 | 32.918/105.478 | 1650 | |||
| Daocheng, Sichuan | 18 (DC) | 4 | Daocheng_1 | DC1 | 28.451/100.345 | 3890 |
| Daocheng_2 | DC2 | 28.451/100.345 | 3890 | |||
| Daocheng_3 | DC3 | 28.451/100.345 | 3890 | |||
| Daocheng_4 | DC4 | 28.452/100.347 | 3890 | |||
| Maoxi, Sichuan | 19 (MX) | 5 | Maoxian_1 | MX1 | 31.694/103.905 | 1940 |
| Maoxian_2 | MX2 | 31.694/103.905 | 1940 | |||
| Maoxian_3 | MX1 | 31.694/103.905 | 1940 | |||
| Maoxian_4 | MX1 | 31.694/103.905 | 1940 | |||
| Maoxian_5 | MX1 | 31.694/103.905 | 1940 | |||
| Heishui, Sichuan | 20 (HS) | 6 | Heishui_1 | HS4 | 32.244/103.153 | 2240 |
| Heishui_2 | HS1 | 32.232/103.154 | 2470 | |||
| Heishui_3 | HS2 | 32.323/103.129 | 2390 | |||
| Heishui_4 | HS3 | 32.323/103.129 | 2390 | |||
| Heishui_5 | HS3 | 32.241/103.153 | 2240 | |||
| Heishui_6 | HS1 | 32.379/103.155 | 2540 |
We did not have the coordinates of the individuals in the first five populations because their sequences are from GenBank. Their references and GenBank accession numbers can be found in text.
Amplification and Sequencing
Polymerase chain reaction (PCR) was used to amplify the sequences of cyt b with the universal primers L14724/H15915 [37]. PCR conditions included an initial denaturation for 5 min at 95°C, 35 cycles of denaturation for 40 s at 94°C, 45 s annealing at 50–60°C, and a 1 min extension at 72°C followed by a final extension at 72°C for 10 min. The amplification was performed in 25 µl reaction volumes with 2.5 µl of 10×EX Taq buffer (Mg2+ Free; TaKaRa Biotech), 1.5 µl dNTP (2.5 mM each), 2.0 µl MgCl2 (25 mM), 1 µl of each primer (10 uM), 0.2 µl EX Taq polymerase (5 U/µl, TaKaRa Biotech), and approximately 200 ng total genomic DNA as template. PCR products were sequenced from both directions with the same PCR primers in an ABI PRISM 3730 DNA sequencer (Applied Biosystems, Inc.).
Genetic Diversity and Protein Sequences
We aligned the sequences in software MEGA 4.0 [38]. Nucleotide saturation was tested by plotting transitions and transversions against the JC69 distance using DAMBE [39]. Haplotype diversity (h) and nucleotide diversity (π) were estimated using DnaSP Ver. 5 [40]. Genetic differentiation between populations was evaluated by estimating pairwise values of FST with the software Arlequin 3.1 [41]. The significance was assessed by 1000 permutations. Populations contain only one sample were excluded in this analysis. Pairwise comparisons were used as genetic distances in the following analyses, such as isolation by distance. McDonald–Kreitman test [42] was used to test the overall selection on cyt b on the website (http://mkt.uab.es) [43]. In this analysis, the numbers of synonymous and nonsynonymous substitutions in cyt b of A. draco and A. latronum (Nos. GU908339-GU908394) were estimated and their ratio was compared for each species with the same ratio for fixed mutations between the species. The cyt b sequences of A. latronum were used here due to A. latronum was the sister taxon to A. draco.
Phylogenetic Analysis
Two different methods of phylogenetic analysis were used to reconstruct phylogenetic relationships among cyt b haplotypes: Bayesian inference (BI) and maximum parsimony (MP). Apodemus latronum (GU908426-GU908428, GU908433, GU908436) was used as outgroup due to it was the sister taxon to A. draco. In order to avoid the possible bias, A. flavicollis (AB032853) and A. sylvaticus (AB033695) were also chosen as outgroups. The best fit model of DNA substitution was obtained using MrModeltest2.2 [44] under the Akaike Information Criterion (AIC). BI was performed with MrBayes 3.1.2 [45]. For BI, we used two different datasets, partitioned and non-partitioned. For partitioned dataset, the nucleotides sequences were partitioned by codon position. In total, three partitions were given; one each for codon positions 1, 2, and 3 cyt b. Posterior distributions were obtained by the Markov Chain Monte Carlo (MCMC) method with one cold chain and three heated chains for 10,000,000 generations and sampled every 1000 generations. The first 25% were discarded as a conservative burn-in and the remaining samples were used to generate a 50% majority rule consensus tree. PAUP* 4.0 b 10 [46] was used to perform MP analysis. A heuristic search strategy was employed with the tree bisection and reconnection (TBR) branch swapping algorithm, random addition of taxa and 1000 replicates per search. Supports for branches in the MP trees were tested by bootstrap analysis with 1000 replicates.
Since the haplotype network could better detect the relationship among haplotypes, we also constructed a median-joining network (MJN) approach to depict relationships among the cyt b haplotypes of A. draco using Network 4.6.0.0 [47].
Landscape Analyses
We used analysis of molecular variance (AMOVA) to estimate the geographical pattern of population subdivision using different grouping options [48]. To test for the correlation between genetic differentiation and geographic distance, we performed a Mantel test [49] and a spatial autocorrelation analysis. In Mantel test, we used the Euclidean distance between sampling localities, based on GPS coordinates, as the geographic distance. Because we did not have the coordinates of the samples from GenBank, we approximated distance using mountains or county locality information listed in the corresponding papers [18], [34], [35]. We used three different datasets to perform the Mantel test. First one included 13 populations (exclude populations that only contain one sample); second one further excluded all the samples from other studies since we did not have their exact coordinates. Third one only contained the populations in clade 1 (Jiajin, Gongga and Erlang Mountains, and Hongaya county). The significance of the Mantel test was determined by 1,000 permutations using the isolation by distance (IBD) web service v3.16 (http://ibdws.sdsu.edu) [50]. The spatial autocorrelation analysis was performed in Allele in Space (AIS) [51]. The spatial autocorrelation algorithm calculates the statistic Ay, the average genetic distance between individuals in distance class y. Thus, Ay can be interpreted as the average genetic distance between pairs of individuals that fall within distance classes [51]. In order to ensure that the arbitrary defined distance class size had no effect on result, analyses were performed using 5, 10 and 15 distance classes, respectively. We used a randomisation procedure consisting of 5,000 replicates to identify distance classes where average genetic distances were significantly larger or smaller than random expectations. Because this algorithm uses genetic and spatial data for every individual, we preformed this analysis on different data set, all individuals and individuals without GenBank samples based on the same reason we explained above.
