Abstract
Walleye dermal sarcoma virus (WDSV) is a complex retrovirus associated with dermal sarcomas in walleye fish. Virus expression is tightly regulated and limited to accessory gene transcripts throughout tumour development. During tumour regression, this regulation is lost and the replication of virus is greatly enhanced. Cultured walleye fibroblasts infected in vitro do not produce significant quantities of infectious virus. Tissue culture cells established by explantation of tumour cells were found to harbour WDSV provirus and to express accessory and structural proteins. The sequence of the provirus showed little variation from a previous WDSV isolate. Retroviral particles were isolated from supernatants from these cells and were able to transfer infection to uninfected walleye fibroblasts. In addition to the virus present in supernatants, much of the virus was cell associated and liberated only by sonication. This virus was found at internal cellular membranes, including mitochondria, and was infectious.
INTRODUCTION
Walleye dermal sarcoma virus (WDSV), a member of the genus Epsilonretrovirus in the family Retroviridae, is aetiologically associated with dermal sarcomas in walleye fish (Sander vitreus) (Martineau et al., 1992, 1991; Walker, 1969; Yamamoto et al., 1985; Yamamoto et al., 1976). The seasonal appearance of disease is characterized by the expression of low levels of the spliced accessory gene transcripts A1 and B during tumour development and high levels of full-length and spliced transcripts during tumour regression (Quackenbush et al., 1997; Rovnak et al., 2001). The regressing tumour contains high levels of infectious virus, which serves in the efficient transfer to adult, uninfected fish during spring spawning. Experimentally, the virus can be purified from whole tumour homogenates and applied by topical, oral or intramuscular routes to fingerling fish for rapid and efficient replication of dermal sarcoma (Bowser et al., 1997).
The imitation of this cycle in tissue culture systems has not been successful to date (Holzschu et al., 2003). Efforts at the production of WDSV in vitro have focused on the establishment of naturally infected walleye cells from tumours and the infection of piscine cell lines, including walleye fibroblasts, with virus from tumours. The cultivation of explanted tumours yields uninfected cultures such as the W12 walleye fibroblast cells (D. Martineau, personal communication) or infected cells without apparent virus expression such as the WC1 line (Kelly et al., 1980). Infections in vitro have also not generated productively infected cells. The pattern of WDSV gene expression in such cells is comparable to that of the developing tumour, with low levels of the A1 and B spliced transcripts and little or no virus production.
Much information has been obtained by expressing recombinant forms of the predicted accessory proteins of WDSV in yeast, tissue culture and transgenic systems. Transcript A1 encodes a retroviral cyclin (rv-cyclin or Orf A protein), with a cyclin box motif but with limited homology to host cyclins (LaPierre et al., 1998). Rv-cyclin induces cell-cycle progression in G1-cyclin-deficient yeast (LaPierre et al., 1998) and is associated with hyperplastic skin lesions in transgenic mice when expressed from a keratin promoter (Lairmore et al., 2000). In cell culture, rv-cyclin co-localizes in the nucleus with proteins necessary for mRNA transcription and processing (Rovnak et al., 2001). It is physically associated with co-activators of transcription and interacts directly with TATA-binding protein associated factor 9 (TAF9) (Rovnak & Quackenbush, 2002, 2006; Rovnak et al., 2005). In walleye fibroblasts, rv-cyclin inhibits transcription from the WDSV promoter independently of cis-acting DNA sequences within the virus long terminal repeat (LTR) (Rovnak & Quackenbush, 2002; Zhang & Martineau, 1999), and its ability to inhibit is dependent upon TAF9 interaction (Rovnak & Quackenbush, 2006). The Orf B protein is localized predominantly in the cytoplasm and at the plasma membrane, where it is physically associated with protein kinase C βII and the receptor for activated C kinase 1 (RACK1) (C. C. Daniels and S. L. Quackenbush, personal communication). Orf C protein localizes in mitochondria where it is associated with a loss of membrane potential resulting in the induction of apoptosis (Nudson et al., 2003).
Continued efforts to derive viable, WDSV-infected explant cells from regressing tumours have now resulted in the establishment of a virus-positive culture of spring tumour explant cells (STECs), which are capable of producing WDSV. This cell culture system will allow the confirmation of previous work and the development of wild-type and recombinant virus for infection of fish.
