Abstract
The identification of patients who will respond to anti-tumor necrosis factor alpha (anti-TNF-α) therapy will improve the efficacy, safety, and economic impact of these agents. We investigated whether killer cell immunoglobulin-like receptor (KIR) genes are related to response to anti-TNF-α therapy in patients with rheumatoid arthritis (RA). Sixty-four RA patients and 100 healthy controls were genotyped for 16 KIR genes and human leukocyte antigen-C (HLA-C) group 1/2 using polymerase chain reaction sequence-specific oligonucleotide probes (PCR-SSOP). Each patient received anti-TNF-α therapy (adalimumab, etanercept, or infliximab), and clinical responses were evaluated after 3 months using the disease activity score in 28 joints (DAS28). We investigated the correlations between the carriership of KIR genes, HLA-C group 1/2 genes, and clinical data with response to therapy. Patients responding to therapy showed a significantly higher frequency of KIR2DS2/KIR2DL2 (67.7% R vs. 33.3% NR; P = 0.012). A positive clinical outcome was associated with an activating KIR–HLA genotype; KIR2DS2 + HLA-C group 1/2 homozygous. Inversely, non-response was associated with the relatively inhibitory KIR2DS2 – HLA-C group 1/2 heterozygous genotype. The KIR and HLA-C genotype of an RA patient may provide predictive information for response to anti-TNF-α therapy.
Keywords: Rheumatoid arthritis, Anti-TNF-alpha therapy, Responder, Killer immunoglobulin-like receptor, Natural killer cells, Human leukocyte antigen-C, Genotype
Introduction
Chronic inflammatory conditions such as rheumatoid arthritis are manifestations of inappropriate immune responses, where activation and regulation are imbalanced.
Anti-inflammatory agents such as TNF-α antagonists can achieve significant results; however, a subset of patients do not respond. The current absence of clinically useful predictors of response continues to impact both the clinical management of patients and associated treatment costs.
The KIR gene cluster, located on chromosome 19q13.4 [1], codes for a family of polymorphic receptors which can be activating (KIR2DS/KIR3DS) or inhibiting (KIR2DL/KIR3DL) in function. KIR are expressed on natural killer (NK) cells and subsets of T-cells and modulate cell responses through recognition of human leukocyte antigen (HLA) class I ligands on target cells.
Under normal conditions, inhibitory receptors KIR2DL2 and KIR2DL3 recognize self-HLA-C1 ligands and KIR2DL1 binds HLA-C2 ligands, acting to prevent NK cell attack on normal cells. In the absence of these inhibiting KIR-HLA signals, NK cell activation may occur through activating receptors such as KIR2DS1 and KIR2DS2 [2, 3]. It is the net balance of activating and inhibiting signals from receptors such as KIR that determine NK cell and T-cell reactivity. An individual’s genotype predisposes to specific KIR/HLA interactions, which may range from activating to relatively inhibitory depending on which type of KIR and HLA is present [4].
Specific KIR-HLA combinations have been associated with differential responses to infection [5–7] and susceptibility to autoimmune disorders including Crohn’s disease [8–10], ulcerative colitis [10, 11], systemic lupus erythematosus [12, 13], type I diabetes [14], and psoriatic arthritis [15]. In the case of autoimmunity, the presence of an activating KIR and/or absence of ligands for inhibitory KIR is commonly associated with disease susceptibility.
In RA, activating KIR2DS2 has been shown to function as a co-stimulatory molecule on CD4+CD28null T-cells [16]. The expansion of this subset correlates with the clinical phenotype of RA, where KIR2DS2 has been associated with an increased likelihood for RA patients to develop rheumatoid vasculitis [17–19].
These studies suggest a role for KIR in determining the immune response in inflammatory disease, which prompts the question of whether a particular KIR predisposition could also influence response to anti-inflammatory treatment.
In this study, we examined the KIR and HLA-C genotypes of RA patients beginning anti-TNF-α treatment and healthy controls. We found that the presence of KIR2DS2/KIR2DL2 was significantly associated with patients who responded to therapy. Further consideration of KIR with HLA-C ligand availability indicated a potentially activating KIR–HLA-C genotype in responding patients relative to non-responders to anti-TNF-α therapy.