Molecular Dating
Divergence times among the observed mitochondrial clades on phylogenetic tree were estimated by BEAST v1.7.1 [52] using a Bayesian coalescence approach. We also used partitioned (by codon positions) and non-partitioned data. For partitioned data, we had two different partitioned datasets here. The first one had two partitions (pos1+pos2, pos3), and the second one had three (pos1, pos2, pos3). We used an uncorrelated lognormal relaxed clock with the GTR+I+G model of substitution. All analyses were performed using a coalescent-constant size process of diversification. The prior for mean mutation rate was specified as a lognormal distribution, with a mean of 0.02, log (standard deviation) of 0.56 and offset of 0.0172. This distribution covered the range from 2% to 8% substitutions per site per Myr (million years), with the 95% highest posterior density (HPD) range of 2.4% to 6.01% per Myr. We used this prior because the standard divergence rate for the mammalian cyt b gene is estimated at 2% per Myr [53], and the evolutionary rate of cyt b in genus Apodemus is estimated at 2.4% per Myr [34], [54]. However, a number of studies [28], [29], [55], [56] indicate that the cyt b gene of rodents in general evolved several times faster than the standard rate. Moreover, Fan et al. [22] applied a rate of 2.4–8% per Myr for a phylogeographic analysis of another field mouse, A. latronum in their study. Monophyly of clades was not enforced, and runs were initiated on random starting trees. The MCMC chain was run for 50 million generations, sampling every 1,000 generations. The convergence of the chains to stationary was checked using Tracer 1.5 [57] and the first 25% were discarded as burn-in. The final molecular clock tree was produced in TreeAnnotator 1.7.1 [58] using the mean as node heights. Finally, Figtree v1.3.1 [59] was used to visualize the results. Although the divergence times were only rough estimates here, it will provide useful information when we discuss A. draco’ s divergence.
Historical Demography
The demographic history was tested by mismatch distributions and neutrality tests based on the cyt b sequences. First, two tests of selective neutrality were calculated for each set (whole sample and each observed phylogenetic clade). Fu’s Fs [60] was calculated in Arlequin 3.1 [41], and Ramos-Onsins and Rozas’s R2 statistics [61] was calculated in DnaSP Ver. 5 [40]. Fu’s Fs and R2 were applied due to the fact that they have been suggested as the most powerful tests for detecting sudden population growth or contractions [61], [62]. Fu’s Fs is more powerful for large population sizes, whereas R2 works well for small ones [61]. The sum of squared deviations (SSD) and the raggedness index (r) [63] were also estimated in Arlequin 3.1 to determine whether the sequences deviated significantly from a model of population expansion. Significance of all the statistics was calculated using 1,000 coalescent simulations with the software Arlequin 3.1 and DnaSP Ver. 5. Mismatch analyses [64] were carried out using DnaSP Ver. 5. The time of possible population expansions (t) was calculated according to τ = 2 ut [65], where τ was the model of the mismatch distribution and u was the mutation rate of the DNA sequence. The value u was derived from u = µk, where μ was the mutation rate per nucleotide and k was the number of nucleotides. We used an estimated evolutionary rate of 2–8% per Myr for cyt b (rationale is provided in previous section).
Results
Sequence Information
We generated 103 cyt b gene sequences each consisting of 1050 sites (GenBank Accession Nos. HM162766 - HM162833, JQ424902 - JQ424910). With sequences from the additional 23 individuals, we recovered 76 haplotypes from 126 individuals, and the overall h was 0.982, while the π was 0.0381 (Table 2). Saturation analysis did not show any evidence of saturation, so our sequences were used for further analyses. We were confident that the cyt b sequences obtained in our study represented the mitochondrial cytochrome b gene since all cyt b sequences could be translated into amino acid sequences without interruption and closely matched the previously published cyt b sequence for A. draco. The McDonald–Kreitman test did not provide overall evidence for positive selection on the cyt b gene [42], [43]. The value of Pn/Ps (polymorphism) was larger than Dn/Ds (divergence), and the difference between them was not significant (Table 3).
Table 2. Genetic diversity indices and demographical analyses for cyt b sequences in Apodemus draco.
| mtDNA clades | n | H | h (SD) | π (SD) | Fu’s Fs | R2 | Mismatch distribution | r | SSD |
| Clade 1 | 56 | 31 | 0.951 | 0.0066 | −12.969 | 0.065 | – | 0.008 | 0.006 |
| Subclade 1A | 28 | 15 | 0.902 | 0.0025 | −8.201 | 0.064 | unimodal | 0.020 | 0.001 |
| Subclade 1B | 18 | 7 | 0.752 | 0.0016 | −2.068 | 0.104 | unimodal | 0.110 | 0.023 |
| Subclade 1C | 10 | 9 | 0.978 | 0.0042 | −4.067 | 0.139 | multimodal | 0.069 | 0.016 |
| Clade 2 | 13 | 3 | 0.500 | 0.0010 | 0.930 | 0.164 | multimodal | 0.679 | 0.173 |
| Clade 3 | 21 | 15 | 0.957 | 0.0050 | −5.779 | 0.081 | multimodal | 0.016 | 0.009 |
| Clade 4 | 30 | 21 | 0.968 | 0.0084 | −6.044 | 0.112 | multimodal | 0.013 | 0.008 |
| Clade 5 | 6 | 6 | 1.000 | 0.0064 | −1.606 | 0.257 | multimodal | 0.071 | 0.046 |
| Total | 126 | 76 | 0.982 | 0.0381 | −8.413 | 0.108 | – | 0.002 | 0.012 |
Number of individuals (n), number of haplotypes (H), haplotype diversity (h), nucleotide diversity (π), Fu’s Fs, Ramos-Onsins and Rozas’s R2 statistics (R2), the shape of mismatch distribution, the sum of squared deviations (SSD) and raggedness indexes (r) are shown. Numbers in bold indicate statistically significant genetic differentiation (P<0.05).
Table 3. The result of McDonald-Kreitman test for cyt b gene of Apodemus draco (N = 126) and A. latronum (N = 68).
| Polymorphism | Divergence | |
| Non-Neutral | 66 | 6 (6.03) |
| Neutral | 243 | 45 (50.94) |
| Ratio | 0.27 | 0.13 (0.12) |
| Neutrality Index | 2.037 (2.294) | |
| χ2 P-value | 0.112 (0.06) | |
Values corrected by Jukes and Cantor (1969) are given in parentheses.
Phylogeographic Structure
Based on the AIC test, the GTR+I+G model was chosen for the cyt b. For partitioned data, different models of sequence evolution were detected (pos1: SYM+I; pos2: HKY+I; pos3: GTR+I+G). The 50% majority consensus tree based on BI was shown in Fig. 2. MP tree recovered the same tree topology and was not shown. The phylogenetic trees based on partitioned and non-partitioned datasets exhibited the same topology.