METHODS
Cell isolation
A tumour taken in the spring of 2003 from a fish collected at the New York State Department of Environmental Conservation Oneida Lake Hatchery was washed extensively in PBS supplemented with excess penicillin, streptomycin and gentamicin. The tumour was then cut into 1 mm cubes under sterile conditions and distributed to multiple plastic tissue culture flasks and allowed to adhere in L-15 Leibovitz's medium with 2 mM l-glutamine, 100 U penicillin ml–1, 100 μg streptomycin ml–1 and 15 % fetal bovine serum at 20 °C. After 48 h, uncontaminated cells were trypsinized and replated in flasks for propagation or on to glass slides for immunofluorescence assays (Cel-Line; Erie Scientific Inc.). After overnight incubation, slides were fixed and incubated with affinity-purified rabbit polyclonal immunoglobulin as described previously (Rovnak et al., 2001). Antibodies reactive to WDSV recombinant Orf A, Orf B, Orf C and capsid were used at 1 : 100 dilutions. The anti-Orf B, anti-Orf C and anti-capsid antisera were the generous gift of Dr Volker Vogt, Department of Molecular Biology and Genetics, Cornell University, USA. Photomicrographs were captured digitally at a magnification of ×200 and images were assembled with Adobe Photoshop 7.0 software.
W12 cells (a WDSV-negative walleye fibroblast cell line established in the laboratory of Paul Bowser at Cornell University, USA, by Daniel Martineau) and rainbow trout gill cells (RTgill-W1; ATCC CRL-2523) were maintained in L-15 Leibovitz's medium with 10 % fetal bovine serum at 20 °C.
DNA isolation and sequence analysis
Purified chromosomal DNA (10 μg) was digested by the indicated restriction endonucleases and subjected to analysis on Southern blots as described previously (Martineau et al., 1991). Three separate fragments representing the entire WDSV sequence were radiolabelled by random priming (Invitrogen) and used together to hybridize with WDSV sequences. Chromosomal DNA (200 ng) was amplified with proofreading DNA polymerase according to the manufacturer's instructions (Phusion Hot Start High-Fidelity DNA Polymerase; New England Biolabs). Two overlapping fragments, representing the entire WDSV sequence, were amplified and cloned: a 5′-proximal fragment from nt –441 to 8463 was cloned by TA cloning into the pCR-XL-TOPO vector (Invitrogen), and a 3′-proximal fragment from nt 6120 to 12 710 was amplified with a 3′ EcoRI site in the primer. This fragment was digested at the 3′ EcoRI site and the internal WDSV EcoRI site at nt 6611 and cloned in pBluescript SK– (Stratagene). The sequences of overlapping 500–1000 bp regions were determined in both directions from both clones at the Macromolecular Resources facility core laboratory at Colorado State University, USA. The results from each were reassembled into contiguous regions and aligned with the existing WDSV sequence (GenBank accession no. L41838). The final assembly has been assigned GenBank accession no. EF428979. Envelope protein sequences were analysed by the Topology prediction of membrane proteins algorithm (TopPred at http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) (Claros & von Heijne, 1994; von Heijne, 1992).
WDSV provirus was detected in infected cells by amplification of gag gene sequences from 1 μg chromosomal DNA with the 5′ primer 5′-GCTTAGGAAAGATTGCCCGCAC-3′ and 3′ primer 5′-TGTCGCATGTTGCCACTCAAG-3′. Cell monolayers were trypsinized and washed with PBS prior to DNA isolation with DNAzol (Invitrogen) according to the manufacturer's instructions. Cloned WDSV plasmid DNA (20 pg) served as a positive PCR control.
Virus particle sedimentation and quantification
Tissue culture supernatants were filtered through a 0.22 μm filter and subjected to centrifugation at 100 000 g for 1 h. Pellets were resuspended in PBS and applied to a 15–60 % sucrose gradient and centrifuged for 16 h at 100 000 g at 7 °C. Gradients were fractionated and the density of individual fractions measured. WDSV genomic RNA content was quantified in individual fractions by real-time quantitative RT-PCR (qRT-PCR) with gag-specific primers as described previously (Getchell et al., 2002). Samples for the reverse transcriptase (RT) assay were brought to 0.5 % Triton X-100 and freeze-thawed three times prior to assay. Virus pellets were resuspended in 100 μl PBS. Trypsinized tissue culture cells, suspended in 100 μl PBS in individual microfuge tubes, were sonicated in a cuphorn device (Virtis Virsonic S3000) for 6 min with 10 s intervals at power setting 2 (33 W). RT activities of triplicate 10 μl samples were assayed in 50 μl reactions with poly(rA) template, poly(dT)12–18 primer and [32P]TTP substrate for 2 h at room temperature. cDNA was bound to DE81 paper (Whatman) and washed, and total incorporated counts were determined by scintillation counting.
