Abstract
Crop production on soils containing sub-optimal levels of nitrogen (N) severely compromises yield potential. The development of plant varieties displaying high N use efficiency (NUE) will optimize N fertilizer use and reduce the environmental damage caused by excess N application. Maize is one of the most important crops cultivated worldwide. Identification of the genotypes with an enhanced NUE in the field is both time and resource consuming and sometime is difficult due to the regulation in the biotechnology programs. Identification of traits associated with adaptation to N limitation at an early vegetative stage which could reflect NUE at maturity is in need. We developed a hydroponic growth system and used it to test two genotypes that were different in their NUE at maturity under N limitation. One genotype SRG-200 showed a higher NUE than the other genotype SRG-100 and we used its hybrid SRG-150 as a reference for NUE. A number of phenotypic, molecular, and metabolic factors were tested using these three genetic lines at an early vegetative stage to determine which of these could be more indicative of predicting improved NUE at an early seedling stage. These include a transcriptional analysis which showed that the higher NUE in SRG-200 genotype is associated with higher transcript levels for the genes involved in nitrate transport, N assimilation, and GS and that the SRG-200 genotype maintained higher sugar content in leaves. Those identified in this study could be useful indicators for selecting promising maize lines at early stages to help develop elite varieties showing an enhanced NUE.
Keywords: Zea mays, nitrogen limitation, NUE, seedlings
Introduction
Nitrogen (N) is an essential element required for many physiological processes during the plant life cycle. Despite the benefits gained by using N fertilizers, a large amount of N fertilizer used for crop production enters aquifers which lead to irreversible damage to our ecosystem (Goyal et al., 2005). Further, in many developing countries, farmers do not have access to sufficient N fertilizer due to the high cost. Improving plant nitrogen use efficiency (NUE) and hence increasing their adaptation under lower nitrogen levels will improve the agronomic value of fertilizers, save energy required for fertilizer production, and minimize damage to the environment.
It is known that plants growing under N limitation conditions undergo adaptive morphological changes including increased root shoot ratio, early senescence, and early transition to flowering (Goyal et al., 2005). Also, plants exposed to N limitation exhibit lower levels of N-containing compounds such as amino acids, proteins, chlorophyll, secondary metabolites, and accumulate anthocyanins (Peng et al., 2007a,b) and starch (Scheible et al., 2004) compared to plants grown under optimal N environment. In addition, there is a correlation between nitrogen and carbon metabolism, where nitrogen limiting conditions decrease root growth, photosynthesis, and increase the C to N ratio (Malamy and Ryan, 2001; Martin et al., 2002 and Wingler et al., 2006). The root system plays an important role in nutrient uptake from the soil, especially at the early seedling stage. A correlation between the Quantitative trait locus (QTLs) for nitrogen uptake and the QTLs for root architecture traits has been proposed (Coque et al., 2008). However, the involvement of the root traits in NUE is not fully understood, due to the difficulty of studying the root system in the field (Garnett et al., 2009). Despite the importance of the roots in providing nutrients for the plants and its effect on plant adaptation under N limitation, studies in this area are limited. At the early stages, plants are dependent on their own roots to uptake the nutrient from the soil, and thus affecting the root system at this stage affects the rest of plant development. Similarly exposing young seedlings to nutrient stress affects later growth. It has been suggested that the vigor of wheat seedling having stronger root growth help the plants to uptake the nitrogen quickly from the soil (Liao et al., 2004). Nitrogen uptake and N remobilization from the leaves to the sink sources such as the seeds and the fruits has been the subject of several studies (Hortensteiner and Feller, 2002; Gregersen et al., 2008; Masclaux-Daubresse et al., 2008; Fan et al., 2009 and Taylor et al., 2010). However, nitrogen translocation from the root to the shoots still requires elucidation especially at the early seedling stage where the N is a limiting factor to produce healthy and productive plants.
Maize (Zea mays) is one of the most important crops cultivated worldwide. Several indicators have been reported for NUE in maize such as chlorophyll content, plant growth, and height, number of the senesced leaves below the ear and kernel number. In addition, other biochemical traits including the activities of some enzymes involved in nitrogen metabolism have been reported to be important for enhanced NUE (Gallais and Hirel, 2004; Medici et al., 2005). The genetic manipulation of key genes involved in nitrogen assimilation such as glutamine synthetase modifies the maize response to nitrogen (Martin et al., 2006). The relationship between nitrate uptake, flux and reduction, and accumulation of reduced nitrogen was studied in two maize hybrids (Reed and Hageman, 1980). Also, the genetic variation for traits relating to nitrogen content of maize stalks were investigated (Rizzi et al., 1991). Additionally, higher photosynthesis has been reported to help in the response of maize plants to N limitation (Khamis et al., 1990; Lu et al., 2001; Vos et al., 2005 and Ding et al., 2005). Using the QTL approaches reported the importance of some traits at maturity stage (Hirel et al., 2001 and Bernard and Habash, 2009). Gallais and Hirel (2004) reported the importance of the GS locus as a good candidate gene which can explain the variation in NUE. Further, manipulation of amino acids pathway could help in increasing plant NUE, for example increases in the cytosolic located GS activity and asparagine synthetase could improve NUE (Lea and Azevedo, 2007). However, it is very time and resource consuming to identify corn genotypes with a high NUE at maturity. Here we are interested in identifying factors, especially molecular and metabolic factors at early seedling stages, which could be more indicative of predicting improved NUE at maturity. In the present work, we report on the physiological, molecular, and metabolic responses of two inbred lines which show different NUE capacities. We used the hybrid SRG-150 as a reference for NUE. We studied the involvement of the root system, nitrogen uptake, translocation, and assimilation in young maize seedlings of these three genotypes. In addition, an analysis of key transcript and metabolite levels was performed to elucidate the different responses of these three maize genotypes to N-limiting conditions at an early seedling stage.
Materials and Methods
Plant materials and growth condition
Seeds of the two elite maize inbred lines SRG-100 and SRG-200 and its hybrid SRG-150 were obtained from Syngenta Biotechnology Inc. NC, USA. They were germinated in turface for 2 days, the seedlings were transplanted to the hydroponic system in nutrient solution containing 4 mM MgSO4, 5 mM KCl, 5 mM CaCl2, 1 mM KH2PO4, 0.1 mM Fe–EDTA, 0.5 mM MES (pH 6.0), 9 μM MnSO4, 0.7 μM ZnSO4, 0.3 μM CuSO4, 46 μM NaB4O7, and 0.2 μM (NH4)6Mo7O2. Seedlings were transferred to a 35-L container contains 25 L of the nutrient solution; the volume and the pH were adjusted weekly by adding a fresh nutrient solution and using phosphoric acid to adjust the pH to 5.5. Different nitrate concentrations as KNO3 were used. Eight seedlings from each genotype were transferred to the same container and grown under the same condition. Plants were grown in growth cabinet (Conviron, MB, Canada) under long day condition, 16 h light (∼400 μmol m−2s−1) at 28°C and 8 h dark at 23°C. Plants were harvested 4 weeks later, including leaves (third to fifth), first stem internodes, and the whole roots. Each biological replicate represent tissues from four plants. Plant harvest was carried out at noon for each replicate. The materials were frozen in liquid nitrogen and stored at −80°C for further analysis. For the greenhouse experiment, plants were grown in a semi hydroponics system, seeds were germinated in a small pots contains L4 Sunshine soil mix supplemented with full nutrients. One week after germination seedlings were transplanted into 20 L pots contain turface soil, a 100% backed calcined clay growth media with grain size between 2.5 and 3.5 mm (Turface MVP_; Profile Products LLC, Buffalo Grove, IL, USA). The plants were subjected to drip fertigation until harvest according to the method previously reported by (Tollenaar and Migus, 1984; Ying et al., 2000).