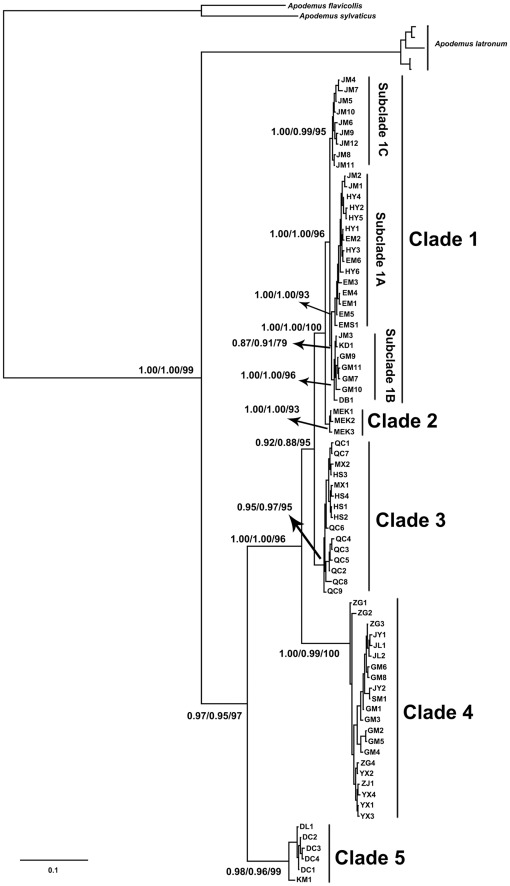
Figure 2. Fifty percent majority rule consensus tree from Bayesian analysis of Apodemus draco based on all the haplotypes of cyt b data.
Numbers represent node supports inferred from Bayesian posterior probability of non-partitioned data and partitioned data, and maximum parsimony bootstrap analyses, respectively. Only values of the main evolutionary clades are shown. The haplotype names can be found in Table 1.
Five major evolutionary clades, based on geographic locations, were identified: Maerkang (clade 2), northern Sichuan (including Qingchuan, Maoxian and Heishui counties, clade 3), southern Sichuan (including Jinyang, Zhaojue, Yuexi, Jiulong and Shimian counties and the Gongga Mountain and Zhongguo, clade 4), and Daocheng county and Yunnan Province (clade 5) (Fig. 2). Haplotypes from Jiajin Mountain, Emei Mountain, Erlang Mountain, Gongga Mountain, and Hongya, Danba and Kangding counties formed clade 1. In this clade, three subclades were identified. Haplotypes from Jiajin Mountain clustered together with samples from Erlang Mountain, Emei Mountain and Hongya county, and then they formed subclade 1A. One haplotype (JM3) from Jiajin Mountain fell into subclade 1B, which was clustered with individuals from Gongga Mountain, Danba and Kangding Counties. Then, the remaining haplotypes from Jiajin Mountain (11 individuals, nine haplotypes) formed subclade 1C. Thus, the haplotypes from Gongga Mountain distributed in both clade 1 and clade 4, and samples from Jiajin Mountain appeared in all the subclades of clade 1.
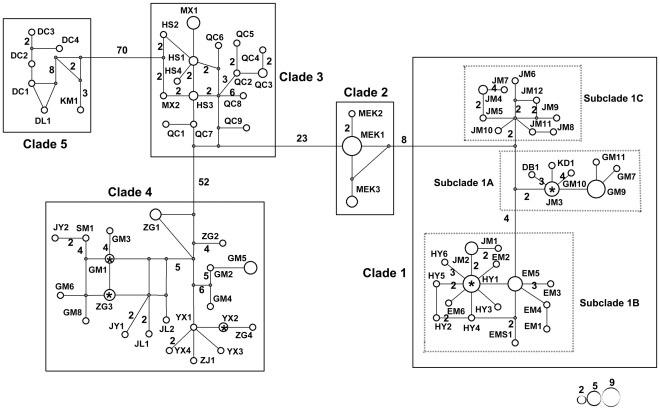
The haplotype network also revealed five clades separated by 9 to 73 mutational steps (Fig. 3), which was consistent with the phylogenetic trees. The subclades within clade 1 only separated by four to six mutational steps, which exhibited very close relationships. Moreover, haplotype YX2 was found at samples both from Yuexi and Zhongguo, HY1 was identified in individuals from Hongya and Erlang Mountain, and JM3 was observed in individuals from Jiajin Mountain and Gongga Mountain.
Figure 3. Median-joining network of all cyt b haplotypes found in Apodemus draco.
The missing haplotypes in the network are represented by gray dots. Circle sizes are proportional to the number of individuals sharing the same haplotypes. Each mutation step is shown as a short line connecting neighboring haplotypes, and numbers of mutations between haplotypes are indicated near branches if it is greater than one. Haplotype designations can be found in Table 1. Evolutionary clades correspond to the five major clades and three subclades in Fig. 2. Haplotypes from different populations are marked as *.
Genetic Variation and Landscape Genetic Analyses
The F ST values for pairwise populations comparisons ranged from −0.026 to 0.985. Of the pairwise values obtained, only 3 of the 78 were nonsignificant (Table 4). The AMOVA analysis showed a very strong differentiation among different populations or evolutionary clades, and showed that among populations or clades variation accounted for 66% (p<0.001) or 86% (p<0.001) of the overall variation. However, when we focused on the populations in clade 1, the result exhibited that among populations variation only accounted for 26% of the overall variation, but 48% of the overall variation was contributed by variation among samples (Table 5).
Table 4. Table 4. Pairwise F ST values among the 13 populations surveyed of Apodemus draco.
| 4 (MEK) | 5 (ZG) | 6 (JM) | 7 (JY) | 9 (HY) | 10 (YX) | 13 (JL) | 15 (GM) | 16 (EM) | 17 (QC) | 18 (DC) | 19 (MX) | |
| 5 (ZG) | 0.959 | – | ||||||||||
| 6 (JM) | 0.700 | 0.907 | – | |||||||||
| 7 (JY) | 0.970 | 0.406 | 0.907 | – | ||||||||
| 9 (HY) | 0.904 | 0.950 | 0.379 | 0.958 | – | |||||||
| 10 (YX) | 0.969 | 0.059 | 0.911 | 0.415 | 0.960 | – | ||||||
| 13 (JL) | 0.981 | 0.470 | 0.918 | -0.026 | 0.972 | 0.560 | – | |||||
| 15 (GM) | 0.379 | 0.426 | 0.287 | 0.386 | 0.312 | 0.416 | 0.412 | – | ||||
| 16 (EM) | 0.919 | 0.961 | 0.421 | 0.970 | 0.058 | 0.970 | 0.980 | 0.348 | – | |||
| 17 (QC) | 0.884 | 0.907 | 0.794 | 0.909 | 0.872 | 0.913 | 0.923 | 0.429 | 0.894 | – | ||
| 18 (DC) | 0.985 | 0.954 | 0.934 | 0.957 | 0.978 | 0.964 | 0.979 | 0.666 | 0.984 | 0.942 | – | |
| 19 (MX) | 0.958 | 0.939 | 0.822 | 0.952 | 0.946 | 0.955 | 0.979 | 0.394 | 0.959 | 0.310 | 0.984 | – |
| 20 (HS) | 0.956 | 0.941 | 0.828 | 0.954 | 0.945 | 0.956 | 0.978 | 0.409 | 0.958 | 0.325 | 0.983 | −0.004 |
Populations contain only one sample are exclude in this analyses.
