Abstract
Walleye dermal sarcoma virus (WDSV) is a complex retrovirus associated with seasonal dermal sarcomas. Developing tumors have low levels of accessory gene transcripts, A1 and B, and regressing tumors have high levels of full-length and spliced transcripts. Transcript A1 encodes a retroviral cyclin (rv-cyclin) with limited homology to host cyclins. The rv-cyclin is physically linked to components of the transcriptional co-activator complex, Mediator, and regulates transcription. In walleye fibroblasts, it inhibits the WDSV promoter independently of cis-acting DNA sequences. The rv-cyclin activates transcription from GAL4 promoters when fused to the GAL4 DNA binding domain. A 30 a.a. activation domain in the carboxy region can be inactivated by single point mutations, and these mutations diminish the ability of the rv-cyclin to inhibit the WDSV promoter. When fused to glutathione S-transferase, the rv-cyclin, its carboxy region, and the activation domain pull down components of transcription complexes from nuclear extracts, and pulldown is lost by mutation of the activation domain.
Keywords: WDSV, rv-cyclin, cdk8, Cyclin C, Sur2, TBP, VP16, Mediator
Introduction
Walleye dermal sarcoma virus (WDSV), a member of the Epsilonretroviruses in the family Retroviridae, is a complex retrovirus etiologically associated with dermal sarcomas in walleye fish (Stizostedion vitreum) (Martineau et al., 1991, 1992; Walker, 1969; Yamamoto et al., 1976, 1985). The seasonal appearance of this disease is characterized by the expression of low levels (1–10 copies/cell) of spliced accessory gene transcripts, A1 and B, during the period of tumor development in the fall and winter and high levels (500–1000 copies/cell) of full-length and spliced transcripts during tumor regression in the spring (Quackenbush et al., 1997; Rovnak et al., 2001).
Transcript A1 encodes a retroviral cyclin (rv-cyclin, Orf A1 protein), with a cyclin box motif but with limited homology to host cyclins (LaPierre et al., 1998). Homologues of the rv-cyclin of WDSV are encoded by walleye epidermal hyperplasia virus types 1 and 2 (WEHV-1 and WEHV-2), but these smaller rv-cyclins do not contain the carboxy-terminal region of the WDSV cyclin (Bowser et al., 1988, 1996; LaPierre et al., 1998, 1999; Quackenbush et al., 1997). The rv-cyclin from WDSV, but not from WEHV-1 or WEHV-2, was able to induce cell-cycle progression in G1-cyclin deficient yeast (LaPierre et al., 1998). The WDSV rv-cyclin was also associated with the induction of hyperplastic skin lesions in transgenic mice when expressed from a keratin promoter (Lairmore et al., 2000).
When expressed in mammalian or piscine cell culture, the WDSV rv-cyclin was localized in the nucleus and concentrated in interchromatin granule clusters (IGCs) (Rovnak et al., 2001), nuclear domains enriched in proteins necessary for mRNA transcription and processing. The first 112 amino acids were necessary for localization in IGCs. The rv-cyclin was found, by co-immunoprecipitation (coIP), to be physically associated with cyclin dependent kinase 8 and cyclin C (Rovnak and Quackenbush, 2002), components of a large, multi-protein complex generally known as Mediator. Mediator, first identified in yeast, regulates activation and repression of transcription (reviewed in Blazek et al., 2005; Conaway et al., 2005; Roeder, 2005). Similar complexes, such as TRAP/SMCC, PC2, CRSP, ARC-L, and DRIP, have been identified in metazoan organisms. These complexes vary in overall subunit composition and this variation corresponds with Mediator function.
In walleye cell culture systems, where WDSV promoter activity is low, exogenous rv-cyclin further inhibited transcription from the WDSV promoter in a luciferase reporter system (Rovnak and Quackenbush, 2002; Zhang and Martineau, 1999), and this inhibition was independent of sequences in the R and U5 regions of the LTR. However, other viral promoters, such as that of SV40, were found to be activated by exogenous rv-cyclin in the same fish cells (Rovnak and Quackenbush, 2002). In select mammalian cells, rv-cyclin activated the WDSV promoter and inhibited SV40, demonstrating its ability to either inhibit or activate transcription in a manner dependent on both the promoter and the cell type (Rovnak and Quackenbush, 2002). The regulation of transcription and the physical association of the rv-cyclin with Mediator components, cdk8 and cyclin C, were dependent upon intact amino- and carboxy-terminal regions of the protein (Rovnak and Quackenbush, 2002).
The ability of the rv-cyclin to activate transcription was tested directly in yeast and cell culture from metazoan organisms with GAL4 DNA binding domain (DBD) fusion proteins. This approach allows the isolation of activation domains (ADs) that can replace the GAL4 AD function in transcription initiation from a GAL4 promoter. Herein we define and characterize an rv-cyclin activation domain (AD) and identify interacting transcription co-activators. The results support a model in which the rv-cyclin of WDSV differentially regulates transcription by the modulation of protein–protein interactions to control the composition of the transcriptional complex.
Results
rv-cyclin inhibition of the WDSV promoter functions independently of specific sequence recognition
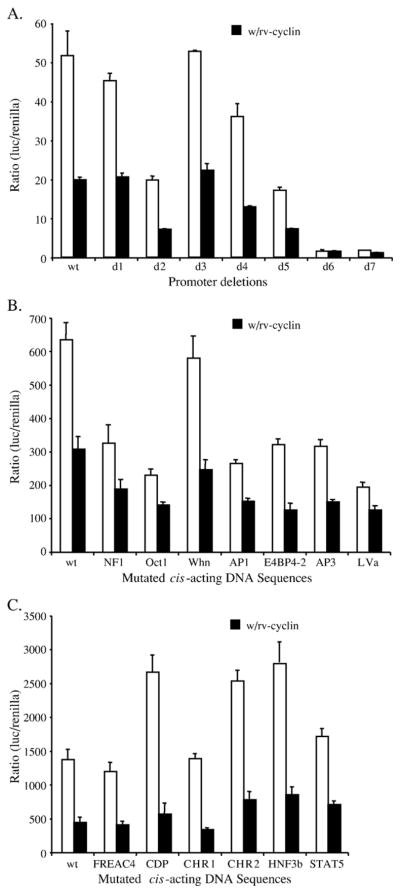
The inhibitory effects of the rv-cyclin on the basal activity of the WDSV promoter in piscine cells were previously determined to be independent of sequence within the R or U5 regions of the LTR (Rovnak and Quackenbush, 2002). The effects of the rv-cyclin on the WDSV promoter were tested with a variety of U3 region mutations (Fig. 1). Progressive, 50-bp deletions from the 5′ end of the U3 region in the pGL3WDSVU3 construct (Hronek et al., 2004) were assessed individually for inhibition of promoter activity by the rv-cyclin (Fig. 1A). HA-tagged rv-cyclin was expressed from the construct, pKH3OrfA (Rovnak et al., 2001). Expression from each functional deleted promoter was subject to greater than 2-fold inhibition when the rv-cyclin was present. Deletions d6 and d7 did not display substantial levels of transcription activity with or without rv-cyclin. These mutations delineated the core promoter of WDSV (Hronek et al., 2004).