NUE calculation
Nitrogen use efficiency was calculated using the following equation as described in Steenbjerg and Jakobsen (1963) and reviewed in Good et al. (2004).
NUE = Sw/N
Sw = Shoot dry weight
N = Shoot nitrogen content
Root analysis
Roots of the three corn genotypes were collected from the plants growing in hydroponic system under different nitrogen levels. Roots were scanned and analyzed using the WinRHIZO software (v. 5.0, Regent Instruments, Inc., QC, Canada).
Chlorophyll and anthocyanin analysis
Chlorophyll analyses were carried out using frozen ground tissues of the expanded leaves. The chlorophyll concentration in the supernatant was determined spectrophotometrically and calculated using the MacKinney equations (Sestak et al., 1971). Total anthocyanin content was determined according to the method described in Kant et al. (2006). Anthocyanins were determined in the methanolic extract by measuring the absorbance at 530 and 657 nm.
Enzymes activity
Nitrate reductase activity and glutamine synthase activity were performed according to the protocols provided by Márquez et al. (2005).
Nitrate determination
Nitrate content was determined calorimetrically according to the methods reported by Cataldo et al. (1975). Briefly 100 mg of the frozen tissue were homogenized with 600 ul of the phosphate buffer. The homogenates were centrifuged at 13,000 rpm for 15 min. Aliquots (0.2 mL supernatant) were pipetted into 50 mL tubes, and mixed thoroughly with 0.8 mL of 5% (w/v) salicylic acid in concentrated sulfuric acid (H2SO4) for 20 min at room temperature (RT). Nineteen milliliter of 2 N NaOH was slowly added to raise the pH above 12. Samples were cooled to RT and the absorbance at 410 nm was determined.
Total amino acid, sugar, and starch analysis
Total amino acids were extracted successively with 80, 50, and 0% ethanol in 10 mM HEPES–KOH (pH 7.4), supernatants were pooled and total amino acids were assayed according to Rosen (1957). Soluble sugars were extracted with 80% ethanol, supernatants were pooled, and total soluble sugars were assayed according to Dubois et al. (1956). For starch quantification, 250 mg fresh weight of maize seedling tissues were ground to a fine powder in liquid nitrogen and extracted with 1 mL 100% methanol by shaking at 70°C for 15 min. The extraction was repeated two times and the insoluble residue was lyophilized overnight, weighed, and the total starch content was determined using the Megazyme kit according to the manufacturer’s instructions.
Real-time PCR analysis
Quantitative real-time PCR was carried out using primers of some genes involved in N metabolism; those genes were chosen based on the sequence availability and its homology to the Arabidopsis sequence. Briefly, total RNA was isolated from plant tissues using TRI-Reagent (Sigma-Aldrich, MO, USA). To eliminate any residual genomic DNA, total RNA was treated with RQ1 RNase-free DNase (Promega, WI, USA). The first strand cDNA was synthesized from total RNA by using the Reverse Transcription System kit (Quanta, MD, USA). Primer Express 2.0 software (Applied Biosystems, CA, USA) was used to design the primers for the target genes (Table A1 in Appendix). Relative quantification (RQ) values for each target gene relative to the internal control tubulin were calculated by the 2CT method (Livak and Schmittgen, 2001).
Metabolite isolation and detection
Maize seedling tissues were extracted and analyzed using the protocol reported in Fiehn et al. (2000). Briefly, 250 mg FW of tissue was extracted with 1 mL 100% methanol with shaking at 70°C for 15 min. Ribitol was added as an internal standard to the samples during extraction, and extracts were separated into polar (methanol/water) and apolar/lipid (chloroform) phases. Polar fractions were vacuum dried and derivatized using methoxyamine and N-Methyl-N-(trimethylsilyl)trifluoroacetamide. The derivatized samples were diluted five-fold with hexane, and then 1 μL was injected into the splitless injection port of a Varian 1200 gas chromatography–mass spectrometry (GC/MS) system (Varian Inc., CA, USA). Chromatography was performed using a 30-m × 0.25-mm Rtx-5MS column (Chromatographic Specialties, ON, Canada). For each run, the gas chromatograph oven was set at 70°C initial temperature, which was held for 5 min, and increased at 5°C min–1 to 310°C, at which it was held for 6 min. The injection temperature was 230°C, and the ion source was kept at 200°C. Mass spectra were recorded at three scans per second with an m/z scanning range of 50–650. The data analysis was performed using the automated mass spectral deconvolution and identification system (AMDIS) software (http://chemdata.nist.gov/mass-spc/amdis).
Statistical analysis
All experiments were conducted under controlled environmental conditions and designed as a completely randomized block. Statistic analyses were performed using SigmaStat (SPSS Inc., Chicago, IL, USA) with an error rate set at α = 0.05. The significance difference between treatments was tested using Tukey’s Honestly Significant Difference Test.
Results
Phenotypic analysis of the three maize genotypes in response to N limitation
Different degree of NUE of the three genotypes
Initially, NUE was tested for the two genotypes, SRG-100 and SRG-200 and used its hybrid SRG-150 as a reference for high NUE at maturity. Plants were grown in a semi hydroponic system described in Echarte et al. (2008) under sufficient and limiting N conditions. The three genotypes attained different NUE under limiting N condition using the equation provided in the Section “Materials and Methods” (Figure 1A). The parent line SRG-100 had a lower NUE compared to the other parent line SRG-200 which was similar to the hybrid SRG-150. A hydroponic system was developed to study their response to N limitation at an early seedlings stage. In this system, maize seedlings were grown hydroponically in a nutrient solution supplemented with different concentrations of potassium nitrate as described in the Section “Materials and Methods.” To determine the limiting N levels, different N concentrations up to 10 mM were tested, and a negative effect on plant growth was observed with the highest N concentration. 3 mM was determined to be the optimal growth condition in this system and 1 mM found to be a limiting N condition with a very significant reduction of biomass at this concentration (details in the next section). It was confirmed that the difference in NUE of the three genotypes under N limitation at this early vegetative stage (Figure 1B) was consistent with the mature NUE index (Figure 1A). Further, for greenhouse plants grown under N limitation the seed yield of the SRG-100 is lower than SRG-200 which is lower than the hybrid SRG1-50 (Figure A1 in Appendix). It should be noted that in the greenhouse, these plants are hand pollinated. Nevertheless the seed yield is as expected when compared to biomass formation.
Figure 1.
Nitrogen Use efficiency (NUE) in the hybrid SRG-150 and its parent lines SRG-100 and SRG-200 under nitrogen limitation condition. (A) at maturity stage in greenhouse experiment; (B) at early seedlings stage (4 weeks old) in hydroponic system (calculated in the plants growing under N limitation). Data are means ± SD (n = 3 each replicate represent four plants). Bars with different letters indicate a significant difference at P ≤ 0.05 (Tukey’s Honestly Significant Difference Test).