Numbers in bold indicate statistically significant genetic differentiation (P<0.05). Populations are numbered as in Table 1.
Table 5. Table 5. Results of hierarchical analyses of molecular variance (AMOVA) conducted using different grouping options.
| Groups compared | No. groups | Source of variation | d.f. | Percentage variation (%) | P value | F statistics |
| All populations | 13 | Among populations | 12 | 66.52 | 0.000 | F CT = 0.665 |
| Among sites within populations | 21 | 13.87 | 0.016 | F SC = 0.414 | ||
| Among samples within sites | 85 | 19.61 | 0.000 | F ST = 0.804 | ||
| Populations in clade 1* # | 4 | Among populations | 3 | 26.35 | 0.019 | F CT = 0.264 |
| Among sites within populations | 14 | 25.49 | 0.017 | F SC = 0.346 | ||
| Among samples within sites | 45 | 48.15 | 0.000 | F ST = 0.518 | ||
| Main clades* | 5 | Among clades | 4 | 86.37 | 0.000 | F CT = 0.864 |
| Among populations within clades | 9 | 7.06 | 0.000 | F SC = 0.518 | ||
| Among samples within populations | 105 | 6.58 | 0.000 | F ST = 0.934 | ||
| Subclades in clade 1* | 3 | Among subclades | 2 | 70.73 | 0.016 | F CT = 0.707 |
| Among populations s within subclades | 3 | 10.76 | 0.000 | F SC = 0.368 | ||
| Among samples within populations | 47 | 18.51 | 0.000 | F ST = 0.815 |
Populations contain only one sample are exclude in all the analyses Main evolutionary clades and subclades in clade 1 are observed in our phylogenetic trees.
# Jiajin Moutain, Erlang Moutain, Gongga Mountain, Hongya county.
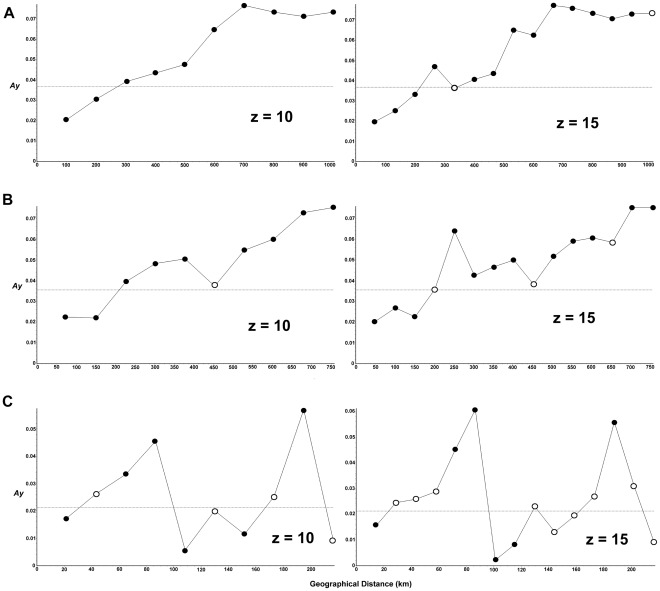
Three different datasets were applied to perform the Mantel test. A significant correlation between genetic (F ST) and geographic distances was detected in first two datasets (13 populations: p = 0.0009, r = 0.4122; 11 populations: p = 0.0006, r = 0.4944), indicating high levels of geographical structure for A.draco. But for the dataset only including 4 populations in clade 1, the result was non-significant (p = 0.3800, r = 0.1831). Spatial autocorrelation analysis (SAA) also found a significant relationship between genetic and geographic distances in different distance classes (5, 10 and 15, only show the results of 10 and 15). In analyses performed using 10 distance classes, both the whole individuals dataset and without GenBank samples dataset were significant smaller than random expectations over the first two shortest distance classes (geographic distance up to ∼200 and 150 km, respectively). Then, when the geographic distance increased (start from 300 and 200 km, respectively), both the datasets yielded significant values of Ay that were higher than random expectations. Similar results were obtained when 5 and 15 distance classes were used for analyses (Fig. 4). Different results were observed when we performed the analyses with individuals only in clade 1. First, it yielded significant smaller value than random expectations at the shortest distance class (∼20 km). Then, significant higher values than random expectations were observed at around 60–80 km. But this dataset showed significant smaller and higher values again at around 100 km and 180 km, respectively (Fig. 4).
Figure 4. Results of spatial autocorrelation analyses of cyt b sequences in Apodemus draco.
Analyses were performed using different distance classes (5, 10 and 15). Only the results of z = 10 and 15 distance classes were shown. Black circles represent values significantly different (p<0.05) from the mean Ay. (a) All individuals. (b) Exclude all the GenBank sequences due to we did not have their coordinates. (c) Individuals in clade 1.
Estimation of Divergence Times
Although we applied three different datasets to estimate the divergence time, we got the very similar results. Therefore, we described and discussed the divergence times based on the three partitions dataset (Table 6). The divergence time between clade 5 and the remaining clades occurred during the Pliocene/Pleistocene boundary at approximately 2.20 Mya, although the range of the 95% HPD interval spanned the period from the late Pliocene through the middle Pleistocene, between 3.80 and 0.87 Mya (Table 6). The divergence between clade 4 and its sister clade and clade 3 and its sister clade occurred in the middle Pleistocene at 1.22 Mya (95% HPD 0.50-2.06 Mya) and 0.55 Mya (95% HPD 0.22-0.96 Mya), respectively. The divergence between clade 1 and clade 2 was the youngest split, which also occurred in the middle Pleistocene at 0.26 Mya (95% HPD 0.09-0.45 Mya). The splits within clade 1 occurred during the late/middle Pleistocene boundary at 0.13 Mya and 0.16 Mya.
Table 6. Result of the Bayesian coalescent-based estimation of divergence time among different evolutionary clades/subclades and the time to the most recent common ancestor (t MRCA) of different evolutionary clades/subclades.