Fig. 1.
(A) Consecutive 50 bp deletions from the 5′ end of the WDSV U3 region (d1–d7) were tested for the ability to drive luciferase expression in walleye fibroblast W12 cells without (white bars) or with expressed rv-cyclin (black bars). (B and C) Assays, as in panel A, of the WDSV U3 region with specific mutations in either the enhancer (B) or core promoter region (C). Functional mutations within the indicated sites were defined previously by DNA footprinting, electrophoretic mobility shift assays, and luciferase assays (Hronek et al., 2004).
In addition to the deletion mutants, point mutations within 13, known cis-acting sites were assayed in the presence of the rv-cyclin. The selected sequences were previously identified by DNA footprinting and electrophoretic mobility shift assays (Hronek et al., 2004). Seven of these sites reside in the enhancer region (bp −441 to −104) (Fig. 1B) and six of the sites are found within the core promoter (bp −103 to −1) (Fig. 1C). Expression of the rv-cyclin reduced WDSV promoter-driven luciferase expression from all of the mutated promoters (Figs. 1B and C). These results suggest that the rv-cyclin does not bind directly to a specific sequence within the enhancer region in order to affect inhibition (Fig. 1A), and it does not interact functionally with the known transcriptional activators that bind the enhancer or core promoter (Figs. 1B and C). The rv-cyclin protein sequence does not contain a predicted DNA-binding domain, and purified recombinant rv-cyclin does not bind to labeled U3 probe in electrophoretic mobility shift assays under conditions established for this probe with known DNA-binding proteins (Hronek et al., 2004; data not shown).
The WDSV rv-cyclin activates transcription in yeast when tethered to a promoter
A yeast two-hybrid screen was employed to identify cellular proteins that interact with rv-cyclin. In preparation for a two-hybrid screen, the rv-cyclin was fused to the GAL4 DBD (pAS2-1A1 – 297) and tested for activation of the lacZ reporter gene from a GAL4 promoter. This DBD-rv-cyclin fusion protein activated transcription, as determined by the appearance of blue colonies in a colony lift assay (Table 1). Fusion of the rv-cyclin with the GAL4 AD did not yield lacZ expression in this system. These results suggest that the rv-cyclin contains an activation domain that can replace the function of the GAL4 AD and excludes its direct interaction with DNA in GAL4 promoter sequences.
Table 1.
Activation of transcription in yeast
| Construct | Colony-lift filter assay | β-galactosidase units (mean ± SD) |
|---|---|---|
| pAS2-1 | − | 0.03 ± 0.01 |
| pAS2-1 A1 – 297 | + | 0.67 ± 0.28 |
| pAS2-1 A95 – 297 | − | 0.02 ± 0.01 |
| pAS2-1 A112 – 297 | − | 0.03 ± 0.01 |
| pAS2-1 A1 – 219 | − | 0.02 ± 0.01 |
| pAS2-1 A112 – 219 | − | 0.03 ± 0.01 |
| pAS2-1 A1 – 112 | − | 0.03 ± 0.00 |
| pAS2-1 A219 – 297 | + | 3.33 ± 1.12 |
| pTD1-1/pVA3-1a | + | 72.68 ± 37.26 |
Positive control DBD and AD vectors.
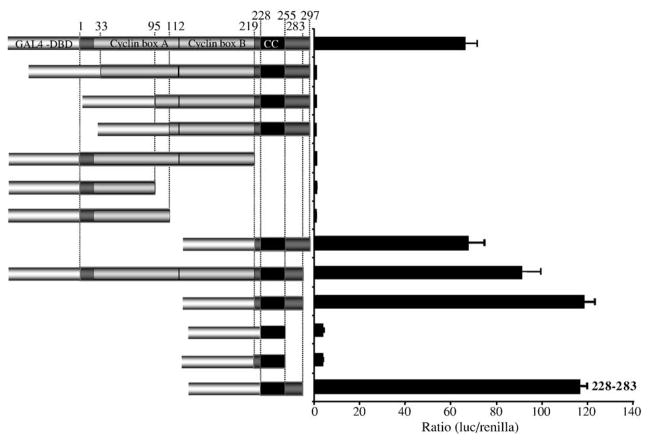
To delineate the region of the rv-cyclin that contained an activation domain, several GAL4 DBD rv-cyclin deletion mutants were constructed. Deletions of a.a. 1–95, 1–112, or 219–297 from the GAL4 DBD-rv-cyclin fusion protein each eliminated activation of the lacZ reporter (Table 1; pAS2-1A95 – 297, pAS2-1A112 – 297, and pAS2-1A1 – 219). Deletions of 1–95 and 1–112 were selected, because these forms are encoded by naturally occurring splice variants of rv-cyclin transcripts, and the amino terminus controls nuclear localization (Quackenbush et al., 1997; Rovnak et al., 2001). The carboxy region is exclusive of the cyclin box motif and contains a predicted coiled-coil domain between positions 228 and 257. Deletion of either the amino-terminal 95 a.a. or the carboxy-terminal 78 a.a. (219–297) blocked cell-specific inhibition of the WDSV promoter (Rovnak and Quackenbush, 2002). To identify the contributions of the amino and carboxy domains, they were fused individually to the GAL4 DBD and results measured in a quantitative assay of β-galactosidase activity (pAS2-1A1 – 112 and pAS2-1A219 – 297; Table 1). Activation of β-galactosidase expression was limited to either the full-length construct or the isolated carboxy region, which had a greater level of expression. Inclusion of the region between a.a. 95 and 219 with the carboxy region completely blocked its ability to activate transcription in this system.
The WDSV rv-cyclin activates transcription in HeLa cells
Results indicating an activation domain within the WDSV rv-cyclin were reproduced in HeLa cells (Fig. 2). Constructs encoding GAL4 DBD-rv-cyclin fusion proteins from the pFA-CMV vector were co-transfected with a luciferase reporter construct, pFR-luc, that contains a minimal promoter with a TATA box and five GAL4-consensus binding sequences. Western blots with anti-DBD were performed to confirm protein expression from all of the constructs tested (not shown). The GAL4 DBD nls sequence confers nuclear localization on the expressed fusion proteins. Consistent with the results in the yeast system, the full-length rv-cyclin (1–297) activated luciferase expression, but amino- or carboxy-truncated forms (33–297, 95–297, 112–297, and 1–219) were unable to activate luciferase expression when tethered to the GAL4 consensus sequences (Fig. 2). Although deletion of the first 33, 95, or 112 a.a. eliminated activation, regions 1–95, 1–112, and 1–219 were not able to activate transcription when fused to the GAL4 DBD. As observed in yeast experiments, the carboxy-terminal 78 a.a.-DBD fusion protein (219–297) was able to activate transcription (Fig. 2), but inclusion of any of the segments between a.a. 33 and 219 blocked this activity. Deletion of the carboxy-terminal 15 a.a., to remove a putative phosphorylation target at serine 285, enhanced activation by both the full-length and carboxy region fusions to a significant degree (P ≤ 0.05) (Fig. 2). A DBD-fusion of just the predicted coiled-coil region (228–255) was not able to activate transcription, and tests with fragments overlapping this region, 219–255 and 228–283, identified the region between 228 and 283 as inclusive of an activation domain. These results confirm a transcription activation function of the rv-cyclin in metazoan organisms. Amino-terminal truncations at positions 33, 95, and 112 would likely disrupt the structure of the first putative cyclin box fold (the region containing the first five alpha helices designated “Cyclin box A”). Such structural disruption may disrupt the function of the fusion proteins that include the region between 33 and 219.