Methods
Patients
Sixty-four unrelated Northern Irish chronic RA patients were included in this study. Each subject was a patient attending the rheumatology department of Musgrave Park Hospital, Belfast, Northern Ireland. All patients fulfilled the American College of Rheumatology 1987 revised criteria for RA [20] and had active disease as indicated by a DAS28 score of >3.2 [21].
There was no significant difference between the responding and non-responding patients with respect to the distribution of age (P = 0.21, unequal variance two-sided t-test) or sex (P = 0.74, two-sided Fishers’ exact test).
Patients were assigned to anti-TNF-α treatment as part of routine clinical practice. Each patient fulfilled the British Society for Rheumatology (BSR) criteria for anti-TNF-α therapy and had failed at least two disease-modifying anti-rheumatic drugs (DMARDS) and had a DAS28 score of >5.1 when originally assessed for treatment [22]. In some cases, DAS28 scores between original assessment and baseline treatment fluctuated, due to time lapsed from assessment to treatment. Forty patients received adalimumab, 15 etanercept, and 9 infliximab. In most cases, methotrexate (MTX) was maintained as a combination therapy, and patients were allowed to continue oral glucocorticoids. The main demographic and clinical features of the patients are shown in Table 1. Thirty-eight patients received a combination of MTX and anti-TNF-α drugs (adalimumab, etanercept, or infliximab). There exists no significant correlation between the anti-TNF-α drug and the treatment outcome in those patients who received MTX (P = 0.35, χ2-test). In the group of patients who did not receive MTX, we observed relatively more patients responding to etanercept than to adalimumab (7 R and 1 NR vs. 8 R and 10 NR, P = 0.08, Fisher’s exact test).
Table 1.
Demographic and clinical characteristics of patients
| Responders | Non-responders | All | |
|---|---|---|---|
| Total | 34 | 30 | 64 |
| Mean age (years) | 58 (range, 35–75) | 54.53 (range, 27–73) | 56.33 (range, 27–75) |
| Woman/man | 28/6 | 26/4 | 54/10 |
| MTX and adalimumab | 9 | 13 | 22 |
| MTX and etanercept | 5 | 2 | 7 |
| MTX and infliximab | 5 | 4 | 9 |
| Adalimumab | 8 | 10 | 18 |
| Etanercept | 7 | 1 | 8 |
| Infliximab | 0 | 0 | 0 |
| Mean pre-DAS28 score | 6.21 (range, 4.08–8.96) | 5.43 (range, 3.75–7.38) | 5.85 (range, 3.75–8.96) |
| Mean post-DAS28 score | 3.64 (range, 1.18–6.15) | 5.61 (range, 3.49–7.27) | 4.53 (range, 1.18–7.27) |
Peripheral blood samples were collected from each patient at time 0. Following 3 months of treatment, the patients were assigned a responder or non-responder status, according to the BSR criterion where clinical response is defined as attaining a DAS28 score improvement of >1.2 or a DAS28 score of <3.2 [22].
A control population of 100 unrelated healthy Northern Irish Caucasians were included for comparison of KIR gene content. The healthy control blood samples were collected at Northern Ireland Blood Transfusion Services, Belfast. All participants in the study gave written informed consent, and the study had local research ethical committee approval from The Queens University, Belfast, Northern Ireland.
Genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using the salting-out method as previously described [23]. DNA was typed for the presence or absence of framework KIR genes: KIR2DL4, KIR3DL2, KIR3DL3, and KIR3DP1, six activatory KIR (KIR2DS1, KIR2DS2, KIR2S3, KIR2DS4, KIR2DS5 and KIR3DS1) and five inhibitory KIR (KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, KIR2DL5). KIR2DP1 was also included in the typing. KIR genotyping was performed using the PCR primers and probes of a KIR PCR-SSOP method [24]. Positive controls of known KIR genotype, collectively incorporating all of the KIR genes, were included in the typing procedure.
HLA-C typing was performed using the PCR-SSOP method. DNA was amplified by PCR using the HLA-C generic primers described by Cereb et al. [25]. A modified version of the HLA-C typing method was used to define the HLA-C1 and C2 groups using probe C293 and C291, respectively [26].
Statistical methods and analysis
The significance of the differences in proportions of responders and non-responders exhibiting a specific genotype was assessed using Fisher’s exact test. Welch’s t-statistics and paired t-tests were used to investigate differences in the DAS28 score. All tests were two-sided unless otherwise stated. Adjustments for multiple testing were made using Holm’s method.