Supernatant and cell-associated virus preparations were normalized for RT activity and a volume equivalent to 3.5 × 106 c.p.m. h–1 was added to 25 cm2 flasks containing 6 × 105 W12 walleye fibroblasts pre-treated with 25 μg DEAE ml–1 for 1 h. The medium was changed after 16 h, cells were split 1 : 2 after 5 days, and supernatants and cells were harvested after an additional 6 day incubation.
For electron microscopy, the confluent cell monolayer was scraped from plates and fixed in 2 % glutaraldehyde in 0.1 M sodium cacodylate, and processed and imaged at the Cornell Integrated Microscopy Center, Cornell University, USA.
RESULTS
Tumour explant cells express WDSV structural and accessory proteins
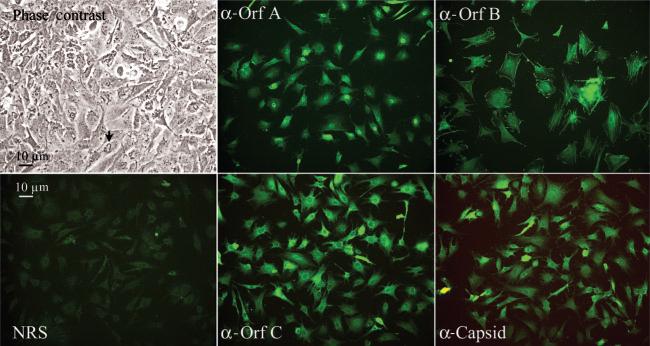
A mixed cell culture from a single, regressing walleye dermal sarcoma from Oneida Lake in New York was established by the explant of sterile tissue fragments into plastic tissue culture vessels. After the appearance of adherent cell types on the plastic, cells were analysed by an immunofluorescence assay (Fig. 1). Four WDSV-specific rabbit antisera were used: anti-Orf A (rv-cyclin), anti-Orf B, anti-Orf C and anti-capsid. Pre-immune serum from one rabbit served as a control. The localization of both Orf A and Orf C matched that previously observed for expressed recombinant forms of these proteins in piscine and mammalian cell culture systems (Nudson et al., 2003; Rovnak et al., 2001). Orf A was concentrated in the nucleus and Orf C in the cytoplasm in structures consistent with mitochondria. The degree of their expression in the population was inconsistent. Orf B protein appeared to be associated with plasma membrane and concentrated at cell ruffles and focal adhesions and along actin stress fibres. Its expression was similar in all cells. Viral capsid appeared at high levels in the cytoplasm of a significant number of cells and could be localized to perinuclear regions in less intensely stained cells. Both phase-contrast microscopy and immunofluorescence indicated a population of mixed cell morphologies. Upon reaching confluence, fluctuating membrane ruffles or fimbriae were apparent on the upper surface of approximately 50 % of cells by phase-contrast microscopy (Fig. 1, arrow).
Fig. 1.
Microscopic observations of STECs. Phase-contrast image of confluent STECs illustrates the apical ruffles common in these cells, indicated by an arrow. Immunofluorescent images were labelled with normal rabbit sera (NRS) or with rabbit antisera reactive to the indicated WDSV proteins and fluoresce-inconjugated goat anti-rabbit IgG. Magnification, ×200.