Fig. 2.
Assay of transcription activation by rv-cyclin and rv-cyclin fragments in HeLa cells. Indicated portions of the rv-cyclin protein were fused to the GAL4 DBD, and their ability to activate luciferase expression from the pFR-luc GAL4 reporter construct measured after 48 h. Boundaries of each segment are indicated by a.a. position at the top. The outlines of the predicted cyclin box folds, here designated ‘Cyclin box A’ and ‘Cyclin box B’ are indicated. ‘CC’ represents the putative coiled-coil region. The smallest active region, a.a. 228 – 283, is indicated.
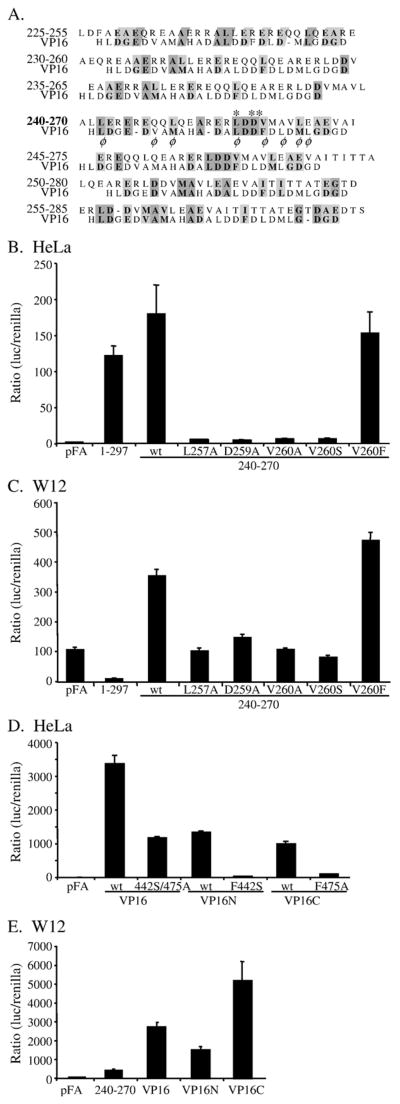
A WDSV AD can be identified by comparison with a known activation domain
In order to further delimit the rv-cyclin AD, sequences from positions 225 to 285 were represented by seven, 31-a.a. increments, that overlap by 26 a.a.. These increments were aligned individually with a prototypic acidic activation domain from HHV1 α-TIF (transcription initiation factor; VP16; a.a. 425–451; Cress and Triezenberg, 1991) (Fig. 3A). The 240–270 increment was found to align best with 5 of 7 characteristic hydrophobic side chains. This 31-a.a. region has a predicted isoelectric point of 4.08. Of particular interest was the alignment of positions 257–260 (LDDV) with positions 439–442 (LDDF) of the VP16 AD. The 240–270 region of the rv-cyclin was cloned into the GAL4 DBD vector, as were constructs with 5 individual point mutations: L257A, D259A, V260A, V260S, and V260F. The 240–270 region activated transcription in HeLa cells to a degree similar to the full-length rv-cyclin fusion protein, and four of the five point mutants eliminated activation by this region (Fig. 3B). Only the conservative mutation of valine to phenylalanine at residue 260 maintained activation by the DBD-240–270 fusion. For comparison, Fig. 3D shows the results from a luciferase assay with DBD fusions of the entire VP16 AD (a.a. 413–490), VP16N (a.a. 413–452; the H1 subdomain), VP16C (a.a. 453–490; the H2 subdomain), and mutations of these constructs: one in VP16N, F442S, one in VP16C, F475A, and both mutations in the full VP16 AD. The activity of the rv-cyclin AD is approximately 1/30 of the VP16 AD and 1/10 that of the VP16N and VP16C subdomains in HeLa cells.
Fig. 3.
(A) Individual alignments of overlapping, rv-cyclin 31 a.a. segments with a prototype acidic activator, VP16 (a.a. 425 – 451; Cress and Triezenberg, 1991). Segments span the rv-cyclin activator region, a.a. 228–283, identified in Fig. 2. Similar and identical residues are shaded. φ designates hydrophobic side chains of the VP16 sequence. Asterisks mark a.a.s L257, D259, and V260 of the rv-cyclin, which were subject to mutation. (B) Luciferase assay of GAL4-DBD vector, pFA, and GAL4-DBD GAL4 fusion proteins containing the full-length rv-cyclin, 1–297, or the wild type (wt) and mutated versions (L257A, D259A, V260A, V260S, and V260F) of the 240–270 a.a. segment in HeLa cells. (C) Luciferase assay of GAL4 fusion proteins as in panel B, but performed in the walleye fibroblast line, W12. (D) Luciferase assay of GAL4 fusions of the entire carboxy region of VP16 (a.a. 413–490) and double mutant VP16 F442S/ F475A, and subdomains, VP16N (413– 452) and VP16C (453–490) and their corresponding individual mutations of F442S and F475A in HeLa cells. (E) Assays of the wild type GAL4 DBD-VP16 fusion constructs in the W12 cell line. Results from the wild type rv-cyclin AD, 240–270, are shown for comparison.
When performed in a walleye fibroblast cell line, W12, the isolated rv-cyclin AD (240–270) activated transcription (Fig. 3C). As in HeLa cells, mutations L257A, D259A, V260A, and V260S reduced the activation to levels comparable to the pFA-CMV control vector, and the conservative V260F mutation maintained activation (Fig. 3C). Interestingly, the full-length rv-cyclin fused to the GAL4 DBD has little apparent activity in these cells. These results distinguish the function of the DBD-fusion, full-length protein in walleye cells from that in HeLa cells by extending the apparent inhibitory region between a.a. 33 and 219 to include the first 33 amino acids or the entire cyclin box region. This result suggests that the intact cyclin box of the rv-cyclin functions to block the activation domain in walleye fibroblast cells.