Results
Within the group of 64 patients (described in Table 1), 34 were responders (53.0%) and 30 were non-responders (47.0%). We compared the profiles of the KIR genes in the groups of responders and non-responders to anti-TNF-α therapy. In the responder population, the frequency of KIR2DS2 and KIR2DL2 (which share high linkage disequilibrium) was significantly higher compared with non-responders (67.7% vs. 33.3%; P = 0.012, Fisher’s exact test). To assess the relevance of the P-value and to reduce the effect of multiple comparisons arising from testing a family of 16 KIR genes, we performed a non-parametric random permutation test as follows [27]. We randomly permuted the class labels of all 64 patients and recomputed the P-value for the difference in proportions of carriers/non-carriers in responders and non-responders. We iterated this procedure 10,000 times to generate the distribution of P-values under the null hypothesis of no association between the presence/absence of KIR2DS2/KIR2DL2 and response to therapy. Among 100,000 permutation-based P-values, only 1.17% are smaller than or equal to the unpermuted P-value of 0.012. This confirms that carriership of KIR2DS2/KIR2L2 and the response to therapy cannot be explained by chance alone.
There was no significant difference between the baseline DAS28 score of patients carrying KIR2DS2/KIR2DL2 and those who did not (P = 0.57, two-sided, unequal variance t-test). The drug regime improved the DAS28 score in both carriers (P = 0.005, paired t-test) and non-carriers of KIR2DS2/KIR2DL2 (P < 1.6 × 10−6, paired t-test). This improvement, however, was significantly larger in patients carrying KIR2DS2/KIR2DL2 (P = 0.04, two-sided, unequal variance t-test).
When compared with healthy controls, the KIR2DS2/KIR2DL2-positive genotype remained significantly more frequent in the responder group (67.6% responders vs. 40.0% controls, P = 0.009). However, carriership of KIR2DS2/KIR2DL2 in the non-responders was not significantly different to healthy controls. The frequencies of all other KIR genes tested were not significantly different between responders, non-responders, or the healthy control groups.
To consider the additional effect of HLA-C zygosity, patients were categorized into four groups similar to a psoriatic arthritis model proposed by Nelson et al. [15]. The genotype groups range from NK cell activating (group I) to inhibiting (group IV) based on KIR-HLA interactions. Nelson’s model considered the presence/absence of both KIR2DS1 and KIR2DS2 with HLA-C zygosity. However, since KIR2DS1 was not informative in our study, we modified Nelson’s model to consider only KIR2DS2 in our interpretation. Thus, the most activating genotype, group I, included patients who were positive for activating KIR2DS2 and were HLA-C homozygous (C1/C1 or C2/C2). Such HLA-C homozygosity limits ligand availability for inhibitory KIR (KIR2DL1 or KIR2DL2/KIR2L3), restricting counteracting inhibition signals. Patients in group II were KIR2DS2 positive and were HLA-C heterozygous (i.e., they had both ligands C1/C2 and therefore relatively more inhibitory receptor functionality due to ligand availability). Group III patients were KIR2DS2 negative and HLA-C homozygous (without the activating receptor KIR2DS2 but limited inhibitory function through homozygosity for the HLA-C ligands of inhibitory KIR). Finally, the most inhibitory genotype group IV patients were KIR2DS2 negative and HLA-C heterozygous. Group IV patients are predisposed to a more inhibiting genotype since they lack KIR2DS2 and carry both HLA-C ligand types, promoting function of all corresponding inhibitory KIR receptors.
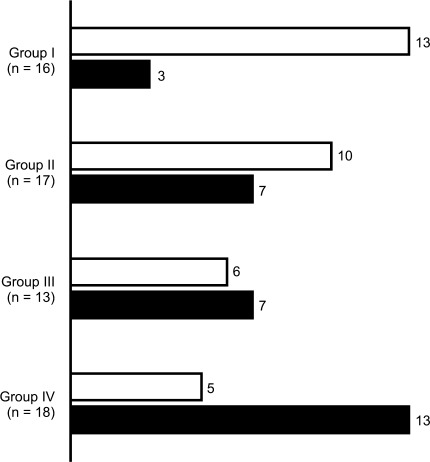
We observed that the ratio of responders to non-responders inverts from groups I to IV (Fig. 1).
Fig. 1.