| Divergence time estimate (Mya) | |||
| Evolutionary clades | Non-partition | Two partitions | Three partitions |
| Clade 1 vs. Clade 2 | 0.2539 (0.0903–0.4742) | 0.2544 (0.096–0.4648) | 0.2566 (0.0942–0.4530) |
| Clade 3 vs. Clade 1 - Clade 2 | 0.5515 (0.1965–0.9945) | 0.5490 (0.2089–0.9809) | 0.5463 (0.2215–0.9607) |
| Clade 4 vs. Clade 1 - Clade 2 - Clade 3 | 1.1659 (0.4652–1.9848) | 1.1958 (0.4755–2.0324) | 1.2180 (0.4971–2.0552) |
| Clade 5 vs. Clade 1 - Clade 2 - Clade 3 - Clade 4 | 2.0905 (0.8492–3.4836) | 2.1829 (0.8575–3.8453) | 2.1952 (0.8733–3.8037) |
| Subclade 1A vs. Subclade 1B | 0.1302 (0.0488–0.2382) | 0.1311 (0.0519–0.2341) | 0.1312 (0.0523–0.2314) |
| Subclade 1C vs. Subclade 1A - Subclade 1B | 0.1612 (0.0590–0.2956) | 0.1628 (0.0653–0.2929) | 0.1635 (0.0644–0.2862) |
| t MRCA | |||
| Clade 1 | 0.1612 (0.0590–0.2956) | 0.1628 (0.0653–0.2929) | 0.1635 (0.0644–0.2862) |
| Clade 2 | 0.0444 (0.0090–0.1086) | 0.0443 (0.0092–0.1085) | 0.0447 (0.0107–0.1071) |
| Clade 3 | 0.1251 (0.0419–0.2474) | 0.1272 (0.0452–0.2450) | 0.1283 (0.0455–0.2404) |
| Clade 4 | 0.2098 (0.0747–0.3887) | 0.2080 (0.0809–0.3881) | 0.2093 (0.0778–0.3720) |
| Clade 5 | 0.2038 (0.0571–0.4302) | 0.2217 (0.0655–0.4540) | 0.2211 (0.0652–0.4558) |
| Subclade 1A | 0.0788 (0.0266–0.1490) | 0.0800 (0.0282–0.1495) | 0.0805 (0.0294–0.1497) |
| Subclade 1B | 0.0681 (0.0181–0.1278) | 0.0622 (0.0189–0.1231) | 0.0625 (0.0206–0.1231) |
| Subclade 1C | 0.0780 (0.0222–0.1541) | 0.0793 (0.0242–0.1528) | 0.0799 (0.0257–0.1554) |
Results based on non-partition and partitioned by each codon were shown (two partitions: pos1+pos2, pos3; three partitions: pos1, pos2, pos3). Mean values are shown in bold and 95% HPD are shown in brackets.
Historical Demography
We did not include the data sets of whole sample and clade 1 in mismatch distribution because the phylogenetic trees indicate they exhibit a high level of population structure. But the non-significant values of Fu’s Fs and Ramos-Onsins and Rozas’s R2 indicated that the whole population maintained a stable population size. The mismatch distribution analysis indicated a ragged and multimodal distribution (Table 2), suggesting a relatively constant population size for the clades 2, 3, 4 and 5, this result was also supported by non-significant values of Fu’s Fs and R2 tests for clades 2 and 5 (Table 2). However, Fu’s Fs for clade 4, and Fu’s Fs and R2 for clade 3 showed significant values. Thus, considering the multimodal distribution in mismatch distribution of clades 3 and 4, it was hard to conclude whether they experienced a population expansion.
The Subclade 1C showed multimodal distribution and non-significant value in R2, but a significant Fu’s Fs value. This inconsistent pattern maybe caused by single sampling region of this subclade. We did not calculate the possible expansion time for clade1, because historical expansion in subclade 1A and subclade 1B was also supported by unimodal mismatch distributions and significant negative values of all the neutrality tests (Table 2). The values of τ were different in subclades 1A and 1B, indicating that clade 1 did not simply experience a single expansion. Therefore, we calculated the possible expansion times for subclades 1A and 1B instead of clade 1. Here, τ = 2.166 for subclade 1A and τ = 0.598 for subclade 1B, generation time was one year, and k was 1050. Thus, subclade 1A and subclade 1B underwent expansions at 0.052-0.013 Mya and 0.014-0.004 Mya, respectively.
Discussion
Sakka et al. [35] studied the phylogeography of four Apodemus species (A.draco, A.latronum, A.peninsulae, and A.agrarius) in the Asian Far East. Results from our analyses were different from this study in several ways. First, in their study, the cyt b haplotypes from Yunnan Province did not cluster with any other haplotypes from Sichuan Province, and they formed a single lineage [35]. However, in our study, individuals from Yunnan Province (populations 2 and 3) clustered together with individuals from Daocheng (population 18), Sichuan Province (Fig. 2). Second, haplotypes from Maerkang, Emei Mountain and Baoxing fell into one lineage [35], but phylogenetic tree in our study clearly showed that individuals from Maerkang (population 4) formed clade 2, and samples from Emei Mountian (population 1) and Jiajin Mountain (population 6) fell into subclade 1A and subclade 1C, respectively (Fig. 2). It is possible that their [35] limited numbers of individuals and sampling sites (individuals: 13 in Yunnan and 22 in Sichuan; sampling sites: 4 in Yunnan and 4 in Sichuan) caused these differences. Based on the topographic complexity and unique geologic history of the SEMTP, it is necessary to use more samples in this region to investigate the relationship between geologic events and local species’ evolutionary history.
Apodemus draco exhibits a high level of population structure in our study. Most individuals from the same or nearby sampling localities clustered together, with the exception of individuals from Gongga Mountain (in both clades 1 and 2) and individuals from Jiajin Mountain (in different subclades of clade 1). The results of multiple landscape analyses further support the geographic structuring of genetic diversity in this species. As shown in Figure 1, some big rivers and mountains would act like barriers, such as Yalong River, Dadu River, and Qionglaishan Mountain and Jiajin Moutain. But other rivers and mountains did not act as barriers, such as Daxiangling Mountain. Hongya County (population 9) and Emei Mountain (population 1) located at right side of Daxiangling Mountain, but their samples were clustered with samples from its left side (populations 6, 11, 12, 15, 16). Therefore, the phylogeographical structure of A. draco was very complicated due to the complex geologic history and topography of the SEMTP. However, an unexpected pattern was observed in clade 4. Individuals from the central part of Sichuan Province, Zhongguo (population 5), clustered together with samples from southern Sichuan Province, including Jinyang, Zhaojue, Yuexi, Jiulong and Shimian Counties (populations 7, 8, 10, 13, 14) and the Gongga Mountain (population 15), whereas the samples from Hongya County (population 9) and Emei Mountain (population 1), which were more close to the sampling sites in southern Sichuan, did not fall into clade 4 (Fig. 1). Further studies are needed to elucidate the unique pattern found in clade 4.
The estimated divergence times of A. draco’s main splits (2.20–0.55 Mya) were congruent with the periods of the Qingzang Movement (3.6–1.7 Mya) and Kun-Huang Movement (1.2–0.6 Mya) [9]–[11]. These two movements were known as the most intense uplift events in the Tibetan Plateau. These events shaped the geologic configuration of this area, making the topography of the plateau very complex [13]. Because small rodents are sensitive to the changing environment, possess limited migration abilities [22], [24], therefore, many phylogenetic and phylogeographic studies [3], [4], [14], [18], [22] have demonstrated that uplift events and environmental complexity played an important role in the diversification of local species. It is reasonable to suggest that the initial divergence of A. draco would have been shaped by the uplift events of the Tibetan Plateau, and high levels of genetic diversity have beeen maintained as a result of these events. Although the divergence times in our study was rough estimates, this conclusion was supported by Sakka et al. [35]’s study. In their work, the divergence time between the main lineages of A.draco corresponded to periods from 1.5–1.7 Mya to 2.0-2.2 Mya, and they further suggested that the complex land conditions could explain the divergence of A. draco in Sichuan Province.