The proportional biomass reduction in the maize plants in relation to NUE
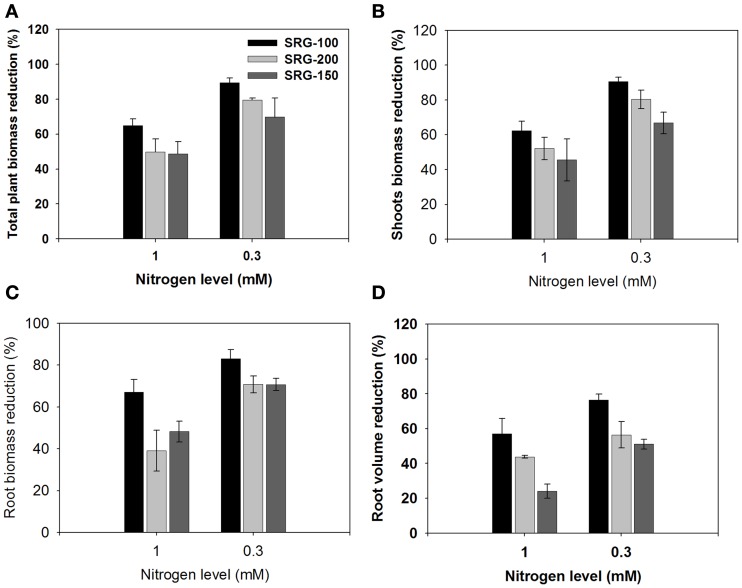
To study the response of the studied genotypes to limiting nitrogen condition, biomass reduction was determined under the different limiting N levels compared to those growing in 3 mM KNO3. Decreasing the nitrogen concentration to 1 mM KNO3 led to a much higher reduction in overall plant biomass in SRG-100 than that in SRG-200 genotype. This reduction was even more severe by reducing the nitrogen level to 0.3 mM nitrate, which resulted in a reduction in overall biomass of 89, 79, and 69% in SRG-100, SRG-200, and SRG-150 genotypes, relative to plants grown at 3 mM (Figure 2A). The shoot to root allocation in the different lines exposed to different nitrate conditions was analyzed to determine the major source for the observed reductions in overall plant biomass. The shoot biomass had the same trend as the total plant biomass in the three genotypes. Maize seedling growth at 1 mM nitrate resulted in a decrease in shoot biomass of 62, 52, and 45%, while growth at 0.3 mM led to 91, 80, and 67% shoot biomass reduction for the SRG-100, SRG-200, and SRG-150 genotypes, respectively (Figure 2B). Interestingly, root biomass was significantly reduced when plants were grown under limiting nitrogen condition (1 mM), and this reduction was significantly higher in the SRG-100 compared to the other genotype SRG-200; 67, 39, and 48% for SRG-100, SRG- 200, and SRG-150, respectively (Figure 2C). This reduction was high as 83% for SRG-100 and 71% for both SRG-200 and SRG-150, when plants were subjected to 0.3 mM nitrate.
Figure 2.
Reduction of shoot and root growth of the maize hybrid; SRG-150 (Dark gray Bars) and its parent line SRG-100 (black bars) and SRG-200 Parent line (light gray bars) relative to the plants of the same genotype growing under optimal N condition. (A) Reduction in the total plant dry biomass. (B,C) shoot and root biomass reduction (D), root volume reduction. Data are means ± SD (n = 3).
The importance of roots in NUE
Roots play an important role in providing the plants with the nutrient from the soil and have an impact in plant NUE (Garnett et al., 2009). The present hydroponic system allowed us to obtain a clean complete root system for more detailed analysis. The root system of the different genetic lines subjected to nitrogen limitation was analyzed using the WinRHIZO system to quantify differences in root volume and root morphology between these lines. Consistent with the reduction in the biomass, the root volume decreased significantly under nitrogen limitation condition compared to 3 mM nitrate, and this decrease was more significant in the SRG-100 compared to the SRG-200 genotype (Figures 2D and 3A,B). Nitrogen limitation caused a reduction in the root volume by 56 and 43% at 1 mM and 76 and 56% at 0.3 mM in SRG-100 and SRG-200 which was close to the values obtained in from the hybrid SRG-150 (24 and 51%; Figure 2D). These results showed that nitrogen limitation caused a reduction in the root growth system in all genotypes under study. However, this reduction was more significant in the less adapted SRG-100 genotype compared to the more adapted genotype, SRG-200 and as expected the hybrid showed less reduction in the root system under N limitation. Further analysis of the different parts of the root system showed that this reduction at low nitrogen concentration occurred at all levels of the root system and it was not due to the inhibition of a particular part of the root system (Figure 3C). Therefore, the SRG-100 genotype was more affected by limiting nitrogen condition compared to the other genotype SRG-200.
Figure 3.
N limitation effect on root system of the maize hybrid SRG-150 and its parents lines SRG-100 and SRG-200. Images were taken by the WinRHIZO system, (A) plants grown at 3 mM; (B) plants grown at 1 mM. (C) Roots length of the different parts of the root system under N limitation, classified based on the diameter (A = 0–0.3; B = 0.3–0.5; C = 0.5–1; D = 1–2; E = 2–5 mm). Data are means ± SD (n = 3). Bars with different letters indicate a significant difference at P ≤ 0.05 (Tukey’s Honestly Significant Difference Test).
Biochemical modification associated with the NUE in the three maize genotypes
Anthocyanin production under N limitation is a better indicator for NUE than chlorophyll reduction
Plants grown at 3 mM nitrate were healthy and retained higher chlorophyll levels compared to plants grown below this concentration (Figure 4A). Below 3 mM nitrate, all genotypes displayed symptoms of N limitation marked by pale leaves that contained up to 50–80% less chlorophyll compared to those plants grown at 3 mM nitrate (Figure 4A). No significant difference was observed in chlorophyll level of the three genotypes under nitrogen limitation (Figure 4A). However, a higher reduction in chlorophyll was observed in the less adapted genotype SRG-100 under 1 mM condition and this reduction was similar in SRG-100 and SRG-200 under sever N limitation (0.3 mM; Figure A2 in Appendix).
Figure 4.
N limitation effects on chlorophyll and anthocyanins content in the three genotypes; SRG-100 (black bars); SRG-200 (light gray bars); and SRG-150 (Dark gray Bars). (A) Total chlorophyll content (Chlorophyll a and b). Data are means ± SD (n = 3 each replicate represent four plants). Bars with different letters indicate a significant difference at P, 0.05 (Tukey’s Honestly Significant Difference Test). (B,C) Induction percentage of the leaves and stem anthocyanins in the plants growing under N limitation relative to the plants of the same genotype growing optimal nitrogen level (3 mM). Data are means ± SD (n = 3).
Anthocyanin has been shown to be induced and involved in the adaptation to the limiting nitrogen condition (Olsen et al., 2008; Peng et al., 2008). Leaf and stem anthocyanin content was determined for all genotypes grown under optimal and limiting N levels. An increase in leaf and stem anthocyanin content was observed in response to the reduction in nitrogen level from 3 to 1 mM. This induction was more pronounced in the SRG-200 and similarly in the SRG-150 genotype, compared to the SRG-100 genotype (Figures 4B,C). The SRG-100 genotype starts to accumulate anthocyanins, when the N level was further reduced to 0.3 mM. However, this level was still significantly less than that observed in the other parent line and the hybrid.