Number of responders (white) and non-responders (black) in the groups I to IV. Groups range from most activating to most inhibiting from groups I to IV, respectively. Patients included in group I are KIR2DS2 positive and HLA-C group 1/2 homozygous (C1/C1 or C2/C2). Group II patients are KIR2DS2 positive and HLA-C group 1/2 heterozygous (C1/C2). Group III patients are KIR2DS2 negative and HLA-C group 1/2 homozygous. Group IV patients are KIR2DS2 negative and HLA-C group 1/2 hetrozygous
A groupwise comparison of the number of responders and non-responders revealed a significant difference between groups I and IV (P = 0.015, adjusted for multiple comparisons using Holm’s correction). No other groupwise comparison revealed a significant difference.
There was no significant difference between the groups with respect to the DAS28 score before therapy. The majority of patients in groups I to III experienced a significant reduction in the DAS28 score after treatment (P = 0.004, P = 0.004, and P = 0.033, respectively, based on paired t-test with Holm’s correction), whereas patients in group IV did not (P = 0.161). Table 2 summarizes the DAS28 scores at baseline, 3 months after therapy, and the improvements in groups I and IV. The mean DAS28 score improvement for patients in group I was significantly larger than that of group IV (P = 0.022).
Table 2.
Patient disease activity scores according to KIR and HLA-C genotype status
| Group I (2DS2+HLA-C homo) | Group IV (2DS2−HLA-C hetero) | |
|---|---|---|
| DAS28 at baseline | 6.05 (range, 4.28–8.96) | 5.64 (range, 3.75–7.38) |
| DAS28 after 3 months | 4.25 (range, 2.11–7.27) | 5.16 (range, 1.95–7.22) |
| DAS28 improvement | 1.80 (range, −1.85 to 3.92) | 0.48 (range, −2.25 to 3.32) |
| # responders/non-responders | 13/3 | 5/13 |
| P-value of improvement | 0.004 | 0.161 |
Group I: KIR2DS2 positive and HLA-C homozygous (C1/C1 or C2/C2), Group IV: KIR2DS2 negative and HLA-C heterozygous (C1/C2)
These results suggest that there exist subgroups of patients characterized by distinct KIR and HLA-C genotype profiles that are associated with a significant difference of response to anti-TNF-α therapy.
Discussion
The influence of the KIR genes in inflammatory disease is supported by a growing body of evidence demonstrating the association of specific KIR genotypes with disease.
Further consideration of the cumulative effect of KIR with HLA ligand availability permits differentiation of subjects to relatively activating or inhibiting genotypes, which ultimately influence the responses of KIR-expressing cells.
The promotion or prevention of NK cell activation crucially influences innate defences, the facilitation of cross talk between cells, and adaptive immunity. NK cell importance in immune regulation is reflected through their interaction with components of the adaptive immune system where they have been shown to eradicate autoreactive T-cells and B-cells [28].
In this study, a KIR-HLA combination that favors NK cell activation through the presence of activating KIR2DS2 and homozygosity for HLA-C1 or HLA-C2 was associated with responders to anti-TNF-α. Historically, NK cell activity in the peripheral blood of RA patients has been reported to be normal [29, 30] or low [31–33]. It is possible that responders could have a different mechanism of disease pathogenesis than non-responders, and it would be interesting to investigate NK cell activity in both groups. The occurrence of an inhibitory profile of NK cells in non-responders is supported by their prevalence in the proposed group IV cohort that are KIR2DS2 negative and HLA-C heterozygous. The general inversion of the numbers of responders and non-responders moving through groups I to IV, perhaps speaks to the continuum of excellent to poor responders observed in clinical practice. In this model, determination of where those patients who gain responses from up-dosing or switching anti-TNF-α therapy are positioned would also be of interest.
Our study has several limitations that impact the interpretation of these findings. In this small cohort of 64 subjects, we provide preliminary data suggesting a possible role for a KIR–HLA-C genotype in response to anti-TNF-α therapy. These findings will require independent validation in a larger population, which will permit further sub-analysis of KIR–HLA-C genotype and response for each anti-TNF-α therapy.