The Tibetan Plateau underwent four or five glaciations during the Quaternary period [66], and the frequency and range in this plateau was less than that in other regions in Asia [66], [67]. According to geologic data, the extensive glacial period (EGP) on the Tibetan Plateau occurred during the middle Pleistocene (about 0.5 Mya), and continued until 0.17 Mya [66], [68], [69]. The last glacial period (LGP) occurred 0.08–0.01 Mya, in which the last glacial maximum (LGM) was 0.021–0.017 Mya [9], [70]. During the EGP, ice coverage could be permanent at high elevations and central regions of the Tibetan Plateau [69], [70], [71]. Conversely, the frequency of glaciers in the east was less than that in the west, and indeed, no large ice sheets were found in the SEMTP, so many continuously ice-free areas were likely to have existed in the SEMTP because of its lower elevation and the complicated geologic configuration [66], [67], [70], [71]. Consequently, many areas located in the SEMTP could provide suitable refugia, allowing A. draco to maintain stable population sizes.
Indeed, some recent phylogeographic studies have indicated that the large mountain ranges on the east edge of the Tibetan Plateau could provide substantial refugia for the local species during the Quaternary [19], [22], [72]. However, this did not mean the major glaciations in the Pleistocene did not have any effect on the evolution of A. draco. The estimated divergence time showed that the split between clade 1 and clade 2 occurred at 0.26 Mya, which fell into the EGP (0.5–0.17 Mya). Thus, it was possible that the largest glaciation further isolated these groups, which could promote the differentiation. Similarly, Zhan et al. [72]’s study showed that the major glaciations during the Pleistocene have had a major impact upon the evolution of the blood pheasant (Ithaginis cruentus).
The IBD and SAA results showed that there was no clear relationship between geographic distance and genetic distance in the central region (clade 1), although there were many huge mountains. It seems that there is a certain connection between the pattern in clade 1 and the regional historical demography. Indeed, two of the subclades in clade 1 experienced population expansions.
Subclade 1A was estimated to have experienced population expansions at about 0.052–0.013 Mya, after the EGP (0.5–0.17 Mya), and probably before the LGM (0.021–0.017 Mya). Most individuals of this subclade were collected from Hongya County (population 9). Its elevation is low, and the sampling site in this county (about 1800 m) was lower than other sampling sites (2450–3700 m) in subclade 1A. Thus, Hongya could have provided suitable habitats for species during the glacial period. During the population expansions, many individuals from nearby areas might have migrated into Hongya County. At the same time, some individuals in these regions could have dispersed into nearby areas. This expansion scenario could explain why several individuals from the Jiajin Mountain (population 6) appeared in subclade 1A.
The expansion of the subclade 1B was inferred to have happened at about 0.014-0.004 Mya, right after the LGM (0.021-0.017 Mya). Most individuals of subclade 1B were collected from Gongga Mountain (population 15). Gongga Mountain located at the eastern margin of the Tibetan Plateau, was an important glaciation centre in the eastern Tibetan Plateau. Here, remains of Quaternary glaciers are still widespread. Glacial accumulation and erosion landforms coexisted, which indicated this region has been glaciated many times [20]. The range and intensity of the Quaternary glaciers in this region were very different in different periods. All contemporary glaciers are located at high elevations (above 3000 m). During the early Pleistocene, no glacier developed in the GMR because the relative low elevation at that time. Glaciers developed during the mid-Pleistocene, but most of them occurred at valley heads or high altitudes [20]. The GMR experienced the most extensive glaciation at about 0.277 Mya [20]. As a result, many small areas in this region were suitable for species to inhabit during different periods. Additionally, many ancient and endemic species inhabited in this region [21], [23] (e.g. Flora: Taxus chinensis, Kingdonia uniflora, Cordyceps sinensis, Cercidiphyllum japonicum; Fauna: Ailuropoda melanoleuca, Panthera uncia, Pantholops hodysoni, Rhinopithecus roxellanae), leading to the conclusion that the GMR acted as a refuge during the cold periods [21], [22].
Another character of Gongga Mountain is its elevation is very large, ranging from less than 1000 m to more than 7000 m, so many types of micro-environments are available [20], [21], [73]. Apodemus draco has been found at elevations from 800 m to 4000 m, and it occupies different microhabitats, such as broad-leaved forests, mixed broadleaf-conifer forests, scrublands, and meadows [23], [74]. Therefore, A. draco could have found available microhabitats in high elevations or simply retreated to lower elevations. Consequently, based on the characteristics of A. draco and the geologic history of the GMR, A. draco could have survived in many parts of the GMR even during the EGP and LGM. This conclusion could explain why individuals from GMR fell into different evolutionary clades (Fig. 2).
Two recent studies found the similar pattern in the blood pheasant (I. cruentus) [73], Sichuan field mouse (A. latronum) and Chinese scrub vole (Neodon irene) [22]. But the Sichuan field mouse and Chinese scrub vole had to find suitable places at high elevations in the SEMTP instead of shifting elevation, because they are only found in high elevations [22], [23], [74]. Therefore, since there were two refugia for A. draco in clade 1 (Hongya county and GMR) in the central region, the discontinuous gene flow among different sampling regions could form the current observed pattern at clade 1.
Conclusion
The present study demonstrated that the genetic differentiation of the South China field mouse’s population in the SEMTP not only was influenced by the complex geologic history and unique topography of the SEMTP, but also was influenced by the major glaciations in the Pleistocene. As a consequence, we hypothesize that its evolutionary history included two stages. First, an initial divergence would have been shaped by the uplift events of the Tibetan Plateau, and high genetic diversity was maintained by environmental and topographic complexity. Then, major glaciations in the Pleistocene added further complexity to its demographic history and genetic structure. The observed genetic differentiation pattern in this work is consistent with that of other recent studies focused on the eastern Tibetan Plateau [8], [19], [22], [72]. Our work, together with the studies from Fan et al. [22] and Zhan et al. [72], indicate that past geologic and climatic events on the eastern Tibetan Plateau could have similar effects on the regional historical demography of many animals in the region.