Nitrate content, enzyme activity, and NUE
Upon nitrate uptake from the soil, plants transport the nitrate into their vascular system to the leaves. Plants assimilate leaf and root nitrate into amino acids through the actions of nitrate reductase and nitrite reductase. This process is dependent on the availability of nitrate in plant tissues. Deficiency in N uptake from the soil and its translocation through the plant might affect this process. To examine nitrate uptake and translocation within the plant, the nitrate content was measured in the different organs. Under limiting N condition, the leaf nitrate content was significantly lower in the SRG-100 genotype compared to the other genotype SRG-200 (Figure 5A), the same trend was observed in the stem but under both limiting and sufficient N conditions (Figure 5B). However, all genotypes had similar root nitrate content (Figure 5C).
Figure 5.
Nitrogen limitation effects on nitrate content and C/N ratio in the three genotypes; SRG-100 (black bars); SRG-200 (light gray bars); and SRG-150 (Dark gray Bars). (A–C) Leaves, stem, and root nitrate content in the plants growing under optimal (3 mM) and limiting (1 mM) N conditions. (D) C/N ratio calculated in the leaves of the three genotypes growing under same condition as in (A). Data are means ± SD (n = 3 each replicate represent four plants). Bars with different letters indicate a significant difference at P ≤ 0.05 (Tukey’s Honestly Significant Difference Test).
Nitrate reduction into nitrite by nitrate reductase is considered the major metabolic control point, limiting the rate of primary N assimilation in roots and leaves. However, under the condition of this study, there was no significant correlation between NR activity and NUE, apart from the roots nitrate reductase activity being higher in the hybrid compared to the other genotypes (data not shown).
Glutamine synthetase controls the conversion of ammonia into glutamine which is eventually used for the synthesis of other amino acids. Assessment of the glutamine synthetase activity of all studied genotypes does not reveal any significant difference when grown under optimal or N-limiting environments (data not shown). This might be due to the multiple glutamine synthesis genes with their mixed patterns of expression resulting in no change in total activity. Similar results have been observed previously (Kant et al., 2008).
Sugar and starch levels as indicators of NUE
Carbohydrates are involved in nitrogen metabolism through providing the energy and the C skeletons required for amino acid synthesis. Generally, the C/N ratio increased upon nitrogen limitation in all studied genotypes. The genotype SRG-100 showed a significantly lower value for the C/N ratio compared to the other parent, SRG-200 and no significant difference was observed between the hybrid and SRG-200 (Figure 5D).
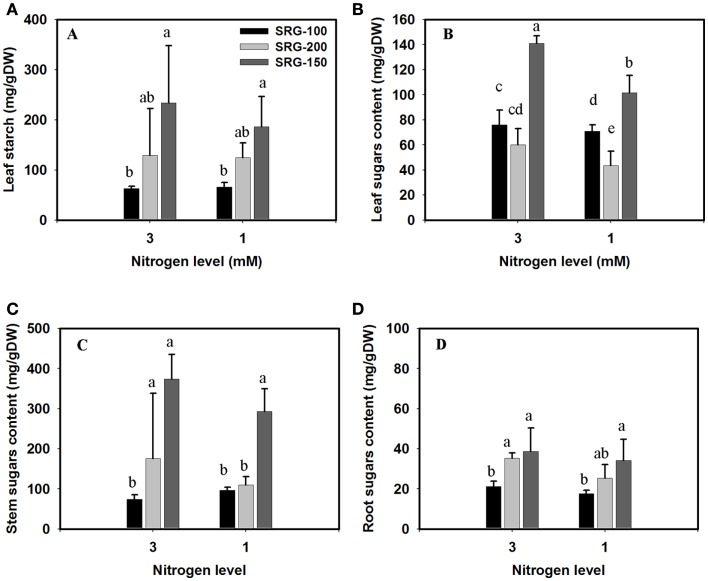
Under our conditions, the overall starch content did not vary significantly with the reduction of the nitrogen in the plant medium from 3 to 1 mM. Consistent with the above results, the less adapted genotypes SRG-100 was characterized by lower leaf starch content and this trend was observed when plants were grown under the optimal and limiting N levels (Figure 6A). However, no significant difference was observed in the starch level in the stem or the roots among the three genotypes. A significant reduction in the sugar content was only observed in the leaves, but not in the stem of the plants growing under N limitation, no significant difference was found between the SRG-100 and SRG-200 (Figures 6B,C). However, the hybrid SRG-150 showed a significantly higher leaf, and stem sugar content compared to its parent lines (Figures 6B,C). In the roots, the sugar content was higher in the SRG-200 and the hybrid SRG-150 compared to the inbred line SRG-100 under both sufficient and limiting N condition (Figure 6D).
Figure 6.
Starch and total sugar content in the three genotypes; SRG-100 (black bars); SRG-200 (light gray bars); and SRG-150 (Dark gray Bars) under optimal (3 mM) and limiting N (1 mM) condition. Starch content in the leaves (A); sugar content in the leaves, stems, and roots (B–D). Data are means ± SD (n = 3 each replicate represent four plants). Bars with different letters indicate a significant difference at P ≤ 0.05 (Tukey’s Honestly Significant Difference Test).
Metabolic attributes to NUE
Metabolic profiling analysis was performed on polar extracts prepared from leaf, stem, and root tissues harvested from maize grown under N-limiting conditions using GC/MS. Of the 100 components detected using this approach, 51 were statistically significantly different in their abundance in the three genotypes analyzed (P < 0.05). We were able to chemically identify a total of 19 of these components using a library of authentic chemical standards analyzed using the same GC/MS instrument. Table 1 shows the fold-ratios for these metabolites present in leaf, stem, and root tissues for the SRG-200 line compared to the SRG-100 line grown under 1 mM nitrate and with the hybrid SRG-150. Similar to SRG-150, the SRG-200 line maintained lower levels of amino acids and organic acids in the leaves and the stem compared to the SRG-100 line. This might be due to the rapid use of those components in those two lines, resulted in a better growth. Interestingly, only the SRG-150 maintained higher levels of alanine, glutamate, and threonine in the roots compared to the SRG-100 line (Table 1). Further, the hybrid SRG-150 had higher carbohydrate levels present in the leaves, stems, and roots compared to the SRG-100 line. Only the SRG-150 maintained higher levels of fructose, glucose, and a C6-sugar in all tissues collected (Table 1).
Table 1.