KIR2DS2 has been associated with vascular complications of RA [16, 18, 19] and with numerous inflammatory conditions and autoimmune outcomes [11, 13–15, 34, 35]. It is therefore not surprising that this activating KIR is implemented in response to anti-inflammatory therapy. It could be argued that patients carrying KIR2DS2 may be subject to a greater disease severity and hence are likely to show a more robust response to therapy. In this study, the baseline DAS28 scores of responders were significantly higher than those of non-responders, but this could not be attributed to carriership of KIR2DS2. The baseline DAS28 scores for patients with and without KIR2DS2 were not statistically different; however, the improvement of the score was significantly larger in the group of patients carrying this gene.
In previous reports, inflammatory cytokine polymorphisms such as TNF-α –308G/A have indicated that a genetic predisposition toward increased TNF-α expression can be associated with non-response [36–39], These findings have been disputed in a recent meta-analysis by Pavy et al. [40], and so, the role of this SNP remains unclear.
A gene expression study of baseline synovial tissue indicated an upregulation of inflammation-related genes in patients who responded to infliximab [41]. In proteomics, a baseline 24-protein signature of elevated serum antibody and cytokine concentrations is reported in RA patients who responded to etanercept [42]. Alternatively, Badot et al. [43] related non-responders to adalimumab with overexpression of cell division and immune regulation genes.
These studies and our current study point to a possible activating profile that may provide an advantage for response to anti-TNF-α therapy.
Acknowledgments
This work was supported by a Co-operative Award in Science & Technology (CAST) grant from the Department of Employment & Learning, Northern Ireland.
Conflict of interest
A patent application (US2010323358) for “the use of KIR genes for predicting response to anti-TNF-α therapy” was deposited by the University of Ulster (CM McGeough, AJ Bjourson, Daniel Berrar). GW and CM have received honoraria and/or grant support from Abbott, Schering-Plough and Wyeth. There has been no financing of the current manuscript by any of the above. RC and PG have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any non-commercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Cathy M. McGeough, Email: cathymcgeough@hotmail.co.uk
Daniel Berrar, Email: dp.berrar@ulster.ac.uk.
Gary Wright, Email: garyd.wright@belfasttrust.hscni.net.
Clare Mathews, Email: clare_matthews@hotmail.com.
Paula Gilmore, Email: gilmore.p@hotmail.com.
Rodat T. Cunningham, Email: drr.cunningham@btinternet.com
Anthony J. Bjourson, Phone: +442870324885, Email: aj.bjourson@ulster.ac.uk
References
- 1.Suto Y, Maenaka K, Yabe T, Hirai M, Tokunaga K, Tadok K, et al. Chromosomal localisation of the human natural killer cell class I receptor family genes to 19q13.4 by fluorescence in situ hybridisation. Genomics. 1996;35:270–272. doi: 10.1006/geno.1996.0355. [DOI] [PubMed] [Google Scholar]
- 2.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 3.Valéz-Gómez M, Reyburn HT, Erskine RA, Stominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immmunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 5.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nature Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 6.Jennes W, Verhryden S, Demanet C, Adjé-Toure CA, Vuylsteke B, Nkengasong JN, et al. Cutting edge: resistance to HIV-1 infection among African Female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 7.Kakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 8.Hollenbach JA, Ladner MB, Saeteurn K, Taylor KD, Mei L, Haritunians T, et al. Susceptibility to Crohn’s disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics. 2009;61:633–671. doi: 10.1007/s00251-009-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HX, Li JC, Liu ZJ. Relationship between expression of inhibitory killer cell immunoglobulin-like receptor HLACw ligand and susceptibility to inflammatory bowel disease. Zhonghua Yi Xue Za Zhi. 2008;88:3108–3111. [PubMed] [Google Scholar]