The activities of the VP16 AD and its subdomains, VP16N and VP16C, were also tested in walleye fibro-blasts (Fig. 3E). All three DBD fusions activate transcription, but VP16C had greater function in these cells relative to its activity in HeLa cells and when compared to the full VP16 AD, VP16N, and the rv-cyclin AD: approximately 2, 3, and 11 times their activation levels, respectively (Fig. 3E).
The results from these reporter assays demonstrate comparable function of the isolated rv-cyclin AD in HeLa and W12 cells. This suggests a common target for contact by the AD in mammalian and piscine cells as well as in yeast. The VP16C activation domain functions distinctively in W12 cells suggesting variations in the milieu of HeLa and W12 cells that can allow the regulation of an activation domain or affect the degree of activation.
Mutations within the AD affect the function of the full-length rv-cyclin
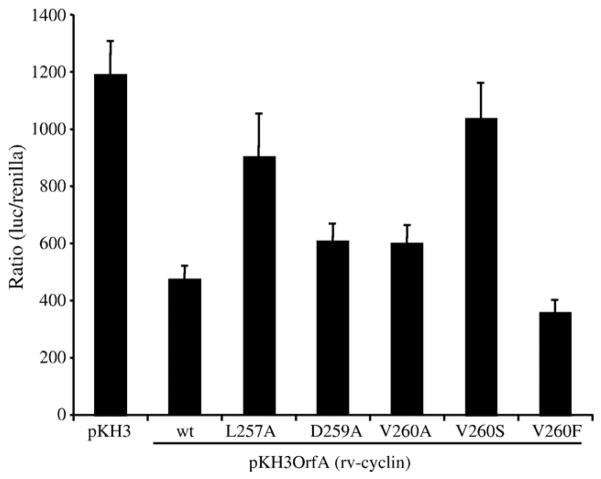
The wild type rv-cyclin significantly inhibited expression from the WDSV promoter to 39% of the activity found in cells that do not express rv-cyclin (set at 100% activity) (Fig. 4, P < 0.0001). Mutations D259A and V260A, which had blocked activation by GAL4-DBD fusion proteins, reduced the inhibitory action of the rv-cyclin, resulting in 50% of the luciferase activity (P < 0.03) from the WDSV promoter alone. The L257A and V260S mutations significantly reduced (P < 0.005) the ability of rv-cyclin to inhibit expression from the WDSV promoter. The L257A and V260S mutations resulted in 76% and 87% activity, respectively, which were not significantly different than control levels. The conservative V260F mutation enhanced (P < 0.02) the ability of the rv-cyclin to inhibit transcription (30% luciferase activity compared with WDSV promoter alone). These results suggest a dependence upon a functional “activation domain” of the rv-cyclin for its ability to inhibit transcription.
Fig. 4.
Effects of wild type (wt) and mutated full-length rv-cyclin on transcription. Luciferase expression in W12 cells from the WDSV U3-luciferase construct, pGL3WDSVU3, co-transfected with control vector pKH3 or an rv-cyclin expression vector, pKH3OrfA (wild type (wt) or mutated forms L257A, D259A, V260A, V260S, and V260F).
rv-cyclin–cellular protein interactions
Previously, WDSV rv-cyclin was physically associated with components of transcription complexes, including RNA Pol II, cdk8, and cyclin C by coIP of transiently expressed, HA-tagged rv-cyclin in HeLa cells (Rovnak and Quackenbush, 2002). Additional coIPs confirmed interactions of rv-cyclin with the transcriptional co-activator complex, Mediator (Fig. 5). Specifically, antibodies to mediator components, Sur2 (MED23), Med6, CRSP70 (MED26), TRAP95 (MED16), TRAP100 (MED24), TRAP150 (TRAP3), TRAP220 (MED1), and TRAP230 (MED12, srb8) were able to coIP expressed, HA-tagged rv-cyclin. Antibodies to transcriptional co-activators, CBP and p300, and antibodies to TBP and TFIIB, which bind TATA box DNA, were also able to coIP the rv-cyclin (Fig. 5). An example of a transcriptional regulatory protein that is not apparently associated with the rv-cyclin is shown by coIP with antibody reactive to p53. Overall, only antibodies to mediator components and to p300 consistently produced robust coIP of the rv-cyclin.
Fig. 5.
Western blot analysis of coIPs from 150 μg HeLa cell lysates. Antibodies were used to precipitate proteins either from control, pKH3-transfected cell lysates (left lane of each pair) or from lysates of HA-tagged, rv-cyclin-transfected cells (pKH3OrfA vector; right lane of each pair). The first two samples were immunoprecipitated with preimmune rabbit sera (NRS), and the second two with rabbit sera reactive to the rv-cyclin protein (rv-cyc). Subsequent pairs were precipitated with antibody reactive to the indicated cellular protein. All Westerns were probed with anti-HA MAb (12CA5). Cdk8, cycC, Sur2, Med6, CRSP70, and TRAP proteins are components of the Mediator co-activator complex. The IP antibodies used for CBP and p300 are specific for these proteins and do not crossreact.
GST pulldown of specific co-activators
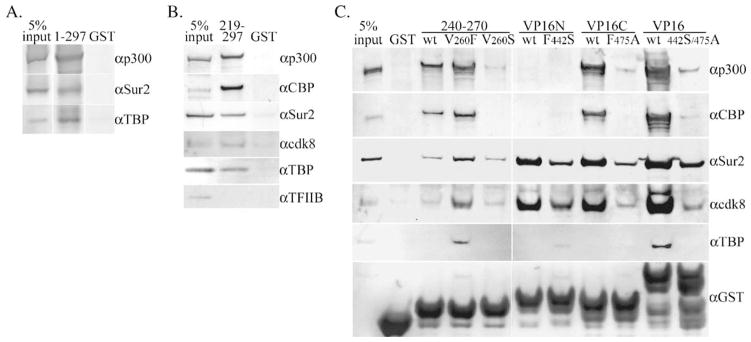
To further assess protein–protein contacts, the rv-cyclin (a.a. 1–297) was expressed as a GST fusion protein and a small amount of soluble protein purified on glutathione sepharose. Purified GST-rv-cyclin or purified GST alone was then rebound to glutathione sepharose and the bound proteins incubated with HeLa cell nuclear extracts. Protein complexes pulled down with the glutathione sepharose were separated by SDS PAGE and Western blots probed successively with antibodies to p300, Sur2, and TBP. All three cellular proteins were pulled down with the GST-rv-cyclin-sepharose only (Fig. 6A).
Fig. 6.