Relative quantification of the different metabolites detected in the three maize Genotypes growing under nitrogen limitation.
| Leaf | Stem | Root | ||||
|---|---|---|---|---|---|---|
| Metabolite | 150/100 | 200/100 | 150/100 | 200/100 | 150/100 | 200/100 |
| P-COMPOUNDS | ||||||
| Phosphate | 1.6 | 1.5 | 1.1 | −1.4 | 2.1 | 1.1 |
| Glucose-6-phosphate | −2.0 | −1.7 | −1.9 | −3.9 | 2.1 | 1.8 |
| N-COMPOUNDS | ||||||
| Alanine | 1.1 | −3.8 | −4.7 | −2.7 | 1.5 | 1.1 |
| Aspartate | −1.9 | −1.6 | −3.2 | −6.2 | 1.4 | 1.1 |
| Glutamate | 1.2 | −1.4 | −2.2 | −2.2 | 1.9 | −1.1 |
| Glycine | −2.2 | −1.7 | −23.7 | −28.8 | −1.4 | −1.7 |
| Isoleucine | −3.9 | −1.4 | −4.8 | −4.7 | 1.4 | 2.1 |
| Serine | −1.7 | −3.1 | −34.8 | −26.1 | 1.1 | −1.4 |
| Threonine | −1.6 | −2.4 | −10.1 | −7.5 | 1.5 | −1.1 |
| Tryptophan | −1.3 | 1.2 | −1.4 | −2.1 | −1.1 | −1.2 |
| Total Amino Acids | −1.1 | −1.5 | −11.0 | −7.8 | 1.2 | −2.3 |
| C-COMPOUNDS | ||||||
| Organic acids | ||||||
| Citrate | 1.4 | −1.2 | −6.2 | −1.9 | 1.2 | 2.7 |
| Malate | −1.7 | −2.7 | −1.3 | −1.5 | 1.2 | 1.8 |
| Succinate | −1.4 | −1.3 | −1.9 | 1.0 | 1.6 | 2.6 |
| Carbohydrates | ||||||
| Fructose | 3.4 | −2.1 | 7.3 | −1.8 | 6.8 | 1.0 |
| Glucose | 2.4 | −6.1 | 3.3 | −3.1 | 12.4 | 2.1 |
| C6-sugar | 3.9 | −2.2 | 3.7 | −3.0 | 4.0 | 1.8 |
| Sucrose | 1.0 | −2.3 | 1.9 | 1.0 | 2.3 | 1.6 |
| Myo-inositol | −1.5 | −3.9 | −1.6 | −3.9 | 1.3 | 1.3 |
| Galactinol | 1.3 | −1.3 | −1.3 | 1.5 | 1.8 | 1.8 |
| Total Sugars | 1.4 | −1.6 | 3.0 | 1.1 | 1.9 | 1.4 |
| Total Starch | 2.8 | 1.9 | −1.4 | −2.5 | 1.0 | −1.2 |
 >+10.0
>+10.0
 +2.0 to +10.0
+2.0 to +10.0
 +1.5 to +2.0
+1.5 to +2.0
1
 −1.5 to −2.0
−1.5 to −2.0
 −2.0 to −10.0
−2.0 to −10.0
 <−10.0
<−10.0
Gene expression correlating with NUE in the three genotypes
To gain insights into the molecular basis underlying the different NUE phenotypes, transcript levels of genes associated with N transport or N assimilation were measured using quantitative RT-PCR. The SRG-200 genotype had a higher expression of the genes involved directly in nitrate transport such as ZmNRT1.1, ZmNRT1.2, ZmNRT2.2, and ZmNRT2.3 in leaves (Figure 7A), ZmNRT1.1 in the stem (Figure 7B), and ZmNRT1.2 and ZmNRT2.3 in the roots (Figure 7C). The expression of the N assimilation genes such as ZmNR1, ZmNR2, ZmNAR1S, and ZmNiR was also higher in leaves of SRG-200 (Figure 7D). However, no significant difference was detected in the expression of those genes in the root tissues (Figure 7E). Glutamine synthetases are coded by a small gene family and convert ammonium into glutamine. In the leaf tissues, the expression level of the ZmGln4, ZmGln5, and ZmGln1.3 was significantly higher in the SRG-200 and SRG-150 genotypes compared to the SRG-100 (Figure 7F). Additionally, SRG-200 showed higher expression of other GS genes such as ZmGln1, ZmGln2, and ZmGs1.4 compared to the other genotype SRG-100 (Figure 7F). However, their expression trend was different in the roots, where only ZmGln5, ZmGs1.3, and ZmGs1.4 showed a higher expression in the hybrid SRG-150 (Figure 7G).
Figure 7.
Quantitative relative gene expression of nitrogen and carbon metabolism genes under N limitation conditions in the three genotypes; SRG-100 (black bars); SRG-200 (light gray bars); and SRG-150 (Dark gray Bars). (A–C) expression of the N transporter genes in leaves, stem, and root of the three genotypes. (D,E) Leaves and roots expression of some nitrate and nitrite reductase genes. (F,G) expression of glutamine synthetase genes in the leaves and roots of the three genotypes. (H) expression of the starch and sugar synthetase genes in the leaves of the plants growing under N limitation. Data are means ± SD (n = 3) and are representative of similar results from three independent experiments.
Expression of genes associated with key metabolic steps in carbohydrate metabolism was also analyzed. Alpha-1,4 glucan- phosphorylase (AGPP), sucrose phosphate synthase1 (SuPP), and the starch synthesis gene (SS) expression showed no significant difference between the two inbred lines and only showed a higher level in the hybrid leaves (Figure 7H).
Discussion
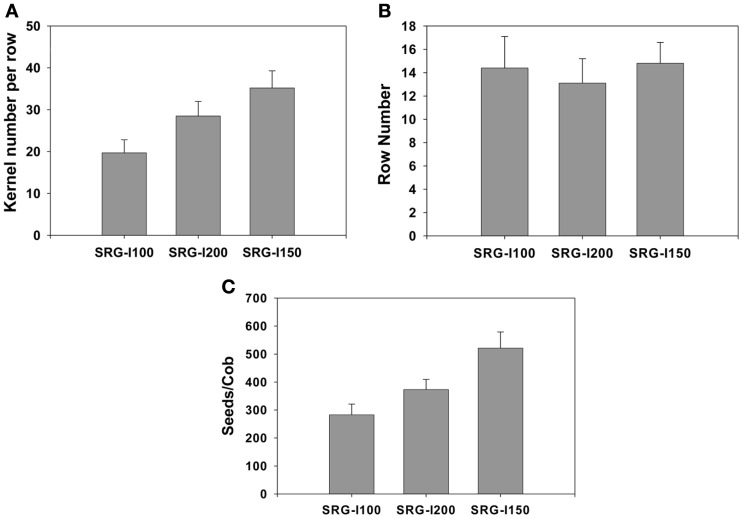
It would be highly advantageous to identify phenotypes at an early vegetative stage that correlate well with NUE at maturity to save time and resources. As a first step, we developed a hydroponic system and used it to test the differences between three genotypes that were different in their NUE at maturity under N limitation. These lines were an elite inbred line SRG-200 that showed a higher NUE than the other elite inbred line SRG-100 and as a reference we included its hybrid SRG-150 which has a higher NUE as expected. In the greenhouse, the kernel number per row was lower in the inbred line SRG-100 compared to the SRG-200 lines (Figure A2 in Appendix). Their NUE at an early vegetative stage (4-week-old) was determined and found it to be similar to that at maturity. A number of factors possibly contributing to NUE were tested using these two genetic lines and its hybrid to determine which of these could be indicative of predicting improved NUE at maturity. Generally, N limitation caused a reduction in a number of traits including chlorophyll content, root and shoot biomass, nitrate content, nitrate reductase activity, and sugar content, while causing an increase in anthocyanin content and the C/N ratio. However, the levels of reduction or induction were very different in the different genotypes exhibiting different NUE. These results are summarized in Tables 2 and 3. Those factors that could be most indicative of predicting improved NUE at maturity are summarized.