Acknowledgments
We are grateful to Dr. Keith A Crandall and three anonymous reviewers for their useful criticism and suggestions for improvement. We would like to thank Drs. Robert Wayne, John Pollinger, Shauna Price and Adam Freedman at University of California, Los Angeles (UCLA), and Pedro Silva at Research Center in Biodiversity and Genetic Resources, University of Porto – CIBIO/UP for their helpful comments on a previous version of this manuscript. We also would like to thank Dr. Graham Slater at UCLA for his help in performing the divergence dating analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was funded by the National Key Technology R&D Program (2012BAC01B06). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 2.Qu YH, Ericson PGP, Lei FM, Li SH. Postglacial colonization of the Tibetan plateau inferred from the matrilineal genetic structure of the endemic red-necked snow finch, Pyrgilauda ruficollis. Molecular Ecology. 2005;14:1767–1781. doi: 10.1111/j.1365-294X.2005.02528.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhang FF, Jiang ZG. Mitochondrial phylogeography and genetic diversity of Tibetan gazelle (Procapra picticaudata): Implications for conservation. Molecular Phylogenetics and Evolution. 2006;41:313–321. doi: 10.1016/j.ympev.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 4.Jin YT, Brown RP, Liu NF. Cladogenesis and phylogeography of the lizard Phrynocephalus vlangalii (Agamidae) on the Tibetan plateau. Molecular Ecology. 2008;17:1971–1982. doi: 10.1111/j.1365-294X.2008.03721.x. [DOI] [PubMed] [Google Scholar]
- 5.Ci HX, Lin GH, Cai ZY, Tang LZ, Su JP, et al. Population history of the plateau pika endemic to the Qinghai-Tibetan Plateau based on mtDNA sequence data. Journal of Zoology. 2009;279:396–403. [Google Scholar]
- 6.Fan ZX, Liu SY, Liu Y, Zeng B, Zhang XY, et al. Molecular phylogeny and taxonomic reconsideration of the subfamily Zapodiane (Rodentia: Dipodidae), with an emphasis on Chinese species. Molecular Phylogenetics and Evolution. 2009;51:447–453. doi: 10.1016/j.ympev.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Qu YH, Lei FM. Comparative phylogeography of two endemic birds of the Tibetan plateau, the white-rumped snow finch (Onychostruthus taczanowskii) and the Hume’s ground tit (Pseudopodoces humilis). Molecular Phylogenetics and Evolution. 2009;51:312–326. doi: 10.1016/j.ympev.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Liu S, Liu Y, Hao H, Zeng B, et al. Phylogeography of the large white-bellied rat Niviventer excelsior suggests the influence of Pleistocene Glaciations in the Hengduan Mountains. Zoological Science. 2010;27:487–493. doi: 10.2108/zsj.27.487. [DOI] [PubMed] [Google Scholar]
- 9.Shi YF, Li JJ, Li BY. Guangzhou: Guangdong Science and Technology Press; 1998. Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic. [Google Scholar]
- 10.Zheng HB, Powell CM, An ZS, Dong GR. Pliocene uplift of the northern Tibet Plateau. Geology. 2000;8:715–718. [Google Scholar]
- 11.An Z, John EK, Warren LP, Stephen CP. Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature. 2001;411:62–66. doi: 10.1038/35075035. [DOI] [PubMed] [Google Scholar]
- 12.Wu YQ, Cui ZJ, Liu GN, Ge DK, Yin JR, et al. Quaternary geomorphological evolution of the Kunlun Pass areas and uplift of the Qinghai-Xizang (Tibet) plateau. Geomorphology. 2001;36:203–216. [Google Scholar]
- 13.Li JJ, Li BY, Wang FB. Geological and ecological studies of Qinghai-Xizang Plateau. Beijing: Science Press; 1981. The process of uplift of the Qinghai-Xizang Plateau. [Google Scholar]
- 14.Yang SJ, Yin ZH, Ma XM, Lei FM. Phylogeography of ground tit (Pseudopodoces humilis) based on mtDNA: Evidence of past fragmentation on the Tibetan Plateau. Molecular Phylogenetics and Evolution. 2006;41:257–265. doi: 10.1016/j.ympev.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Kuhle M. The Pleistocene glaciation of Tibet and onset of ice age: an autocycle hepothesis. Geographical Journal. 1988;17:581–595. [Google Scholar]
- 16.Lehmkuhl F, Owen LA. Late Quaternary glaciation of Tibet and the bordering mountains: a review. Boreas. 2005;34:87–100. [Google Scholar]
- 17.Ehlers J, Gibbard PL. The extent and chronology of Cenozoic Global glaciation. Quaternary International. 2007;164–165:6–20. [Google Scholar]
- 18.Liu XM, Wei FW, Li M, Jiang XL, Feng ZJ, et al. Molecular phylogeny and taxonomy of wood mice (genus Apodemus Kaup, 1829) based on complete mtDNA cytochrome b sequences, with emphasis on Chinese species. Molecular Phylogenetics and Evolution. 2004;33:1–15. doi: 10.1016/j.ympev.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Qu Y, Lei F, Zhang R, Lu X. Comparative phylogeography of five avian species: implications for Pleistocene evolutionary history in the Qinghai-Tibetan plateau. Molecular Ecology. 2010;19:338–351. doi: 10.1111/j.1365-294X.2009.04445.x. [DOI] [PubMed] [Google Scholar]
- 20.Su Z, Shi YF, Zheng BX. Quaternary glacial remains on the Gongga Mountain and the division of glacial period. Advance in Earth Sciences, 2002;17:639–647. [Google Scholar]
- 21.Liu GQ, Zhang XJ. Conservation and utilization on biological resource of Hailuogou Glacier and Forest National Park. Journal of West China Forestry Science. 2006;35:147–153. [Google Scholar]
- 22.Fan ZX, Liu SY, Liu Y, Zhang XY, Yue BS. How Quaternary geologic and climatic events in the southeastern margin of the Tibetan Plateau influence the genetic structure of small mammals: inferences from phylogeography of two rodents, Neodon irene and Apodemus latronum. Genetica. 2011;139:339–351. doi: 10.1007/s10709-011-9553-5. [DOI] [PubMed] [Google Scholar]
- 23.Hu JC, Wang YZ, Cheng HX, Cheng EY, Cheng K, et al. Volume 2. Chengdu: Sichuan Publishing House of Science & Technology; 1984. Sichuan Fauna Economica. [Google Scholar]
- 24.Fedorov VB, Goropashnaya AV, Boeskorov GG, Cook JA. Comparative phylogeography and demographic history of the wood lemming (Myopus schisticolor): implications for late Quaternary history of the taiga species in Eurasia. Molecular Ecology. 2008;17:598–610. doi: 10.1111/j.1365-294X.2007.03595.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang YZ, Hu JC. Beijing: China Forestry Press; 1999. Sichuan Mammal Primitive Colors Illustrated Handbook. [Google Scholar]
- 26.Xia WP. A study on Chinese Apodemus with a discussion of its relations to Japanese species. Acta Theriologica Sinica. 1984;4:93–98. [Google Scholar]
- 27.Liu Q, Chen P, He K, Kilpatrick CW, Liu SY, et al. Phylogeographic study of Apodemus ilex (Rodentia: Muridae) in Southwest China. PLoS ONE. 2012;7:e31453. doi: 10.1371/journal.pone.0031453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galbreath KE, Cook JA. Genetic consequences of Pleistocene glaciations for the tundra vole (Microtus oeconomus) in Beringia. Molecular Ecology. 2004;13:135–148. doi: 10.1046/j.1365-294x.2004.02026.x. [DOI] [PubMed] [Google Scholar]