Pulldown of proteins from 75 μg of HeLa nuclear extracts by GST fusion proteins and glutathione sepharose. (A) Western blot of pulldown by GST-rv-cyclin (1–297) probed and reprobed with the indicated antibodies specific for p300, Sur2, and TBP. (B) Pulldown by GST fused to the carboxy region of the rv-cyclin (219–297) and probed successively for p300, CBP, TBP, TFIIB, cdk8, and Sur2. Antibodies for the detection of CBP and p300 detect unique regions and are not crossreactive. (C) Pulldowns by wild type (wt) GST-rv-cyclin AD (240–270) and mutated forms, V260S and V260F, and by VP16 subdomain fusions, GST-VP16N and GST-VP16C and GST-VP16 AD and corresponding mutated forms, F442S, F475A, and double mutation, 442S/475A, in the VP16 AD. The input GST fusion proteins were detected with anti-GST (αGST; bottom panel). ‘GST’ indicates pulldown with purified GST only. Input represents 5% of total extract used for each pulldown.
The carboxy-terminal region (a.a. 219–297) was similarly expressed as a GST fusion protein, and the purified GST fusion or purified GST alone was rebound to glutathione sepharose and incubated with HeLa cell nuclear extracts. Bound protein complexes were analyzed by Western blot with antibodies to CBP, p300, Sur2, cdk8, TBP, and TFIIB (Fig. 6B). CBP, p300, Sur2, TBP, and cdk8 were identified in the GST-rv-cyclin-sepharose pulldowns and not with GST-sepharose alone. TFIIB was not visible in the pulldown fraction. Signals from CBP and p300 were most robust.
After identification of the rv-cyclin AD (a.a. 240–270), a GST-AD fusion construct was prepared as were GST-AD constructs containing the V260S and V260F mutations. GST fusions of the VP16 AD, and subdomains VP16N and VP16C, and their corresponding mutations, F442S and F475A, were also prepared. The GST-rv-cyclin AD pulled down p300, CBP, Sur2, and cdk8 from HeLa nuclear extracts (Fig. 6C). The V260F mutation yielded greater signal for these proteins and also allowed pulldown of TBP. The V260S mutation reduced or eliminated p300 and CBP pulldown, and had less affect on Sur2 and cdk8 pulldown, compared to wild type. This suggests that a principle component of the AD function is an interaction with CBP and/or p300.
Comparative assays with GST-VP16N, GST-VP16C, and GST-VP16 AD demonstrated their different capacities for the pulldown of co-activators (Fig. 6C): VP16N pulled down Sur2 and cdk8 well, but not CBP, p300, or TBP; VP16C pulled down p300, CBP, Sur2, and cdk8 well. The full VP16 AD pulled down all of the assayed proteins. The VP16 mutants, F442S and F475A, demonstrated no or reduced capacity for all interactions. Pulldown results for the GST-VP16 AD, GST-VP16 subdomains, and their mutants were comparable to those previously published (Hardy et al., 2002; Ikeda et al., 2002). The capacity of the rv-cyclin AD to pull down CBP and p300 was most comparable to that of the VP16C subdomain. The pulldown of cdk8 by a fragment of the rv-cyclin, exclusive of its cyclin box domain, and the similar pulldown of cdk8 by VP16 ADs further support an association of the rv-cyclin with cdk8 by virtue of its interaction with Mediator proteins as opposed to its cyclin homology (Holzschu et al., 2003; Rovnak and Quacken-bush, 2002).
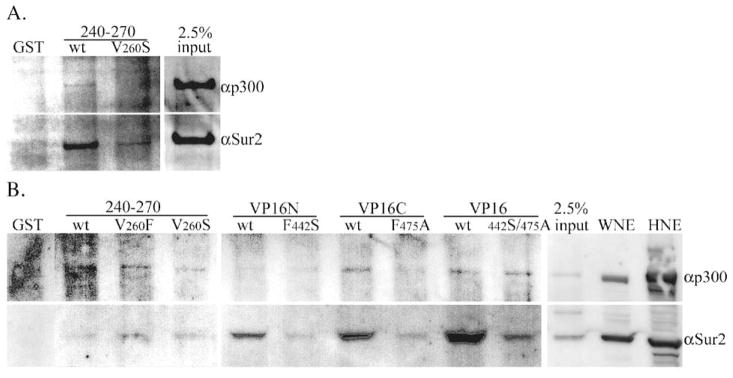
GST pulldown from walleye nuclear extracts
Efforts to pull down and identify proteins from walleye cell nuclear extracts were limited by the crossreactivity of specific antibodies across genera. Comparisons between human genes and genes from the pufferfish (Takifugu rubripes) indicate the degree of conservation of these proteins. Pufferfish and human p300 are 67% identical, CBP: 62%, Sur2: 90%, and TBP: 64%. Sequences among the fish species, pufferfish, and zebrafish (Danio rerio), share greater identities, and functional domains within the individual proteins have very high degrees of conservation from fish to humans. The antibodies to human p300 and Sur2 were able to detect appropriate size bands in walleye extracts (Fig. 7). The antibodies used to detect human CBP and TBP were unable to detect these proteins in walleye extracts (not shown). Both Sur2 and p300 were detected in pulldowns from W12 cell nuclear extracts with GST-rv-cyclin AD, although pulldown of p300 was much less robust than from HeLa extracts (Fig. 7A). The GST-rv-cyclin AD with the V260S mutation was negative for the pulldown of p300 and Sur2 from W12 extracts. Similar results were obtained with nuclear extracts from regressing tumors: the V260F and V260S mutations reduced pull-down of p300, and the V260F mutation enhanced pull-down of Sur2 (Fig. 7B). This assay included the VP16N, VP16C, and VP16 AD GST fusions, which could clearly pull down walleye Sur2, but also pulled down much less walleye p300 when compared to their pulldown of p300 from HeLa extracts (Fig. 7B). As in luciferase assays, there was a clear difference between the results from HeLa cells versus walleye cells, specifically, reduced pulldown of p300 with both rv-cyclin and VP16 GST-fusion proteins.
Fig. 7.
Pulldown of proteins from 150 μg of walleye cell nuclear extracts by GST fusion proteins and glutathione sepharose. (A) Western blot of pulldown from W12 cell nuclear extract by wild type (wt) and mutated (V260S) GST-rv-cyclin AD (240–270) probed with antibodies that crossreact with walleye homologs of p300 and Sur2. (B) Pulldowns from nuclear extracts prepared from regressing walleye dermal sarcomas by wild type (wt) GST-rv-cyclin AD (240–270) and mutated forms, V260S and V260F and by VP16 subdomain fusions, GST-VP16N and GST-VP16C and GST-VP16 AD and corresponding mutated forms, F442S, F475A, and double mutation, 442S/475A, in the VP16 AD. ‘GST’ indicates pulldown with purified GST only. Input represents 2.5% of total extract used for each pulldown. 2 μg each of nuclear extracts from W12 cells (WNE) and HeLa cells (HNE) was loaded in the last two lanes and demonstrates crossreactivity of antisera to walleye proteins, p300 and Sur2.