Tumour explant cells harbour clonal WDSV provirus
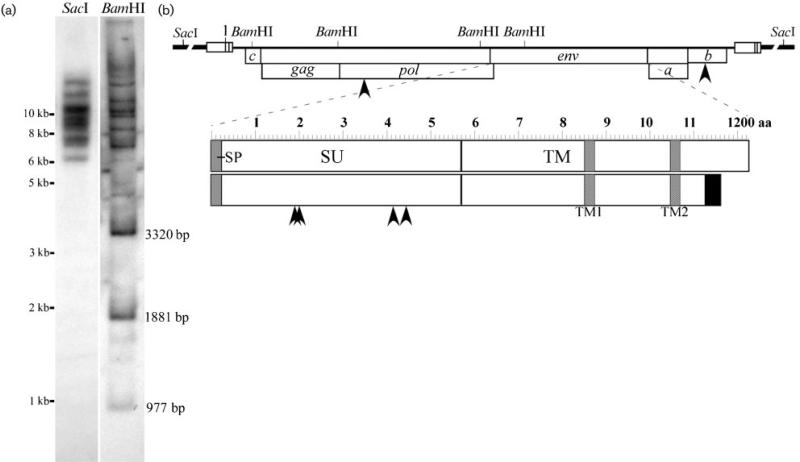
High-molecular-mass chromosomal DNA was isolated from STECs and 10 μg aliquots were digested with BamHI or SacI restriction endonuclease and analysed by Southern blot with a mixture of probes representative of the entire WDSV provirus (Fig. 2a). A SacI cut site is not present in the WDSV sequence and the appearance of eight distinguishable bands indicated that at least eight separate proviruses at different insertion sites were present in these cultures (Fig. 2b). BamHI cuts four times in the WDSV sequence and yielded the three predicted internal fragments of 977, 1881 and 3320 bp. Sixteen additional fragments were distinguishable. These corresponded to variable lengths of the 5′- and the 3′-terminal fragments joined to chromosomal DNA at different insertion sites, indicating at least eight proviral copies.
Fig. 2.
(a) Southern blot analysis of STEC chromosomal DNA. SacI liberates whole provirus with adjacent chromosomal DNA. BamHI liberates three internal fragments of fixed sizes as indicated and two external fragments with adjacent host DNA from each provirus. (b) Diagram of the WDSV provirus with adjacent chromosomal DNA (broken line). Open reading frames in frames 1 and 2 are indicated with arrowheads to locate amino acid mutations. The env reading frame is expanded to identify point mutations as well as the revised C terminus with variant sequence (black box) and smaller size. Shaded boxes indicate predicted transmembrane domains, TM1 and TM2, in the transmembrane region (TM), as well as the predicted signal peptide (SP) at the N terminus of the surface region of the protein (SU).
Variations in the WDSV sequence
Overlapping DNA fragments, representative of the entire WDSV genome, were PCR amplified with a proofreading DNA polymerase enzyme from STEC chromosomal DNA and cloned and sequenced. The resulting sequence was aligned with the existing WDSV sequence derived from a tumour from a fish from Oneida Lake in 1990 (Holzschu et al., 1995; Martineau et al., 1991). Overall, the WDSV genome was found to be stable. There were no nucleotide changes within the LTR or upstream untranslated region of the genome. A total of 27 point mutations out of 12 706 bp was identified. Five of these variations altered the amino acid sequences of predicted proteins (Fig. 2b). One mutation was found in pol at nt 3068, generating a proline to alanine substitution at aa 35 (P35A) (N terminus of the WDSV RT, as determined by Fodor & Vogt, 2002), four mutations in the surface (SU) region of env at nt 6539 (G189E), 6575 (T201I), 7219 (N416D) and 7302 (M443I), and one mutation in orf b at nt 10 923 (S118P). No mutations were identified in the regions of gag or env previously found to be variable in isolates from distinct geographical locations (Zhang et al., 1996). The extended time between isolations suggests strong selective forces and high-fidelity replication of the WDSV genome.
A significant sequence variation was identified in the transmembrane region of env with insertions of single guanine and cytosine residues between nt 9340 and 9347 (9440GGGgACCc9347) (Fig. 2b). The resulting frame shift altered 37 aa and generated a stop codon at nt 9451 compared with the previous stop at nt 9649. This shortened the predicted size of the envelope protein by 63 aa (1162 vs 1225 aa). Because of the size of this change and the guanine-rich site, this region was resequenced in both the new and original WDSV molecular clones and both contained the guanine and cytosine insertions found in the new sequence. This result indicated that the insertions did not represent a variation from the previous WDSV sequence and that the envelope protein varied significantly from previous predictions. The envelope protein has at least two, and possibly five, membrane-spanning regions as determined by analysis with software for membrane protein structure predictions (Claros & von Heijne, 1994; von Heijne, 1992). The two most likely transmembrane helices lie between aa 850 and 874 and between aa 1045 and 1069 with an intervening cytoplasmic loop (Fig. 2b).
