Abstract
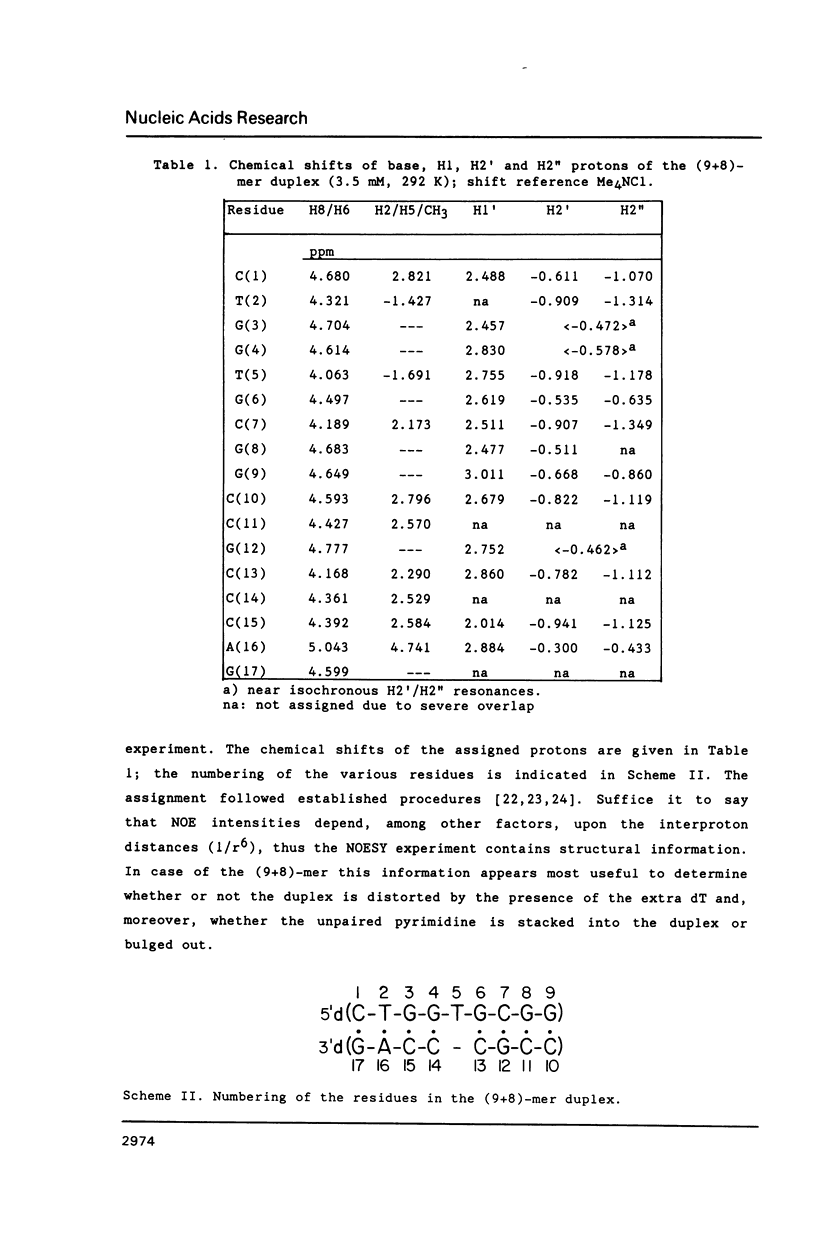
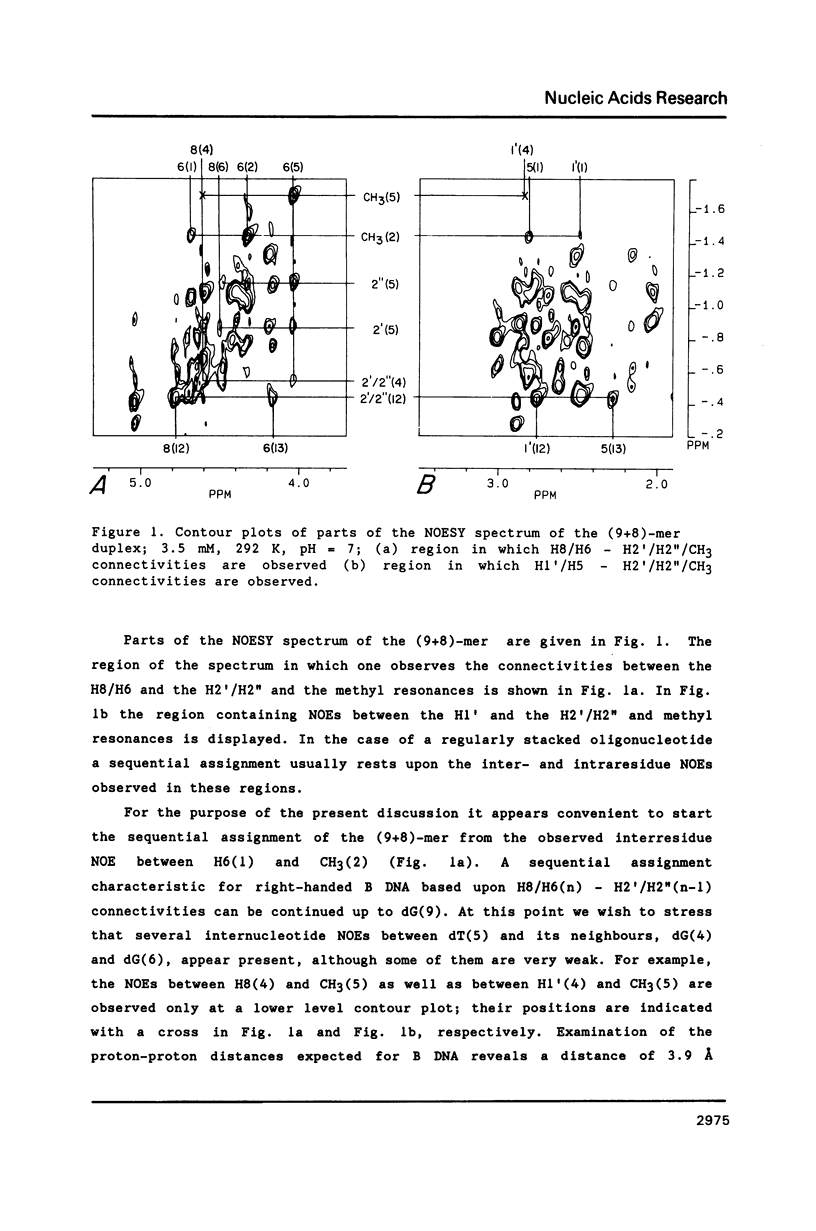
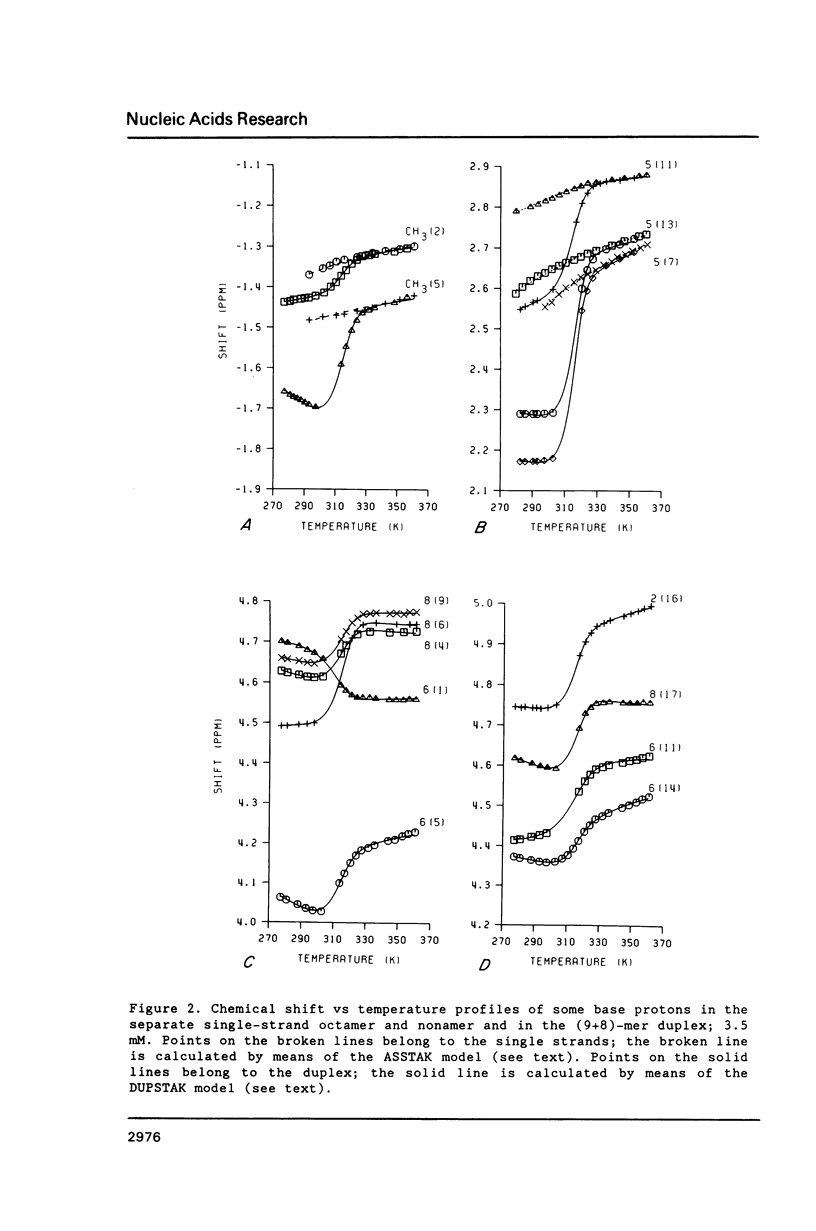
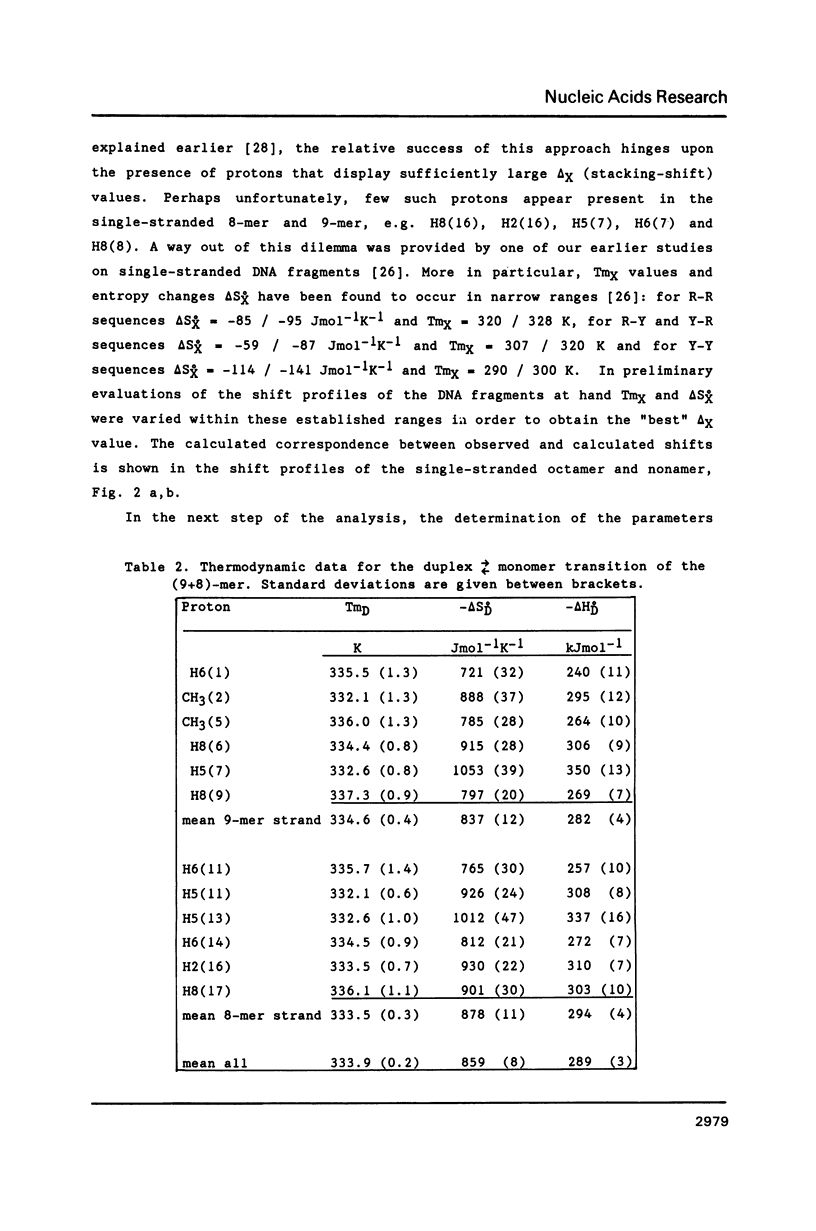
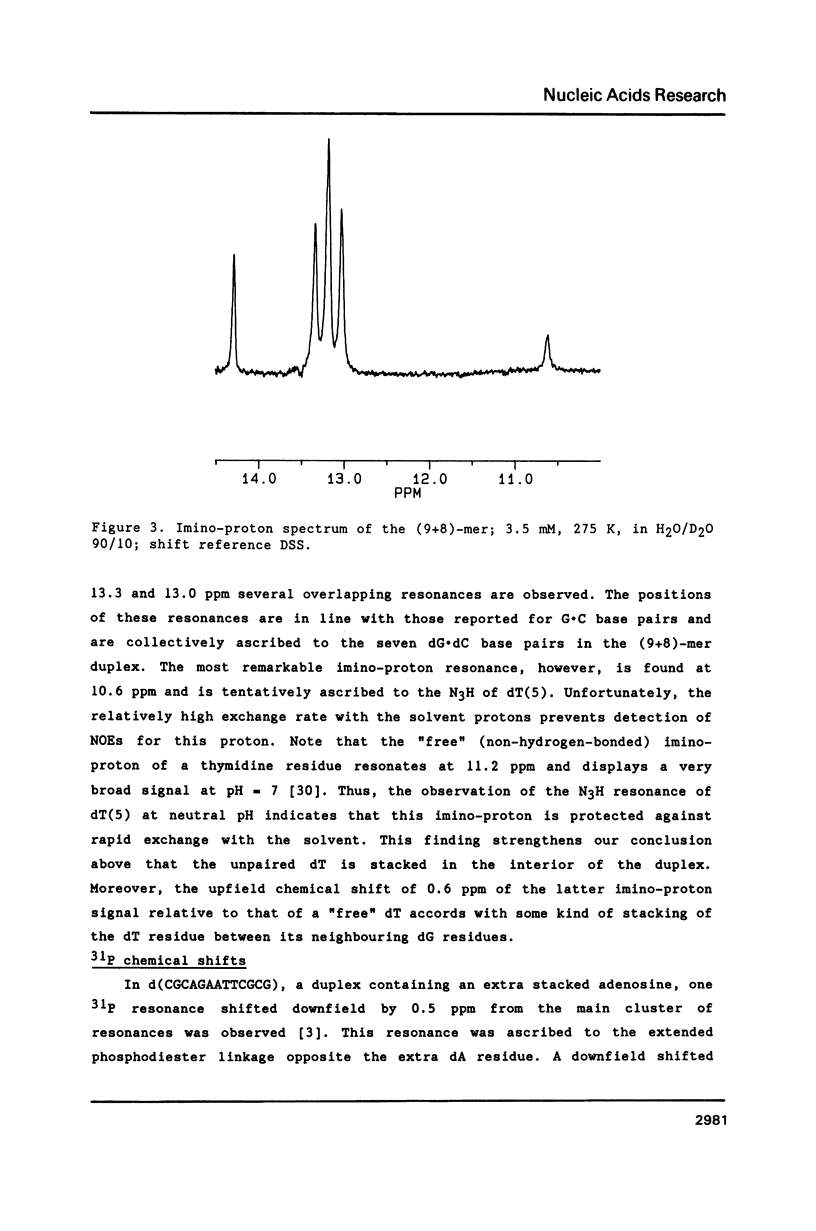
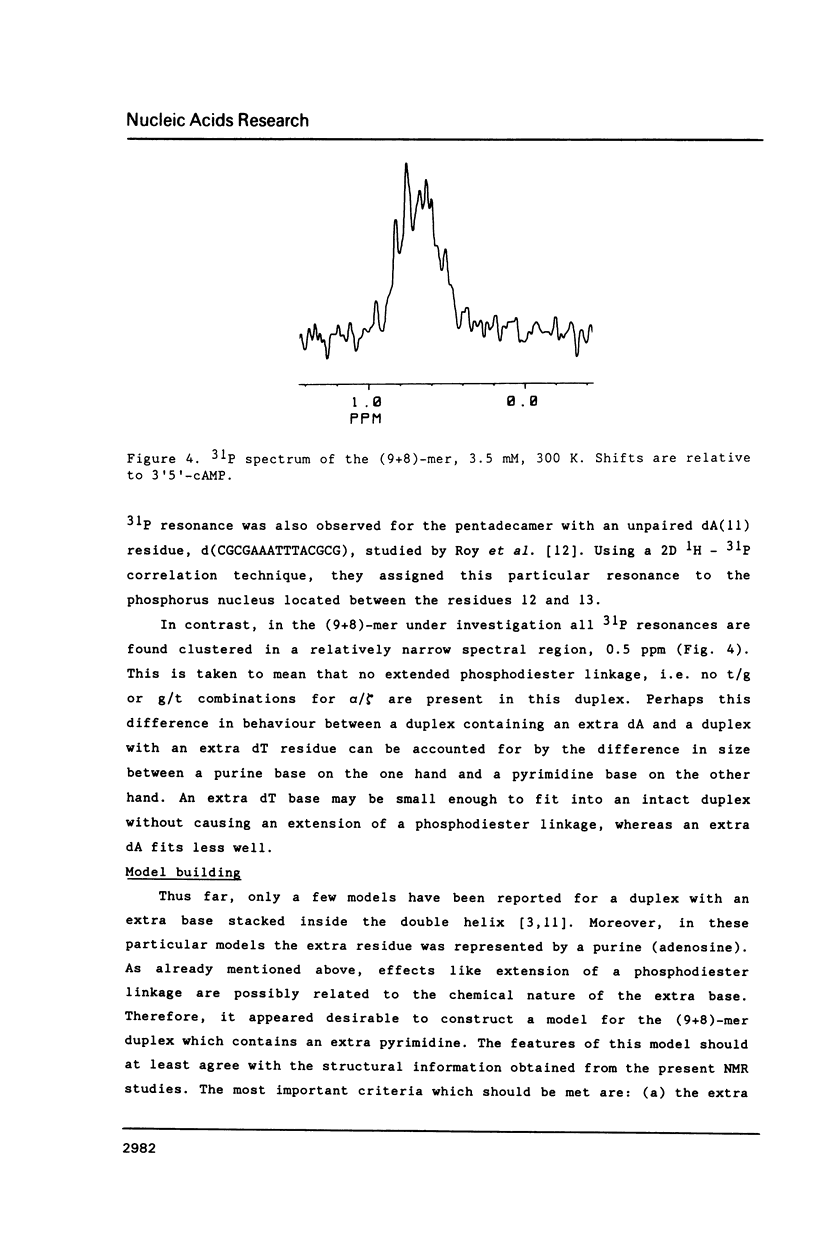
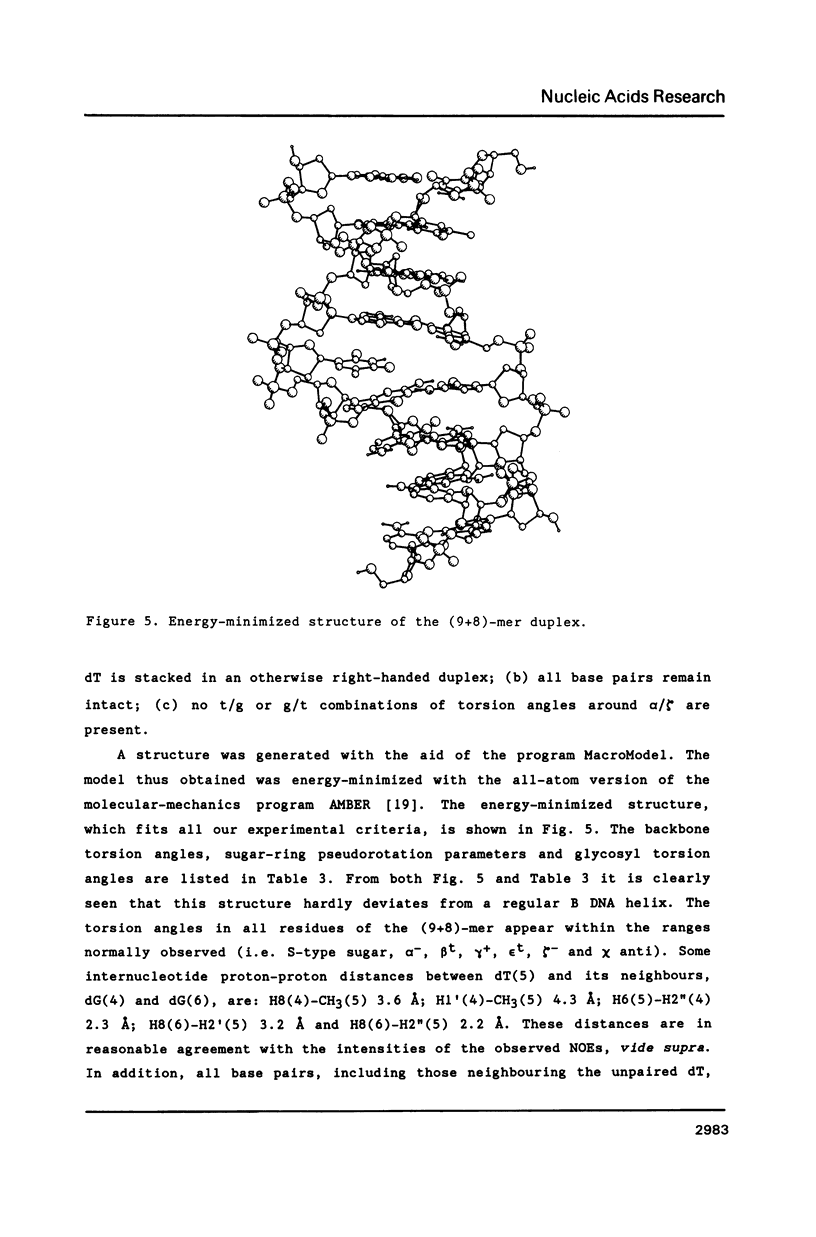
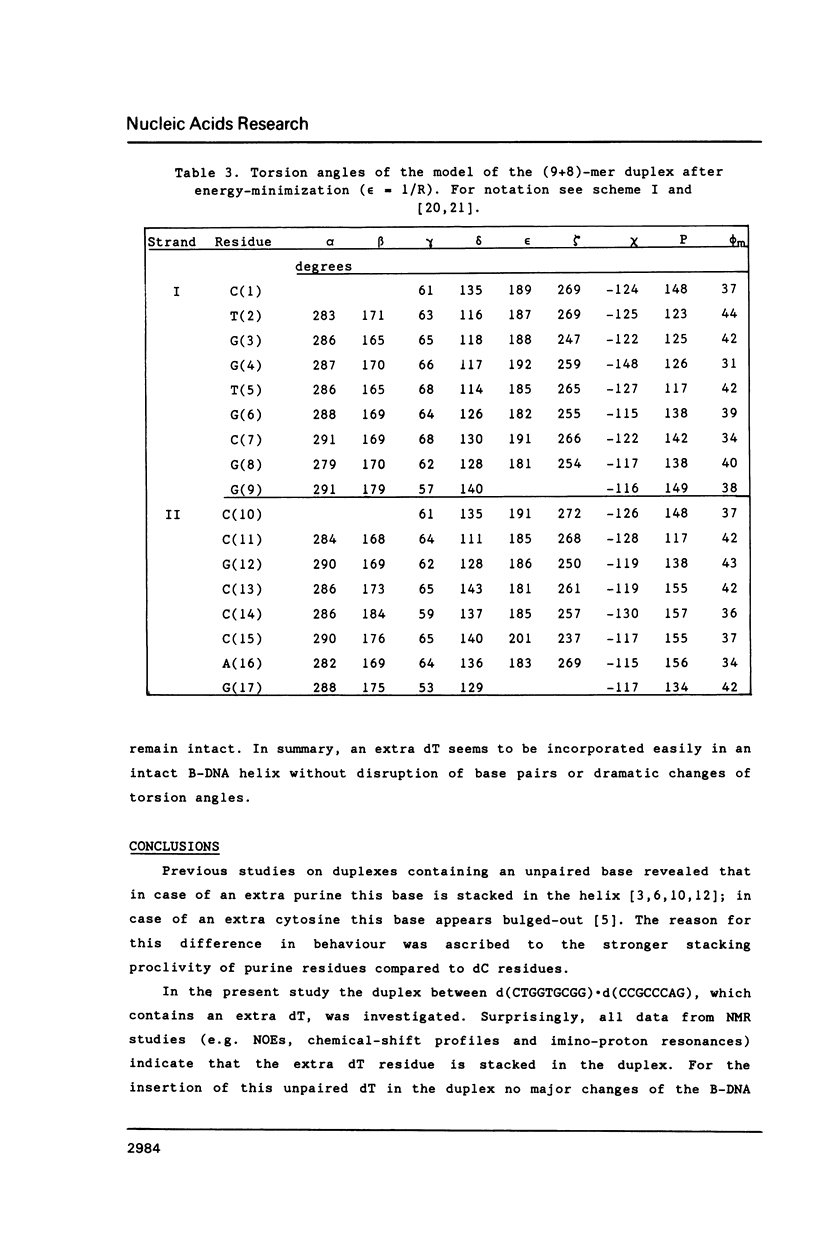
NMR and model-building studies were carried out on the duplex d(CTGGTGCGG).d(CCGCCCAG), referred to as (9+8)-mer, which contains an unpaired thymidine residue. Resonances of the base and of several sugar protons of the (9+8)-mer were assigned by means