Discussion
The results of these studies suggest a common, direct mechanism for both the activation and inhibition of transcription by the rv-cyclin. This mechanism is dependent upon the sequence within the defined activation domain and requires its physical interaction with co-activators of transcription, Mediator, represented by Sur2 and cdk8, and the histone acetyl transferase proteins, CBP and p300. In the context of the larger carboxy region and of the full-length rv-cyclin or with the specific mutation, V260F, the observed protein–protein interactions are expanded to include pull-down of TBP. The data do not identify or confirm a specific protein targeted for binding by the rv-cyclin, but the results do indicate a direct protein–protein interaction and function of the rv-cyclin at the transcription complex.
Comparisons with the activation domains of VP16 served to delineate the rv-cyclin AD and to validate the protein–protein interactions identified by GST pulldowns from HeLa cells. Specific CBP or p300 binding motifs within VP16C have not been identified, nor have specific sites within CBP and p300 been defined for VP16 contact. The interactions of VP16 with Mediator have been defined (Ito et al., 1999; Mittler et al., 2003), but no component of VP16 is known to interact directly with Sur2 or cdk8. The use of the VP16 AD constructs also served in the identification of cell-type or species variations in protein levels and protein–protein interactions. Specifically, the interactions of the GST-VP16, GST-VP16C, and the GST-rv-cyclin constructs with p300 were greatly reduced in nuclear extracts from the walleye cell line, W12. In addition, the levels of p300 in tumor nuclear extracts appeared to be reduced relative to W12 extracts and in comparison to Sur2 expression. The inclusion of sufficient quantities of p300 in complexes bound by either VP16 or rv-cyclin may profoundly effect the outcome of their action on transcription.
The functions of AD-containing proteins are also defined by domains other than the AD. In the case of VP16, the region 1–412 interacts with Oct-1 POU transcriptional activators and with HCF-1 (host cell factor) in order to provide specificity, stability, and cooperativity for activation of transcription at immediate early genes of HHV1 (Wysocka and Herr, 2003). The rv-cyclin, excluding its activation and coiled-coil domains, contains a predicted cyclin box motif (LaPierre et al., 1998). This motif consists of two repeats of five alpha helical regions. These regions each define a predicted cyclin box fold, a structure characterized by low sequence but high structural homology (Noble et al., 1997). The rv-cyclin box has no apparent association with cis-acting sequences in the WDSV promoter or with the DNA-binding proteins known to contact these sequences. The efforts to identify such elements were limited to inhibition assays and might not yet include all possible transcription factors. Also, the ability of purified rv-cyclin to bind promoter DNA does not address possible cooperative effects with host cell proteins, as is the case with VP16 and Oct-1. Although associations of the rv-cyclin with cis-acting sequences cannot yet be ruled out entirely, the data to date suggest that the regulation of gene expression by rv-cyclin is mediated by a general control of the composition of the transcription complex not through a specific transcription factor.
The cyclin box motif is common to cyclins and to the transcriptional regulatory proteins, pRb and TFIIB. The cyclin box fold is capable of interactions with a variety of proteins such as the cyclin dependent kinases (cyclins), E2F (pRb), and TBP and TBP-bound DNA (TFIIB). These interactions serve to regulate cell cycle and gene expression (reviewed in Noble et al., 1997). The protein interactions of the WDSV cyclin box fold are unknown, but the data do not support direct interaction with known cyclin dependent kinases per se. The protein interactions of pRb and TFIIB are quite distinct, so there is no clear indication that the divergent rv-cyclin should mimic one of these two proteins or interfere with their function. Of the two, pRb does model as a regulated repressor of transcription without DNA binding. Disruption or alteration of the TFIIB-TBP-DNA ternary complex by the rv-cyclin would certainly lead to repression of transcription initiation, but it is difficult to envision such a mechanism for activation. Identification of specific interactions of the cyclin box folds of the rv-cyclin awaits further experimentation.
In spite of its AD, the role of the rv-cyclin in the control of WDSV expression appears to be inhibition. The apparent function of the cyclin box motif, in the context of a GAL4 DBD-rv-cyclin fusion protein, was to block AD function. This was observed with the segment 95–219 in yeast, with 33–219 in HeLa cells, and with the entire cyclin box motif in walleye fibroblasts. The latter result suggests a role for the cyclin box motif in the inhibition of WDSV transcription in these cells, and the loss of full inhibition with the V260S mutation indicates the requirement for both the cyclin motif and a functional AD for inhibition of transcription. The results in yeast and in HeLa cells may reflect a requirement for two, intact cyclin box folds for structural stability. An example of this is the requirement, in the structure of the pRb pocket domain, for an intact A-box fold for the stable folding of the B-box (Lee et al., 1998). Intramolecular interactions between each cyclin box fold of the rv-cyclin may be required to yield a stable structure with the appropriate orientation of the AD, but AD inhibition by an intact cyclin box motif indicates distinctive, species- or tissue-specific protein–protein interactions that block AD function in walleye fibroblasts.
The coiled-coil domain (a.a. 228–255) is predicted to promote rv-cyclin multimers and may function in ways unrelated to its overlap of AD 240–270. It does not, however, contain a second rv-cyclin AD (Fig. 2), and loss of half of the coiled-coil motif (a.a. 228–240) from the defined AD did not impair its activity, indicating independence of the AD from multimerization. Such multimers may, however, be significant for putative cyclin box interactions.
A working model that explains the inhibition or activation of specific promoters can be based on differential contacts of the rv-cyclin with co-activators or co-repressors. The isolated rv-cyclin AD shares attributes with powerful viral transcriptional activators, such as VP16, that are involved with the selective expression of virus genes. Without a specific promoter association, the rv-cyclin may sequester co-activators or co-repressors away from promoters. Alternatively, the association with Mediator components suggests function via a complex known to regulate activated transcription of a subset of cellular genes. Its association specifically with cdk8 and cyclin C, components of a mobile Mediator subcomplex, suggests possible control of Mediator assemblies at the promoter, either by stabilization of complexes or by inclusion or exclusion of certain subcomplexes.
In the context of the tumor model, low levels of spliced transcripts encoding two WDSV proteins, rv-cyclin (Orf A) and Orf B, are the only detectable viral transcripts during the period of tumor growth in infected animals. The lack of full virus expression is likely to be a critical component of immune evasion by the virus during tumor growth. The outcome of this evasion is the successful production of a mass of cells carrying provirus. At the appropriate time, during spawning and close fish-to-fish contact, virus expression shifts radically to virus production, and transmission is maximized by the regression of the tumor mass and the release of infectious virus to the environment. A specific role for the rv-cyclin in controlling virus expression is easy to envision, yet its ability to enhance gene expression suggests a role in host cell proliferation (in conjunction with the Orf B protein) as well as a possible contribution to the shift in virus expression.