- 10.Wilson TJ, Jobin M, Jobin LF, Portela P, Salim PH, Rosito MA, et al. Study of killer immunoglobulin-like receptor genes and human leukocyte antigens class I ligands in a Caucasian Brazilian population with Crohn’s disease and ulcerative colitis. Hum Immunol. 2010;71:293–297. doi: 10.1016/j.humimm.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Jones DC, Edgar RS, Ahmad T, Cummings JR, Jewell DP, Trowsdale J, et al. Killer Ig-like receptor genotype and HLA ligand combinations in ulcerative colitis susceptibility. Genes Immun. 2006;7:576–582. doi: 10.1038/sj.gene.6364333. [DOI] [PubMed] [Google Scholar]
- 12.Ho YF, Zhang YC, Jiao YL, Wang LC, Li JF, Pan ZL, et al. Disparate distribution of activating and inhibitory killer cell immunoglobulin-like receptor genes in patients with systemic lupus erythematosus. Lupus. 2010;19:20–26. doi: 10.1177/0961203309345779. [DOI] [PubMed] [Google Scholar]
- 13.Pellett F, Siannis F, Vukin I, Lee P, Urowitz MB, Gladmann DD. KIRs and autoimmune disease: studies in systemic lupus erythematosus and scleroderma. Tissue Antigens. 2007;69(Suppl 1):106–108. doi: 10.1111/j.1399-0039.2006.762_6.x. [DOI] [PubMed] [Google Scholar]
- 14.Van der slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52:2639–2642. doi: 10.2337/diabetes.52.10.2639. [DOI] [PubMed] [Google Scholar]
- 15.Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in Psoriatic arthritis. J Immunol. 2004;173:4273–4276. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- 16.Namekawa T, Snyder MR, Yen JH, Goehring BE, Liebson PJ, Weyand CM, et al. Killer cell activating receptors function as costimulatory molecules on CD4+CD28null T-cells clonally expanded in rheumatoid arthritis. J Immunol. 2000;165:1138–1145. doi: 10.4049/jimmunol.165.2.1138. [DOI] [PubMed] [Google Scholar]
- 17.Martens PB, Goronzy JJ, Schaid D, Weyand CM. Expansion of unusual CD4+ T cells in severe rheumatoid arthritis. Arthritis Rheum. 1997;40:1106–1114. doi: 10.1002/art.1780400615. [DOI] [PubMed] [Google Scholar]
- 18.Majorczyk E, Pawlik A, Łuszczek W, Nowak I, Wiśniewski A, Jasek M, et al. Associations of killer cell immunoglobulin-like receptor genes with complications of rheumatoid arthritis. Genes Immun. 2007;8:678–683. doi: 10.1038/sj.gene.6364433. [DOI] [PubMed] [Google Scholar]
- 19.Yen JH, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, et al. Major histocompatibility complex class-I recognising receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193:1159–1167. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 21.Prevoo ML, van’t Hof MA, Kuper HH, van Leeuween MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts: development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 22.British Society for Rheumatology (2001) Guidelines for prescribing TNF-blockers in adults with rheumatoid arthritis. http://www.rheumatology.org.uk/guidelines/clinicalguidelines. Accessed 21 Apr 2010
- 23.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol. 2005;14:135–142. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Cereb N, Maye P, Lee S, Kong Y, Yang SY. Locus specific amplification of HLA class I genes from genomic DNA: locus specific sequences in the first and third introns of HLA-A, -B, and C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 26.Williams F, Meenagh A, Patterson C, Middleton D. Molecular diversity of the HLA-C gene identified in a Caucasian population. Hum Immunol. 2002;63:602–613. doi: 10.1016/S0198-8859(02)00408-1. [DOI] [PubMed] [Google Scholar]
- 27.Ringnér M, Edén P, Johansson P (2003) Classification of expression patterns using artificial neural networks. In: Berrar D, Dubitzky W, Granzow M (eds) A practical approach to microarray data analysis. Kluwer Academic, Boston/Dordrecht/London
- 28.Takeda K, Dennert G. The development of autoimmunity in C57BL/6 mice correlates with the disappearance of natural killer type 1-positive cells: evidence for their suppressive action on bone marrow stem cell proliferation, B cell immunoglobulin secretion, and autoimmune symptoms. J Exp Med. 1993;117:155–164. doi: 10.1084/jem.177.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neighbour PA, Grayzel AI, Miller AE. Endogenous and interferon-augmented natural killer cell activity of human peripheral blood mononuclear cells in vitro. Studies of patients with multiple sclerosis, systemic lupus erythematosus or rheumatoid arthritis. Clin Exp Immunol. 1982;49:11–21. [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong RD, Panayi GS. Natural killer cell activity in inflammatory joint disease. Clin Rheumatol. 1983;2:243–249. doi: 10.1007/BF02041398. [DOI] [PubMed] [Google Scholar]