Virus particle isolation
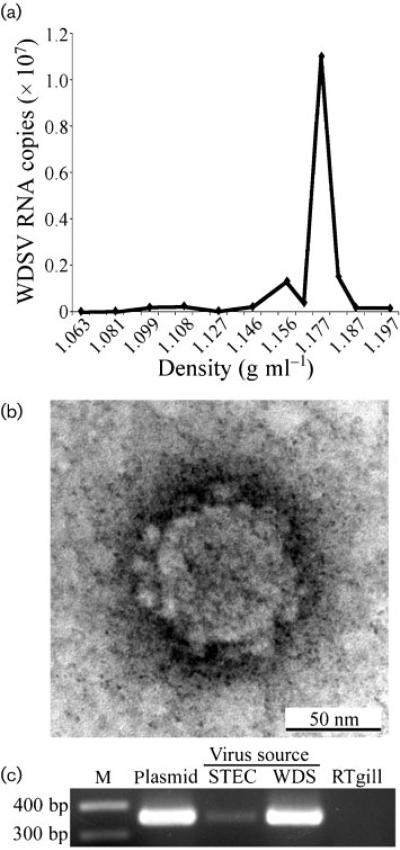
Filtered STEC culture supernatants were subjected to ultra-centrifugation in order to pellet virus particles. The pellet was resuspended and banded on a sucrose density gradient. The density of gradient fractions was determined and their WDSV RNA content measured by real-time qRT-PCR. The peak RNA fraction had a density of 1.17 g ml–1, characteristic of retroviral particles (Fig. 3a). Pelleted material was also subjected to transmission electron microscopy (TEM), which revealed particle morphologies consistent with those previously observed for WDSV (Fig. 3b; Martineau et al., 1991). The pellet material was also tested for in vitro infection of a rainbow trout gill cell line, RTgill-W1. Cell-free homogenate of a regressing tumour served as a positive control. Cells were split once and DNA was harvested 1 week later and subjected to PCR to detect WDSV provirus. The virus-infected STECs were positive for the predicted 354 bp amplicon, indicating the establishment of the WDSV provirus in the infected cultures (Fig. 3c).
Fig. 3.
(a) Sucrose-density gradient sedimentation of virus from culture supernatants. WDSV-specific RNA sequence was measured by qRT-PCR and plotted against the gradient fraction density. (b) TEM micrograph of virus pellet. Magnification, ×300 000. (c) Amplicons of the 354 bp gag sequence from 1 μg DNA from RTgill cells infected with STEC virus pellet (STEC) or WDS tumour homogenate (WDS). Controls comprised 20 pg WDSV clone DNA (Plasmid) and 1 μg RTgill DNA. M, Molecular mass marker.
Cell association of WDSV
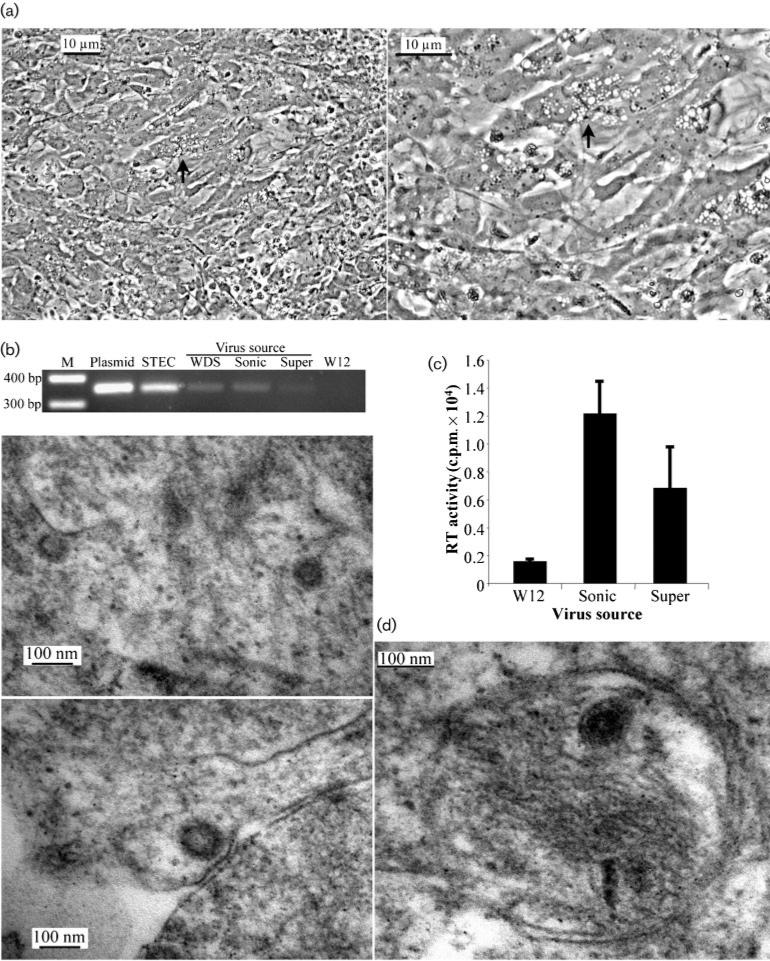
STECs frequently demonstrated vacuolization upon reaching confluence in subculture (Fig. 4a). The appearance of a cytopathic effect was similar to that associated with in vitro infection by spumaretroviruses, which bud internally and remain cell-associated (Delelis et al., 2004). In order to assay possible intracellular virus and compare it with virus released from cells, particles were collected from culture media by ultracentrifugation after 1 week of incubation and the corresponding monolayer cells were trypsinized, washed, suspended in PBS and subjected to sonication to release any cell-associated virus. Equivalent amounts of cell-associated and supernatant virus preparations, normalized by their RT activity, were then assayed for infectious virus particles by incubation with uninfected walleye fibroblast (W12) cells. Cultures were split at 4 days post-infection and supernatant and cells harvested 1 week later for virus assays. Cells infected by either the cell-associated or supernatant virus preparation were positive for provirus as determined by PCR amplification of proviral DNA (Fig. 4b), and there was no significant difference between the levels of RT activity in the supernatants of either infected culture (Fig. 4c). These results demonstrated that intracellular WDSV particles are comparable to virus released from cells for the infection of fish cell cultures.