Table 2.
Percentage of the induction (+) and the reduction (−) of some parameters in the three genotypes growing under N limitation relative the plants of the same genotype growing under optimal N condition.
| Genotype | SRG-100 | SRG-200 | SRG-150 |
|---|---|---|---|
| NUE | * | ** | *** |
| Chlorophyll | − − − − | − − − − | − − − − |
| Anthocyanins | + | ++++ | ++++ |
| Total dry biomass | − − − − | − − − | − − − |
| Shoot biomass | − − − − | − − − | − − |
| Root volume | − − − − | − − − | − − − |
| C/N Ratio | ++ | ++++ | ++++ |
The number of positive or negative indicates the induction or the reduction of the trait relative to the SRG-100 genotype.
Table 3.
Summary of the relative gene expression level of N and C metabolism genes in the three genotypes growing under N limitation, the number of positive or negative indicates the higher or lower level of the trait relative to the SRG-100 genotype.
| SRG-100 | SRG-200 | SRG-150 | |
|---|---|---|---|
| Adaptation to N limitation | * | ** | *** |
| LEAVES N TRANSPORTER GENES | |||
| ZmNRT1.1 | + | +++++ | ++ |
| ZmNRT1.2 | + | ++++++ | ++ |
| ZmNRT2.2 | + | +++ | + |
| ZmNRT2.3 | + | ++ | + |
| STEM N TRANSPORTER GENES | |||
| ZmNRT1.1 | + | ++++ | ++ |
| ZmNRT1.2 | + | + | + |
| ZmNRT2.2 | + | + | + |
| ZmNRT2.3 | + | + | + |
| ROOTS N TRANSPORTER GENES | |||
| ZmNRT1.1 | + | + | + |
| ZmNRT1.2 | + | ++ | − |
| ZmNRT2.2 | + | − | − |
| ZmNRT2.3 | + | +++ | + |
| LEAVES GLUTAMINE SYNTHASE GENES | |||
| ZmGln1 | + | +++ | + |
| ZmGln2 | + | +++ | + |
| ZmGln4 | + | +++++ | +++ |
| ZmGln5 | + | ++++++ | ++ |
| ZmGS1.3 | + | ++++++++ | ++ |
| ZmGS1.4 | + | ++ | + |
| ROOTS GLUTAMINE SYNTHESIS GENES | |||
| ZmGln1 | + | + | + |
| ZmGln2 | + | + | + |
| ZmGln4 | + | + | + |
| ZmGln5 | + | + | ++ |
| ZmGS1.3 | + | + | + |
| ZmGS1.4 | + | + | +++ |
| LEAVES NITRATE AND NITRITE REDUCTASE GENES | |||
| ZmNR1 | + | +++++ | ++ |
| ZmNR2 | + | +++ | + |
| ZmNAR1S | + | +++++++ | +++ |
| ZmNiR | + | ++ | + |
| ZmF-NiR | + | + | + |
| ROOTS NITRATE AND NITRITE REDUCTASE GENES | |||
| ZmNR1 | + | + | + |
| ZmNR2 | + | + | + |
| ZmNAR1S | + | + | + |
| ZmNiR | + | + | + |
| ZmF-NiR | + | + | + |
| Carbon metabolism genes | |||
| ZmAGPP | + | + | + |
| ZmSuPP | + | + | + |
| ZmSS | + | + | +++ |
Phenotypically, higher NUE correlates with smaller biomass reduction especially in shoot, root volume reduction and higher anthocyanin induction under limiting N condition
Nitrogen limitation caused a reduction in shoot and root biomass. This reduction was significantly higher in the SRG-100 parent line compared to the other parent SRG-200 and its hybrid SRG-150. In addition, a reduction in the root volume and root diameter was detected in the roots of the SRG-100 parent but not in the other line SRG-200. These results are consistent with the fact that a larger root system might help in increasing nitrogen uptake and help the plant to transport and assimilate more nitrogen (Coque et al., 2008) and thus increase the use efficiency of nitrogen.
The induction of anthocyanins in response to nitrogen limitation has been previously reported. Anthocyanins play a role in scavenging reactive oxygen species produced from the plant cell upon the exposure to stress (Gould, 2004). Thus, possibly less anthocyanin production makes the cells more sensitive to stress. The more poorly adapted parent SRG-100 was unable to accumulate anthocyanins to levels observed in the SRG-200 parent and the hybrid SRG-150 grown under limiting N condition. Similar results have been reported in the Arabidopsis nla mutant previously characterized in our laboratory that was less adapted under nitrogen limitation (Peng et al., 2007a,b, 2008).
Biochemically, higher NUE correlates with higher nitrate content in leaves and stem, higher sugar content especially in stem and higher starch content in leaves
Nitrogen use efficiency in plants depends on the uptake, transport, and assimilation of nitrogen within different tissues. N limitation caused a reduction in root nitrate content. However, the similar levels of the root nitrate content in the three genotypes indicate that the SRG-100 genotype is able to uptake the nitrate from the medium. The low leaf nitrate content in leaves of the SRG-100 genotype might be an indicator for a limited capacity to transport nitrate from roots to the leaves. This assumption is supported by the finding that the nitrate transporter genes, ZmNRT1-1, ZmNRT1.2, ZmNRT2.2, and ZmNRT2-3 were expressed at a lower level in the SRG-100 especially compared to the other parent line SRG-200 under limiting nitrogen condition (Figure 7). Consistent with this notion, the expression of several genes involved in the subsequent steps for nitrogen assimilation were low in SRG-100 leaves, including the nitrite reductase gene ZmNiR, and the glutamine synthetase genes (Figure 7). It has been reported that nitrate and nitrite play a role as a signaling molecule (Crawford, 1995; Stitt, 1999; Wang et al., 2000, 2007). Therefore, it is possible that the altered nitrate exhibited by the plant lines tested under N limitation could lead to significant differences in signal transduction networks that could influence many fundamental processes related to N metabolism beyond the usual role of these substrates for N assimilation. Further work will be required to delineate the signal transduction pathway differences between these two lines to gain insights into pathways contributing toward the hybrid’s enhanced ability to adapt to N limitation.
Despite the significant difference between the two parents (SRG-100 and SRG-200) in nitrate content, both genotypes possess similar nitrate reductase activity. This could be due to the fact that nitrate reductase activity is regulated by different factors including, nitrate, light, and sugars such as sucrose and inhibited by some amino acids such as glutamine (Vincentz et al., 1993; Morcuende et al., 1998). In addition, GS activity did not reflect the differences between the NUE of these genotypes. However, we observed differences among the three genotypes at the transcription level of the nitrate reductase and glutamine synthetase genes, although these differences did not correlate with the enzyme levels. There are a number of other mechanisms which have been shown to be important that can explain this phenomenon including feedback regulation (Fan et al., 2006).