- 29.Oshida T, Abramov A, Yanagawa H, Masuda R. Phylogeography of the Russian flying squirrel (Pteromys volans): implication of refugia theory in arboreal small mammal of Eurasia. Molecular Ecology. 2005;14:1191–1196. doi: 10.1111/j.1365-294X.2005.02475.x. [DOI] [PubMed] [Google Scholar]
- 30.Hafner JC, Upham NS, Reddington E, Torres CW. Phylogeography of the pallid kangaroo mouse, Microdipodops pallidus: a sand-obligate endemic of the Great Basin, western North America. Journal of Biogeography. 2008;35:2102–2118. doi: 10.1111/j.1365-2699.2008.01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouat C, Tatard C, Bâ K, Cosson J, Dobigny G, et al. Phylogeography of the Guinea multimammate mouse (Mastomys erythroleucus): a case study for Sahelian species in West Africa. Journal of Biogeography. 2009;36:2237–2250. [Google Scholar]
- 32.Kryštufek B, Bryja J, Bužan EV. Mitochondrial Phylogeography of the European ground squirrel, Spermophilus citellus, yields evidence on refugia for steppic taxa in the southern Balkans. Heredity. 2009;103:129–135. doi: 10.1038/hdy.2009.41. [DOI] [PubMed] [Google Scholar]
- 33.Tang LZ, Wang LY, Cai ZY, Zhang TZ, Ci HX, et al. Allopatric divergence and phylogeographic structure of the plateau zokor (Eospalax baileyi), a fossorial rodent endemic to the Qinghai-Tibetan Plateau. Journal of Biogeography. 2010;37:657–668. [Google Scholar]
- 34.Suzuki H, Sato JJ, Tsuchiya K, Luo J, Zhang YP, et al. Molecular phylogeny of wood mice (Apodemus, Muridae) in East Asia. Biological Journal of the Linnean Society. 2003;80:469–481. [Google Scholar]
- 35.Sakka H, QUÉRÉ JP, Kartavtseva I, Pavlenko M, Chelomina G, et al. Comparative phylogeography of four Apodemus species (Mammalia: Rodentia) in the Asian Far East: evidence of Quaternary climatic changes in their genetic structure. Biological Journal of the Linnean Society. 2010;100:797–821. [Google Scholar]
- 36.Sambrook J, Russell DW. New York: Cold Spring Harbor Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 37.Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 40.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 41.Excoffier L, Laval G, Schneider S. Arlequin ver.3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature, 351, 1991;652–654 doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 43.Egea R, Casillas S, Barbadilla A. Standard and generalized McDonald–Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Research 36, 2008;W157–W162 doi: 10.1093/nar/gkn337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nylander JAA. Evolutionary Biology Centre, Uppsala University; 2004. Mrmodeltest V2. [Google Scholar]
- 45.Ronquist FR, Huelsenbeck JP. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 46.Swofford DL. Sinauer Associates, Sunderland, Massachusetts; 2002. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). [Google Scholar]
- 47.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 48.Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- 50.Jensen J, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genetics. 2005;6:13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller MP. Alleles in space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- 52.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analyses by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avise JC. The history and purview of phylogeography: a personal reflection. Molecular Ecology. 1998;7:371–379. [Google Scholar]
- 54.Suzuki H, Filippucci MG, Chelomina GN, Sato JJ, Serizawa K, et al. A biogeographic view of Apodemus in Asia and Europe inferred from Nuclear and Mitochondrial gene sequences. Biochemical Genetics. 2008;46:329–346. doi: 10.1007/s10528-008-9149-7. [DOI] [PubMed] [Google Scholar]
- 55.Jaarola M, Searle JB. Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Molecular Ecology. 2002;11:2613–2621. doi: 10.1046/j.1365-294x.2002.01639.x. [DOI] [PubMed] [Google Scholar]
- 56.Brunhoff C, Galbreath KE, Fedorov VB, Cook JA, Jaarola M. Holarctic phylogeography of the root vole (Microtus oeconomus): implications for late Quaternary biogeography of high latitudes. Molecular Ecology. 2003;12:957–968. doi: 10.1046/j.1365-294x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- 57.Rambaut A, Drummond AJ. Molecular evolution, phylogenetics and epidemiology website. Tracer ver. 1.5. 2007;7 Available: http://tree.bio.ed.ac.uk/software/tracer/. Accessed 2012 May. [Google Scholar]
- 58.Drummond AJ, Rambaut A. BEAST website. BEAST ver. 1.7.1. 2008;7 Available: http://beast.bio.ed.ac.uk/Main_Page. Accessed 2012 May. [Google Scholar]
- 59.Rambaut A. Molecular evolution, phylogenetics and epidemiology website. FigTree ver. 1.3.1. 2008;7 Available: http://tree.bio.ed.ac.uk/software/figtree/. Accessed 2012 May. [Google Scholar]
- 60.Fu YX. Statistical test of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- 62.Ramirez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A. Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics. 2008;179:555–567. doi: 10.1534/genetics.107.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harpending HC. Signature of ancient population growth in a low resolution mitochondrial DNA mismatch distribution. Human Biology. 1994;66:591–600. [PubMed] [Google Scholar]
- 64.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers A, Harpending HM. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 66.Zheng B, Xu Q, Shen Y. The relatonship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan plateau: review and speculation. Quaternary international. 2002;97–98:93–101. [Google Scholar]
- 67.Zeng ZY, Song ZM, Deng XZ, Sun QZ. A study of the ungulate fauna in Sichuan. Journal of Sichuan University (Natural Science Edition) 1994;31:540–545. [Google Scholar]
- 68.Zhang D, Feng QL, Jian MB. Eco-environmental effects of the Qinghai-Tibet plateau uplift during the Quaternary in China. Environmental Geology. 2000;39:1352–1358. [Google Scholar]
- 69.Shi YF. Characteristics of late Quaternary monsoonal glaciation on the Tibetan plateau and in East Asia. Quaternary International. 2002;97–98:79–91. [Google Scholar]
- 70.Yang X, Rost KT, Lehmkuhl F, Zhou Z, Dodson J. The evolution of dry lands in northern China and in Republic of Mongolia since the Last Glacial Maximum. Quaternary international. 2004;118–119:69–85. [Google Scholar]
- 71.Shi YF, Zheng BX, Li SJ. Last glaciation and maximum glaciation in Qinghai-Xizang (Tibet) plateau. Journal of Glaciology and Geocryology. 1990;12:1–15. [Google Scholar]
- 72.Zhan X, Zheng Y, Wei F, Bruford MW, Jia C. Molecular evidence for Pleistocene refugia at the eastern edge of the Tibetan Plateau. Molecular Ecology. 2011;20:3014–3026. doi: 10.1111/j.1365-294X.2011.05144.x. [DOI] [PubMed] [Google Scholar]
- 73.Song MK. The climate characters of the Gong-ga Mountain. Exploration of Nature. 1987;6:145–148. [Google Scholar]
- 74.Luo ZX, Chen W, Gao W. 6 Rodentia II Cricetidae). Beijing: Science Press; 2000. Fauna Sinica (Mammalia Vol. [Google Scholar]