Fig. 4.
(a) Phase-contrast micrograph illustrating the vacuolization observed in confluent STECs (arrow) at a magnification of ×200 (left) and ×400 (right). (b) Amplicons of the 354 bp gag sequence from DNA of W12 cells infected with virus from WDS tumour homogenate (WDS), sonicated STECs (Sonic) or STEC supernatant virus (Super). Controls comprised 20 pg WDSV clone DNA (Plasmid), 1 μg STEC DNA (STEC) and 1 μg W12 DNA (W12). (c) RT activity of supernatants from W12 cells (W12) or cells infected with virus from STEC sonicate (Sonic) or STEC supernatant (Super). (d) TEM micrographs illustrating intracellular WDSV in STECs at a magnification of ×70 000 (left panels) and ×100 000 (right panel). M, Molecular mass marker.
Analysis of the WDSV envelope sequence for internal membrane retention signals identified a consensus arginine-based endoplasmic reticulum (ER) localization signal, IRRER (aa 880–884) (Michelsen et al., 2005). This signal lies in the predicted cytoplasmic loop between aa 874 and 1045 (Fig. 2b). Cell-associated WDSV particles were examined by TEM in STECs scraped from a confluent monolayer. Viral particles, consistent with WDSV, were observed within cells and adjacent to internal membranes or cisternae, including mitochondrial membranes (Fig. 4d). The apparent sizes of these particles ranged from 75 to 100 nm and may represent forms prior to and after budding.
DISCUSSION
The propagation of infectious virus in vitro is essential for the efficient analysis of genetic variants and for their production for assessment in an animal model. This component of the experimental WDSV tumour model system has provided a particular challenge. In spite of established procedures for the isolation of virus from tumours by sonication (Martineau et al., 1990a), the actual cell association of the virus remained unclear, and the need to apply homogenization techniques to tissue culture systems was not appreciated. The appearance of vacuolization in confluent STEC cultures, whilst suggestive of viral cytopathic effect, has not yet been associated directly with virus particle morphogenesis. Transmission of infection with preparations of intracellular virus indicates the formation of infectious particles within cells, which would require budding from internal membranes into cellular compartments, akin to virus production observed with foamy viruses.