It has been reported that N-limiting conditions decreases photosynthesis, and increases the carbon to N ratio (Malamy and Ryan, 2001; Martin et al., 2002 and Wingler et al., 2006). A co-ordination of N and C metabolism is required since N assimilation demands C skeletons in the form of 2-oxoglutarate (Lancien et al., 2000). Also, it was previously reported that plants growing under limiting nitrogen conditions required more allocation of the sugars from the leaves to the roots to support its growth under this condition (Paul and Stitt, 1993). In our study, the SRG-200 genotypes maintained higher sugar content in leaves similar to those of the hybrid SRG-150 and suggesting a higher sugar amount was available to be translocated from leaves to roots in these lines. The more adapted hybrid SRG-150 and its parent SRG-200 had 2.8 and 1.9 times higher starch content, respectively, than the less adapted parent SRG-100 under N limitation (Table 1), suggesting the importance of the starch level in the adaptation to N limitation. The lower starch content in the leaves of the SRG-100 genotype could be due to either a lower level of starch synthesis or a higher level of starch degradation. Our data revealed similar transcript levels of the sugar and SS genes in the two parent lines SRG-100 and SRG-200 (Figure 7). Thus, it is likely that the variation in starch and sugar content between the SGR-100 and the other line SRG-200 which is similar to the hybrid SRG-150, was mainly due to a defect in sugar transport from the leaves to the roots to support root growth under N limitation and possibly, in differences in mechanisms controlling starch degradation pathways during growth under low N environments.
At the transcription level, higher NUE correlates with higher expression of some nitrate transporter genes and some key genes in N and C metabolism
Nitrogen use efficiency in plants depends on the uptake, transport, and assimilation of the nitrogen within their different tissues. We found that the transcription levels of some nitrate transporter genes and the glutamine synthetase genes of the SRG-100 genotype were significantly lower than those in the other parent SRG-200 and the hybrid SRG-150, indicating that this parent SRG-100 is less efficient in multiple metabolic steps that could be controlled by the nitrate level status which may subsequently affects the downstream components leading to an overall inferior ability for N assimilation during N limitation. Expressions of some of these genes in all the genotypes are summarized in Table 3. Despite the similarity between the SRG-200 and SRG-150 genotypes, it appeared that they possess different mechanisms to maintain a higher NUE. Given the level of heterosis in the hybrid, this is not surprising. While SRG-200 had a higher expression of some nitrate transporter genes and nitrate assimilation genes, SRG-150 had a higher expression of both N assimilation and C metabolic genes. The active carbon metabolism as observed in the hybrid SRG-150 might be involved directly or indirectly through N assimilation to promote plant growth under N limitation.
From all the above we could see a differences among the three genotypes in NUE at the seedlings level which is consistent with the NUE level at maturity stage. The observed differences in these traits could be involved directly or indirectly in affecting the NUE. The higher NUE in the hybrid compared to the two inbred lines could indicate an additive effect for this trait.
To conclude, a number of factors were examined that are associated with various aspects of metabolism. The SRG-200 genotypes with the higher NUE maintained higher sugar content in leaves similar to those of the hybrid SRG-150. In addition it has 1.9 times higher starch content compared to the less adapted genotype, SRG-100 under N limitation. suggesting the importance of the starch level in the adaptation to N limitation. Transcription analysis showed that the higher NUE in the SRG-200 genotype is associated with higher transcripts in the genes involved in nitrate transport, N assimilation, and GS. However, no difference was observed in the expression of genes associated with key metabolic steps in carbohydrate metabolism. We can make the following conclusions. First, even with just three genotypes a number of significant differences in phenotypes, metabolites, and transcript levels could be detected. Second, it is unlikely that one indicator will be a good predictor of the final trait. More inbred lines need to be tested to confirm that some of these traits are indeed good indicators. Third, many of these can be tested for in a cost effective fashion making the screening of larger numbers of lines easily feasible. Therefore, this study indicates that it is feasible to develop biomarkers that can help in the selection of improved crop genetics for NUE.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Eric M. Lyons, University of Guelph for using his facilities to carry the root analysis.
Appendix
Table A1.
Primers sequences used in performing the real-time PCR.
| Primer Name | Forward primer sequence | Reverse primer sequence | Accession number |
|---|---|---|---|
| ZmTub2 | 5′GCGCCTGTCTGTTGACTATGG3′ | 5′GGGATGGGTACACGGTGAAA3′ | gi|195610153 |
| ZmNRT1.1 | 5′CTGTCTGGCACCGTGATTGT3′ | 5′CGTAGCTGACTGCCCACCTAA3′ | gi|37778585 |
| ZmNRT1.2 | 5′TGTTCTCGGCGTGGTGAA3′ | 5′CCTCTGTACCTGACGGAGCAA3′ | gi|63397127 |
| ZmNRT2.2 | 5′CTCAGGTCGCTGCAGAAGAAG3′ | 5′CGTACCACATGACCTTGGAGAA3′ | gi|49472846 |
| ZmNRT2.3 | 5′CTTCTTCACCACGTCCAGCTACT3′ | 5′GCCATGATGCCCATGTTCTC3′ | gi|63397156 |
| ZmGln1 | 5′GACGGCGGGTTTGAAGAGA3′ | 5′TCATGGCGCAGAGAAAGGTT3′ | gi|162459402 |
| ZmGln2 | 5′GACGGCGGGTTTGAAGAGA3′ | 5′TCATGGCGCAGAGAAAGGTT3′ | gi|434333 |
| ZmGln4 | 5′AGGGCAGTGGGAGTTCCAA3′ | 5′AGACCTGGTCGCCTGAAGATATAC3′ | gi|162463754 |
| ZmGln5 | 5′CCGTCCGTCGGCATCTC3′ | 5′CCTCTCAAGAATGTAGCGAGCAA3′ | gi|162459550 |
| ZmGs1.3 | 5′AGGGCAGTGGGAGTTCCAA3′ | 5′ACCTGGTCGCCTGAAGAAATAC3′ | gi|434327 |
| ZmGs1.4 | 5′GCCAATTCCCACCAACAAGA3′ | 5′GACCTCAGGGCTGCTGAAGA3′ | gi|434329 |
| ZmNR1 | 5′CGAGATCGGCCACTTCGA3′ | 5′AACTTAGGGTGCTCGTTCTTGAA3′ | gi|168518 |
| ZmNR2 | 5′GAGTGGACGAGCATGTCATGA3′ | 5′CGCAATGGTCTCGCTGTCT3′ | gi|22376 |
| ZmNAR1S | 5′CAAGGTGTGGTACGTCATCGA3′ | 5′GAACCCAACGCTGTACTTCCA3′ | gi|168516 |
| ZmNiR | 5′CCTGCCGAGGAAATGGAA3′ | 5′TACGCGAGGTCGTTGATGTG3′ | gi|623203 |
| ZmF-NiR | 5′CCTGCCGAGGAAATGGAA3′ | 5′TACGCGAGGTCGTTGATGTG3′ | gi|195615807 |
| ZmAGPP | 5′CCAGGGTTGGAAGGAACGT3′ | 5′CGGTCGGGACTCTTGGAA3′ | gi|558364 |
| ZmSuPP | 5′GAACATGGCGACACCGATCT3′ | 5′CGGACGATCACGGTCTTGT3′ | gi|162460833 |
| ZmSS | 5′CGGACGGCTACACCAACTACTC3′ | 5′TTGCACTGCCGCTTTCC3′ | gi|45825439 |
Figure A1.