Materials and methods
Plasmid construction and site-directed mutagenesis
All rv-cyclin sequences, whether full-length or partial fragments, were derived from the pKH3OrfA construct (Rovnak et al., 2001). Full-length subclones were prepared directly from the BamHI and EcoRI insertion sites of the rv-cyclin in the pKH3 vector when possible. When necessary, and for the cloning of all subfragments, PCR primers were designed to obtain the desired fragment sequence and bestow restriction sites and appropriate reading frame for each vector system. WDSV rv-cyclin-GAL4 DBD fusions were generated in the pAS2-1 vector (Clontech), and GAL4 AD fusions were generated in the pACT2 vector (Clontech). DBD fusions for assays in metazoan tissue culture cells were generated in the pFA-CMV vector (Stratagene). Glutathione S-tranferase (GST) fusion proteins were generated in the pGEX-4T vector (Pharmacia). The VP16 AD (a.a. 413–490) and mutated VP16 AD (F442S and F475A) were expressed from the vectors, pGEX-2T-VP16 and -VP16M, generous gifts from Andrew P. Rice, Baylor College of Medicine, Department of Molecular Virology and Microbiology. The VP16 AD subregions, VP16N (a.a. 413–452; H1) and VP16C (a.a. 453–490; H2) and their corresponding mutations, VP16N F442S and VP16C F475A, were derived by PCR from pGEX2T-VP16 and -VP16M and cloned in pGEX-4T. Point mutations in the rv-cyclin were made with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions in the context of the pFA-CMV expression construct. The resulting mutations and the sequences of all constructs were confirmed by sequence analysis.
Yeast β-glactosidase assays
WDSV rv-cyclin GAL4-DBD and -AD fusion constructs were introduced into Saccharomyces cerevisiae strain Y190A by lithium acetate transformation (Schiestl and Gietz, 1989) and plated on medium lacking histidine, leucine, or tryptophan in the presence of 25 mM 3-amino 1,2,4-triazole (3-AT). The plates were incubated at 30 °C for 5 days and colonies were screened for β-galactosidase activity using a colony lift assay (Breeden and Nasmyth, 1985). Quantitative β-galactosidase assays were performed according to procedures outlined in the Clontech Yeast Protocols Handbook using o-nitrophenyl β-D-galactopyra-noside (ONPG) as substrate. Vectors pTD1-1 and pVA3-1 served for expression of positive control DBD and AD proteins.
Protein expression
GST fusion proteins were expressed in BL21 (DE3)-RIL cells (Stratagene) by stimulation with 1 mM IPTG for 2 h. Bacteria were lysed in a French press, and soluble proteins were purified by affinity chromatography on glutathione Sepharose (Amersham Pharmacia). Fusion proteins with rv-cyclin a.a. 219–297 and 240–270 and VP16 ADs were largely soluble when expressed.
Reporter-gene constructs and luciferase assays
The pGL3-U3 reporter vector contains the WDSV U3 region cloned upstream of the luciferase gene (Hronek et al., 2004; Rovnak and Quackenbush, 2002). The site-directed mutations in pGL3-U3 were previously described (Hronek et al., 2004). HeLa cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal bovine serum at 37 °C in 5% CO2. W12 cells, a walleye fibroblast cell line, were maintained in HEPES buffered minimal essential media (pH 7.4) with Hank’s salts at 20 °C. Cells were seeded into 24-well plates in a volume of 1 ml media supplemented with 10% fetal bovine serum (Gibco), 4 mM glutamine, 100 units penicillin ml−1, and 100 μg streptomycin ml−1 and transfected with 0.2–0.3 μg of a luciferase reporter vector and 0.0001 μg of pRL-TK (Promega) using FuGENE6 (Roche) according to the manufacturers’ suggestions. Experiments were performed using the dual-luciferase reporter assay system (Promega). Cell lysates were harvested 48 h (HeLa) or 72 h (W12) after transfection with passive lysis buffer and then centrifuged at 20,000 × g for 5 min at 4 °C. Luciferase activities were obtained with a Turner Designs TD-20/20 or a Beckman Coulter LD400 luminometer. Luciferase activity from the reporter vector was normalized for transfection efficiency with values from the co-transfected pRL-TK vector. All transfections were performed in quadruplicate. Student’s t test and 95% confidence intervals based on a t distribution were used for statistical analyses. A P value less than 0.05 was considered significant.
Co-immunoprecipitation
Co-immunoprecipitation (coIP) was performed as previously described (Rovnak and Quackenbush, 2002) with minor modifications. Briefly, HeLa cells were transfected with control vector, pKH3, or the HA-rv-cyclin vector, pKH3OrfA. Cells were lysed at 48 h post-transfection with IP buffer (1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA pH, 1 mM EGTA, 2 μg/ml leupeptin and aprotinin, 1 μg/ml pepstatin 0.2 mM sodium ortho-vanadate, and 0.2 mM PMSF) and pre-cleared with protein G sepharose (Pharmacia Biotech). 150 μg equivalents of cell lysates at 1 μg/μl IP buffer was incubated with 0.2–1 μg of precipitating antibody for 1 h prior to the addition of 30 μl protein G sepharose suspension (50:50) and rotated overnight at 4 °C. Protein G pellets were washed 4 times with IP buffer, suspended in 10 μl loading buffer, and heated for 10 min at 70 °C. The entire soluble fraction was separated under denaturing conditions in a 4–12% polyacrylamide gel, and Western blots were probed with mouse monoclonal (MAb) reactive to HA (12CA5, Roche) and detected with goat anti-mouse IgG-peroxidase conjugate and TMB colorimetric substrate (Kierkegaard and Perry Labs). 10 μg of cell lysates was loaded in control lanes.
GST pulldown
Nuclear extracts from HeLa and walleye cells were prepared by hypotonic lysis and KCl extraction of nuclei as described by Mayeda and Krainer (1999). Extracts were diluted to 2 μg/μl in D100 buffer (10 mM HEPES pH 8.0, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 0.5% NP-40) and pre-adsorbed with 10 μg purified GST protein and 50 μl glutathione sepharose CL-4B per mg total protein extract overnight at 4 °C. 75 μg (HeLa) or 150 μg (walleye) aliquots of pre-adsorbed extracts were rotated overnight with 2.5 μg equivalents of glutathione sepharose-bound GST fusion proteins (10–20 μl of a 10% suspension) in siliconized microfuge tubes. Relative quantities of bound input fusion proteins were confirmed by Western analysis with anti-GST antibody (Pierce). Glutathione sepharose-bound proteins were pelleted for 8 s in an Eppendorf microfuge and washed 5 times with 250 μl D100 buffer. The final pellets were suspended in 8 μl SDS PAGE loading dye, heated, and the soluble portion separated on 4–12% gradient gels, and Western blots probed with the indicated antibodies. Multiple proteins were detected sequentially after treating blots with strip buffer (30 mM Tris–HCl (pH6.8), 150 mM NaCl, 1 M β-mercaptoethanol, 0.2% SDS) for 1 h at 45 °C.