Predictions of the topology of the WDSV envelope protein identify a transmembrane helix between aa 850 and 874, ending on the cytoplasmic side of the membrane. A consensus arginine-based ER retention signal is immediately adjacent. These signals can direct the localization of membrane proteins to a variety of internal membranes (Michelsen et al., 2005). Foamy viruses utilize consensus KKXX motifs for intracellular ER retention and budding. Herpesviruses utilize arginine-based signals for the localization of gB envelope proteins to the inner nuclear membrane (reviewed by Michelsen et al., 2005). Examination of intracellular particles by electron microscopy identified possible membrane-bound viral forms with electron-dense cores in mitochondria (Fig. 4), as well as smaller particles with less-condensed cores, which may be preassembled particles prior to budding.
The cell association of WDSV may be essential for its stability in the environment. During regression, the tumour masses are degrading and cells and tumours are shed from the surface of the fish. The continued tight association of virus particles with cells in the aquatic environment would serve to stabilize them prior to attachment to and infection of naive animals. In addition, tumour masses are ingested by other fish, which is an efficient route of infection. Both cutaneous and oral WDSV infections have been demonstrated experimentally in walleye fingerlings (Bowser et al., 1997).
During the development of dermal sarcoma, viral mRNA expression is extremely limited. Such regulation may be critical to immune evasion by the virus during this period. The outcome is the production of a mass of cells harbouring WDSV provirus. During spawning, the virus expression pattern shifts to virus production, and transmission is maximized by regression of the tumour mass and release of the viral load into the environment. The mechanisms of tumour regression are as yet unresolved. Although the Orf C protein has been demonstrated to induce apoptosis through its association with mitochondria (Nudson et al., 2003), a role for immune rejection of the newly virus-laden tumour has not been excluded. The results presented here suggest a mechanism for continued immune evasion, even during regression: the retention of mature particles within cellular compartments would further limit the stimulation of and recognition by host immune mechanisms. Lymphocytic infiltration of most tumours is similar to that of surrounding dermis; high density infiltration is only associated with the onset of tumour necrosis (Martineau et al., 1990b).
Sequence comparisons between the STEC provirus from Oneida Lake in spring 2003 and the original molecular clone from Oneida Lake in spring 1990 identified significant differences in the size and sequence of env as a result of a sequencing correction. More significant to the nature of WDSV is the apparent stability of the genome over time. Analyses of segments of gag and env sequences showed no variation in the gag sequence among all WDSV isolates from Oneida Lake (Zhang et al., 1996). Variation was observed in a select portion of the SU region of env, and two clades were identified based on these variations. One of these two clades showed no variation from the original clone. The STEC virus most likely represents this clade, as there were no mutations noted within the selected SU sequence. There are clear sequence variations between WDSV isolates from distinct geographical regions of North America. These variants are from walleye populations that have been isolated since the last ice age (~10 000 years) (Zhang et al., 1996). Genomic stability is driven by strong selective constraints, but two mechanisms of retroviral replication could further explain the paucity of WDSV variation: high-fidelity reverse transcription and significant virus replication as provirus by host DNA polymerases. Such replication occurs during the expansion of the tumour cell population and reduces the number of RT replication cycles required to produce infectious virus. In this way, WDSV has similarities to the complex, oncogenic deltaretroviruses (Cann & Chen, 1990).
ACKNOWLEDGEMENTS
We thank Paul Bowser and Greg Wooster for supplying tumours and Volker Vogt for rabbit antisera. This work was supported by the National Institutes of Health, grant CA095056, from the National Cancer Institute to S. L. Q.
Footnotes
The GenBank/EMBL/DDBJ accession number of the sequence determined in this paper is EF428979.