- 31.Combe B, Pope R, Darnell B, Talal N. Modulation of natural killer cell activity in the rheumatoid joint and peripheral blood. Scand J Immunol. 1984;20:551–558. doi: 10.1111/j.1365-3083.1984.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 32.Karsh J, Dorval G, Osterland CK. Natural cytotoxicity in rheumatoid arthritis and systemic lupus erythematosus. Clin Immunol Immunopathol. 1981;19:437–446. doi: 10.1016/0090-1229(81)90086-6. [DOI] [PubMed] [Google Scholar]
- 33.Aramaki T, Ida H, Izumi Y, Fujikawa K, Huang M, Arima K, et al. A significantly impaired natural killer cell activity due to a low activity on a per-cell basis in rheumatoid arthritis. Mod Rheumatol. 2009;19:245–252. doi: 10.1007/s10165-009-0160-6. [DOI] [PubMed] [Google Scholar]
- 34.Boyton RJ, Smith J, Ward R, Jones M, Ozerovitch L, Wilson R, et al. HLA-C and killer cell immunoglobulin-like receptors in idiopathic bronchiectasis. Am J Resp Crit Care Med. 2006;173:327–333. doi: 10.1164/rccm.200501-124OC. [DOI] [PubMed] [Google Scholar]
- 35.Momot T, Koch S, Hunzelmann N, Krieg T, Ulbricht K, Schmidt RE, et al. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004;50:1561–1565. doi: 10.1002/art.20216. [DOI] [PubMed] [Google Scholar]
- 36.Mugnier B, Balandraud N, Darque A, Roudier C, Roudier J, Reviron D. Polymorphism at position –308 of the tumour necrosis factor alpha gene influences outcome of infliximab therapy in rheumatoid arthritis. Arthritis Rheum. 2003;48:1849–1852. doi: 10.1002/art.11168. [DOI] [PubMed] [Google Scholar]
- 37.Balog A, Klausz G, Gál J, Molnár T, Nagy F, Ocksovszky I, et al. Investigation of the prognostic value of TNF-alpha gene polymorphism among patients treated with infliximab, and the effects of infliximab therapy on TNF-alpha production and apoptosis. Pathobiology. 2004;71:274–280. doi: 10.1159/000080062. [DOI] [PubMed] [Google Scholar]
- 38.Criswell LA, Lum RF, Turner KN, Woehl B, Zhu Y, Wang J, et al. The influence of genetic variation in the HLA-DRB1 and LTA-TNF regions on the response to treatment of early rheumatoid arthritis with methotrexate or etanercept. Arthritis Rheum. 2004;50:2750–2756. doi: 10.1002/art.20469. [DOI] [PubMed] [Google Scholar]
- 39.Martinez A, Salido M, Bonilla G, Pascual-Salcedo D, Fernandez-Arguero M, de Miguel S, et al. Association of the major histocompatibility complex with response to infliximab therapy in rheumatoid arthritis patients. Arthritis Rheum. 2004;50:1077–1082. doi: 10.1002/art.20154. [DOI] [PubMed] [Google Scholar]
- 40.Pavy S, Toonen EJ, Miceili-Richard C, Barrera P, van Reil PL, Criswell LA, et al. Tumour necrosis factor alpha-308G->A polymorphism is not associated with response to TNF alpha blockers in Caucasian patients with rheumatoid arthritis: systematic review and meta-analysis. Ann Rheum Dis. 2010;69:1022–1028. doi: 10.1136/ard.2009.117622. [DOI] [PubMed] [Google Scholar]
- 41.Van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Rustenburg F, Baggen JM, Verweij CL, et al. Responsiveness to anti-tumour necrosis factor alpha therapy is related to pre-treatment tissue inflammation levels in rheumatoid arthritis patients. Ann Rheum Dis. 2008;67:563–566. doi: 10.1136/ard.2007.081950. [DOI] [PubMed] [Google Scholar]
- 42.Hueber W, Tomooka BH, Baltliwalla F, Li W, Monach PA, Tibshirani RJ, et al. Blood autoantibody and cytokine profiles predict response to anti-tumor necrosis factor therapy in rheumatoid arthritis. Arthritis Res Ther. 2009;11:R76. doi: 10.1186/ar2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badot V, Galant C, Nzeusseu Toukap A, Theate I, Maudoux AL, Van den Eynde BJ, et al. Gene expression profiling in the synovium identifies a predictive signature of absence of response to adalimumab therapy in rheumatoid arthritis. Arthritis Res Ther. 2009;11:R57. doi: 10.1186/ar2678. [DOI] [PMC free article] [PubMed] [Google Scholar]