Analysis of protein sequences
Protein sequences used for comparative alignments were retrieved from the NCBI nr database and included the HHV1 (HSV1) alpha gene Trans-Inducing Factor (alpha-TIF, VP16, Vmw65, accession no. NP_044650) and WDSV rv-cyclin (ORF-A protein, no. AAA99528).
The following accession numbers for human and for T. rubripes (pufferfish) protein sequences were retrieved from NCBI nr and Fugu genome (SIN prefix) databases: p300 (nos. NP_001420 and SINFRUP00000072827); CBP (CREB-Binding Protein, nos. AAC51331 and SIN-FRUP00000072827); Sur2 (Suppressor of Ras 2, MED23, Cofactor Required for SP1 transcriptional activation subunit 3 (CRSP3), Vitamin D3 Receptor Interacting Protein p130 (DRIP130), or Activator Recruited Cofactor protein p130 (ARC130), nos. NP_004821 and SINFRUP00000080368 + SINFRUP00000074783 + SINFRUP00000056892 (3 contiguous fragments)); and TBP (TATA Binding Protein, nos. AAV38463 and AAR24283 (Fugu)).
All protein sequences were aligned with MacVector multiple sequence alignment software (Accelrys) using the Clustal W algorithm (Thompson et al., 1994) with the BLOSUM 30 matrix and an open gap penalty of 10.0 and extend gap penalty of 0.1.
Antibodies
The antibodies used for Western blots and immunoprecipitation included affinity purified rabbit anti-rv-cyclin (anti-Orf A) (Rovnak et al., 2001), MAb to Sur2 (Pharmingen), MAbs to TBP (ITBP20) and Transcription Factor IIB (TFIIB8) (Covance), and Santa Cruz MAb to CBP (C-1), rabbit antibodies reactive to p53 (FL-393) and p300 (N-15), goat antibodies to cdk8 (C-19), cyclin C (C-19), CRSP 70 (C-19), Med6 (E-20), and to thyroid hormone receptor proteins (TRAP) 95 (L-15), 100 (C-16), 150 (C-18), 220 (C-19), and 230 (A-18).
Acknowledgments
This work was supported by the National Institutes of Health grant, RO1 CA095056, from the National Cancer Institute and by the American Cancer Society research project grant, RPG-00-313-01-MBC.
References
- Blazek E, Mittler G, Meisterernst M. The Mediator of RNA polymerase II. Chromosoma. 2005;113 (8):399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- Bowser PR, Wolfe MJ, Forney JL, Wooster GA. Seasonal prevalence of skin tumors from walleye (Stizostedion vitreum) from Oneida Lake, New York. J Wildl Dis. 1988;24:292–298. doi: 10.7589/0090-3558-24.2.292. [DOI] [PubMed] [Google Scholar]
- Bowser PR, Wooster GA, Quackenbush SL, Casey RN, Casey JW. Comparison of fall and spring tumors as inocula for experimental transmission of walleye dermal sarcoma. J Aquat Anim Health. 1996;8:78–81. [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Conaway JW, Florens L, Sato S, Tomomori-Sato C, Parmely TJ, Yao T, Swanson SK, Banks CA, Washburn MP, Conaway RC. The mammalian Mediator complex. FEBS Lett. 2005;579 (4):904–908. doi: 10.1016/j.febslet.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251 (4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Hardy S, Brand M, Mittler G, Yanagisawa J, Kato S, Meisterernst M, Tora L. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J Biol Chem. 2002;277 (36):32875–32882. doi: 10.1074/jbc.M205860200. [DOI] [PubMed] [Google Scholar]
- Holzschu D, Lapierre LA, Lairmore MD. Comparative pathogenesis of epsilonretroviruses. J Virol. 2003;77 (23):12385–12391. doi: 10.1128/JVI.77.23.12385-12391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hronek B, Meagher A, Rovnak J, Quackenbush SL. Identification and characterization of cis-acting elements residing in the walleye dermal sarcoma virus promoter. J Virol. 2004;78:7590–7601. doi: 10.1128/JVI.78.14.7590-7601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Stuehler T, Meisterernst M. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells. 2002;7 (1):49–58. doi: 10.1046/j.1356-9597.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- Lairmore MD, Stanley JR, Weber SA, Holzschu DL. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proc Natl Acad Sci USA. 2000;97 (11):6114–6119. doi: 10.1073/pnas.110024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Holzschu DL, Bowser PR, Casey JW. Sequence and transcriptional analyses of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: evidence for a gene duplication. J Virol. 1999;73 (11):9393–9403. doi: 10.1128/jvi.73.11.9393-9403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391 (6670):859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Martineau D, Renshaw R, Williams JR, Casey JW, Bowser PR. A large unintegrated retrovirus DNA species present in a dermal tumor of walleye Stizostedion vitreum. Dis Aquat Org. 1991;10:153–158. [Google Scholar]
- Martineau D, Bowser PR, Renshaw RR, Casey JW. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66 (1):596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. In: Haynes S, editor. Methods in Molecular Biology; RNA–Protein Interaction Protocols. Vol. 118. Human Press, Inc; Totowa, NJ: 1999. [DOI] [PubMed] [Google Scholar]
- Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22 (24):6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble MEM, Endicott JA, Brown NR, Johnson LN. The cyclin box fold: protein recognition in cell-cycle and transcription control. TIBS. 1997;22:482–487. doi: 10.1016/s0968-0004(97)01144-4. [DOI] [PubMed] [Google Scholar]
- Quackenbush SL, Holzschu DL, Bowser PR, Casey JW. Transcriptional analysis of walleye dermal sarcoma virus (WDSV) Virology. 1997;237:107–112. doi: 10.1006/viro.1997.8755. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579 (4):909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus cyclin interacts with components of the Mediator complex and the RNA polymerase II holoenzyme. J Virol. 2002;76:8031–8039. doi: 10.1128/JVI.76.16.8031-8039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Casey JW, Quackenbush SL. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin) Virology. 2001;280:31–40. doi: 10.1006/viro.2000.0731. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16 (5–6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. Virus associated with epidermal hyperplasia in fish. Natl Cancer Inst Monogr. 1969;31:195–207. [PubMed] [Google Scholar]
- Wysocka J, Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci. 2003;28 (6):294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, MacDonald RD, Gillespie DC, Kelly RK. Viruses associated with lymphocystis and dermal sarcoma of walleye (Stizostedion vitreum vitreum) J Fish Res Board Can. 1976;33:2408–2419. [Google Scholar]
- Yamamoto T, Kelly RK, Nielsen O. Morphological differentiation of virus-associated skin tumors of walleye (Stizostedion vitreum vitreum) Fish Pathol. 1985;20:361–372. [Google Scholar]
- Zhang Z, Martineau D. Walleye dermal sarcoma virus: OrfA N-terminal end inhibits the activity of a reporter gene directed by eukaryotic promoters and has a negative effect on the growth of fish and mammalian cells. J Virol. 1999;73 (10):8884–8889. doi: 10.1128/jvi.73.10.8884-8889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]