of a NOESY experiment. Interresidue NOEs between dG(4) and dT(5) as well as between dT(5) and dG(6) provided evidence that the extra dT is stacked into the duplex. Thermodynamic analysis of the chemical shift vs temperature profiles yielded an average TmD value of 334 K and delta HD of -289 kJmol-1 for the duplex in equilibrium random-coil transition. The shapes of the shift profiles as well as the thermodynamic parameters obtained for the extra dT residue and its neighbours again indicate that the unpaired dT base is incorporated inside an otherwise intact duplex. This conclusion is further supported by (a) the observation of an imino-proton resonance of the unpaired dT; (b) the relatively small dispersion in 31P chemical shifts (approximately 0.5 ppm) for the (9+8)-mer, which indicates the absence of t/g or g/t combinations for the phosphate diester torsion angles alpha/zeta. An energy-minimized model of the (9+8)-mer, which fits the present collection of experimental data, is presented.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Evans D. H., Morgan A. R. Characterization of imperfect DNA duplexes containing unpaired bases and non-Watson-Crick base pairs. Nucleic Acids Res. 1986 May 27;14(10):4267–4280. doi: 10.1093/nar/14.10.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. H., Morgan A. R. Extrahelical bases in duplex DNA. J Mol Biol. 1982 Sep;160(1):117–122. doi: 10.1016/0022-2836(82)90134-6. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Westerink H. P., van der Marel G. A., van Boom J. H. Conformational analysis of a hybrid DNA-RNA double helical oligonucleotide in aqueous solution: d(CG)r(CG)d(CG) studied by 1D- and 2D-1H NMR spectroscopy. J Biomol Struct Dyn. 1983 Oct;1(1):131–149. doi: 10.1080/07391102.1983.10507430. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Wobble dG X dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- Hartel A. J., Lankhorst P. P., Altona C. Thermodynamics of stacking and of self-association of the dinucleoside monophosphate m2(6)A-U from proton NMR chemical shifts: differential concentration temperature profile method. Eur J Biochem. 1982 Dec 15;129(2):343–357. doi: 10.1111/j.1432-1033.1982.tb07057.x. [DOI] [PubMed] [Google Scholar]
- Miller M., Kirchhoff W., Schwarz F., Appella E., Chiu Y. Y., Cohen J. S., Sussman J. L. Conformational transitions of synthetic DNA sequences with inserted bases, related to the dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1987 May 11;15(9):3877–3890. doi: 10.1093/nar/15.9.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. The B and Z forms of the d(m5C-G)3 and d(br5C-G)3 hexamers in solution. A 300-MHz and 500-MHz two-dimensional NMR study. Eur J Biochem. 1986 Oct 1;160(1):131–139. doi: 10.1111/j.1432-1033.1986.tb09949.x. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Quignard E., Fazakerley G. V., Teoule R., Guy A., Guschlbauer W. Consequences of methylation on the amino group of adenine. A proton two-dimensional NMR study of d(GGATATCC) and d(GGm6ATATCC). Eur J Biochem. 1985 Oct 1;152(1):99–105. doi: 10.1111/j.1432-1033.1985.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of the base sequence on the conformational behaviour of DNA polynucleotides in solution. Eur J Biochem. 1987 Jul 1;166(1):87–101. doi: 10.1111/j.1432-1033.1987.tb13487.x. [DOI] [PubMed] [Google Scholar]
- Roy S., Sklenar V., Appella E., Cohen J. S. Conformational perturbation due to an extra adenosine in a self-complementary oligodeoxynucleotide duplex. Biopolymers. 1987 Dec;26(12):2041–2052. doi: 10.1002/bip.360261206. [DOI] [PubMed] [Google Scholar]
- Roy S., Weinstein S., Borah B., Nickol J., Appella E., Sussman J. L., Miller M., Shindo H., Cohen J. S. Mechanism of oligonucleotide loop formation in solution. Biochemistry. 1986 Nov 18;25(23):7417–7423. doi: 10.1021/bi00371a025. [DOI] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Crothers D. M. Proton nuclear magnetic resonance studies on bulge-containing DNA oligonucleotides from a mutational hot-spot sequence. Biochemistry. 1987 Feb 10;26(3):904–912. doi: 10.1021/bi00377a035. [DOI] [PubMed] [Google Scholar]