REFERENCES
- Bowser PR, Wooster GA, Earnest-Koons K. Effects of fish age and challenge route in experimental transmission of walleye dermal sarcoma in walleyes by cell-free tumor filtrates. J Aquat Anim Health. 1997;9:274–278. [Google Scholar]
- Cann AJ, Chen ISY. Virology. 2nd edn. Raven Press; New York: 1990. Human T-cell leukemia virus types I and II. [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Delelis O, Lehmann-Che J, Saib A. Foamy viruses – a world apart. Curr Opin Microbiol. 2004;7:400–406. doi: 10.1016/j.mib.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Fodor SK, Vogt VM. Characterization of the protease of a fish retrovirus, walleye dermal sarcoma virus. J Virol. 2002;76:4341–4349. doi: 10.1128/JVI.76.9.4341-4349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getchell RG, Wooster GA, Sutton CA, Casey JW, Bowser PR. Dose titration of walleye dermal sarcoma (WDS) tumor filtrate. J Aquat Anim Health. 2002;14:247–253. doi: 10.1577/1548-8667(2002)014<0247:DTOWDS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Holzschu DL, Martineau D, Fodor SK, Vogt VM, Bowser PR, Casey JW. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschu D, Lapierre LA, Lairmore MD. Comparative pathogenesis of epsilonretroviruses. J Virol. 2003;77:12385–12391. doi: 10.1128/JVI.77.23.12385-12391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RK, Miller HR, Nielsen O, Clayton JW. Fish cell culture: characteristics of a continuous fibroblastic cell line from Walleye (Stizostedion vitreum vitreum). Can J Fish Aquat Sci. 1980;37:1070–1075. [Google Scholar]
- Lairmore MD, Stanley JR, Weber SA, Holzschu DL. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proc Natl Acad Sci U S A. 2000;97:6114–6119. doi: 10.1073/pnas.110024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau D, Bowser PR, Wooster GA, Armstrong GA. Experimental transmission of a dermal sarcoma in fingerling walleyes (Stizostedion vitreum vitreum). Vet Pathol. 1990a;27:230–234. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- Martineau D, Bowser PR, Wooster GA, Forney JL. Histologic and ultrastructural studies of dermal sarcoma of walleye (Pisces: Stizostedion vitreum). Vet Pathol. 1990b;27:340–346. doi: 10.1177/030098589002700506. [DOI] [PubMed] [Google Scholar]
- Martineau D, Renshaw R, Williams JR, Casey JW, Bowser PR. A large unintegrated retrovirus DNA species present in a dermal tumor of walleye Stizostedion vitreum. Dis Aquat Organ. 1991;10:153–158. [Google Scholar]
- Martineau D, Bowser PR, Renshaw RR, Casey JW. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen K, Yuan H, Schwappach B. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 2005;6:717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudson WA, Rovnak J, Buechner M, Quackenbush S. Walleye dermal sarcoma virus Orf C is targeted to the mitochondria. J Gen Virol. 2003;84:375–381. doi: 10.1099/vir.0.18570-0. [DOI] [PubMed] [Google Scholar]
- Quackenbush SL, Holzschu DL, Bowser PR, Casey JW. Transcriptional analysis of walleye dermal sarcoma virus (WDSV). Virology. 1997;237:107–112. doi: 10.1006/viro.1997.8755. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus cyclin interacts with components of the mediator complex and the RNA polymerase II holoenzyme. J Virol. 2002;76:8031–8039. doi: 10.1128/JVI.76.16.8031-8039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus retroviral cyclin directly contacts TAF9. J Virol. 2006;80:12041–12048. doi: 10.1128/JVI.01425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Casey JW, Quackenbush SL. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin). Virology. 2001;280:31–40. doi: 10.1006/viro.2000.0731. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Hronek BW, Ryan SO, Cai S, Quackenbush SL. An activation domain within the walleye dermal sarcoma virus retroviral cyclin protein is essential for inhibition of the viral promoter. Virology. 2005;342:240–251. doi: 10.1016/j.virol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Walker R. Virus associated with epidermal hyperplasia in fish. Natl Cancer Inst Monogr. 1969;31:195–207. [PubMed] [Google Scholar]
- Yamamoto T, MacDonald RD, Gillespie DC, Kelly RK. Viruses associated with lymphocystis and dermal sarcoma of walleye (Stizostedion vitreum vitreum). J Fish Res Board Can. 1976;33:2408–2419. [Google Scholar]
- Yamamoto T, Kelly RK, Nielsen O. Morphological differentiation of virus-associated skin tumors of walleye (Stizostedion vitreum vitreum). Fish Pathology. 1985;20:361–372. [Google Scholar]
- Zhang Z, Martineau D. Walleye dermal sarcoma virus: OrfA N-terminal end inhibits the activity of a reporter gene directed by eukaryotic promoters and has a negative effect on the growth of fish and mammalian cells. J Virol. 1999;73:8884–8889. doi: 10.1128/jvi.73.10.8884-8889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Du Tremblay D, Lang BF, Martineau D. Phylogenetic and epidemiologic analysis of the walleye dermal sarcoma virus. Virology. 1996;225:406–412. doi: 10.1006/viro.1996.0616. [DOI] [PubMed] [Google Scholar]