Grain yield of the two maize inbred lines, SRG-100, and SRG-200 and its hybrid SRG-150 growing under greenhouse condition in the presence of 4 mM Potassium nitrate. Plants were hand pollinated with the same pollen grains.
Figure A2.
Reduction in chlorophyll content in three maize genotypes, SRG-100, SRG-200, and SRG-150 growing under N limitation. Values are relative to the same genotypes growing at sufficient N (3 mM).
References
- Bernard S. M., Habash D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620 10.1111/j.1469-8137.2009.02823.x [DOI] [PubMed] [Google Scholar]
- Cataldo D. A., Haroon M., Schrader T. E., Youngs V. L. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant Anal. 6, 71–80 10.1080/00103627509366547 [DOI] [Google Scholar]
- Coque M., Martin A., Veyrieras J. B., Hirel B., Gallais A. (2008). Genetic variation for N-remobilization and postsilking N-uptake in a set of maize recombinant inbred lines. 3. QTL detection and coincidences. Theor. Appl. Genet. 117, 729–747 10.1007/s00122-008-0815-2 [DOI] [PubMed] [Google Scholar]
- Crawford N. M. (1995). Nitrate, nutrient and signal for plant growth. Plant Cell 7, 859–868 10.2307/3870042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Wang K. J., Jiang G. M., Biswas D. K., Xu H., Li L. F., Li Y. H. (2005). Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Ann. Bot. 96, 925–930 10.1093/aob/mci244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Gilles K., Hamilton J., Rebers P., Smith F. (1956). Colorimetric method for determination of sugars and related substrances. Anal. Chem. 28, 350–356 10.1021/ac60111a017 [DOI] [Google Scholar]
- Echarte L., Rothstein S., Tollenaar M. (2008). The response of leaf photosynthesis and dry matter accumulation to nitrogen supply in an older and a newer maize hybrid. Crop Sci. 48, 656–665 10.2135/cropsci2007.06.0366 [DOI] [Google Scholar]
- Fan S. C., Lin C. S., Hsu P. K., Lin S. H., Tsay Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761 10.1105/tpc.109.067603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Gordon-Weeks R., Shen Q., Miller A. J. (2006). Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J. Exp. Bot. 57, 1333–1340 10.1093/jxb/erj110 [DOI] [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Trethewey R., Willmitzer L. (2000). Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 72, 3573–3580 10.1021/ac991142i [DOI] [PubMed] [Google Scholar]
- Gallais A., Hirel B. (2004). An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 55, 295–306 10.1093/jxb/erh006 [DOI] [PubMed] [Google Scholar]
- Garnett T., Conn V., Kaiser B. N. (2009). Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ. 32, 1272–1283 10.1111/j.1365-3040.2009.02011.x [DOI] [PubMed] [Google Scholar]
- Good A. G., Shrawat A. K., Muench D. G. (2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605 10.1016/j.tplants.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Gould K. S. (2004). Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 5, 314–320 10.1155/S1110724304406147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal S. S., Tischner R., Basra A. S. (2005). Enhancing the Efficiency of Nitrogen Utilization in Plants. Haworth: Haworth Press, Inc., 489 [Google Scholar]
- Gregersen P. L., Holm P. B., Krupinska K. (2008). Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. (Stuttg.) 10, 37–49 10.1111/j.1438-8677.2008.00114.x [DOI] [PubMed] [Google Scholar]
- Hirel B., Bertin P., Quillere I., Bourdoncle W., Attagnant C., Dellay C., Gouy A., Cadiou S., Retailliau C., Falque M., Gallais A. (2001). Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 125, 1258–1270 10.1104/pp.125.3.1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortensteiner S., Feller U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53, 927–937 10.1093/jexbot/53.370.927 [DOI] [PubMed] [Google Scholar]
- Kant S., Bi Y. M., Weretilnyk E., Barak S., Rothstein S. J. (2008). The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol. 147, 1168–1180 10.1104/pp.108.118125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Kant P., Raveh E., Barak S. (2006). Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na1 uptake in T. halophila. Plant Cell Environ. 29, 1220–1234 10.1111/j.1365-3040.2006.01502.x [DOI] [PubMed] [Google Scholar]
- Khamis S., Lamaze T., Lemoine Y., Foyer C. (1990). Adaptation of the photosynthetic apparatus in maize leaves as a result of nitrogen limitation: relationships between electron transport and carbon assimilation. Plant Physiol. 94, 1436–1443 10.1104/pp.94.3.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancien M., Gadal P., Hodges M. (2000). Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol. 123, 817–824 10.1104/pp.123.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea P. J., Azevedo R. A. (2007). Nitrogen use efficiency. 2. Amino acid metabolism. Ann. Appl. Biol. 151, 269–275 10.1111/j.1744-7348.2007.00200.x [DOI] [Google Scholar]
- Liao M., Fillery I. R. P., Palta J. A. (2004). Early vigorous growth is a major factor influencing nitrogen uptake in wheat. Funct. Plant Biol. 31, 121–129 10.1071/FP03060 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu C., Zhang J., Zhang Q., Li L., Kuang T. (2001). Modification of photosystem II photochemistry in nitrogen deficient maize and wheat plants. J. Plant Physiol. 158, 1423–1430 10.1078/0176-1617-00501 [DOI] [Google Scholar]
- Malamy J. E., Ryan K. S. (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127, 899–909 10.1104/pp.010406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez A. J., Betti M., García-Calderón M., Estivill G., Credali A., Pajuelo P., Orea A., Clemente M. T., Pajuelo E., Galván F. (2005). Nitrate and Ammonium Assimilatory Enzymes. Lotus Japonicus Handbook (Dordrecht: Springer; ), 315–328 [Google Scholar]
- Martin A., Lee J., Kichey T., Gerentes D., Zivy M., Tatout C., Dubois F., Balliau T., Valot B., Davanture M., Terce-Laforgue T., Quillere I., Coque M., Gallais A., Gonzalez-Moro M. B., Bethencourt L., Habash D. Z., Lea P. J., Charcosset A., Perez P., Murigneux A., Sakakibara H., Edwards K. J., Hirel B. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T., Oswald O., Graham I. A. (2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Orsel M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. (Stuttg.) 10, 23–36 10.1111/j.1438-8677.2008.00097.x [DOI] [PubMed] [Google Scholar]
- Medici L. O., Pereira M. C. B., Lea P. J., Azevedo R. A. (2005). Identification of Maize lines with contrasting responses to applied nitrogen. J. Plant Nutr. 28, 903–915 10.1081/PLN-200055586 [DOI] [Google Scholar]
- Morcuende R., Krapp A., Hurry V., Stitt M. (1998). Sucrose feeding leads to increased rates of nitrate assimilation, increased rates of 2-oxoglutarate synthesis and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta 206, 394–409 10.1007/s004250050415 [DOI] [Google Scholar]
- Olsen K. M., Lea U. S., Slimestad R., Verheul M., Lillo C. (2008). Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 165, 1491–1499 10.1016/j.jplph.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Paul M. J., Stitt M. (1993). Effects of nitrogen and phosphorus deficiencies on levels of carbohydrates, respiratory enzymes and metabolites in seedlings of tobacco and their response to exogenous sucrose. Plant Cell Environ. 16, 1047–1057 10.1111/j.1365-3040.1993.tb00900.x [DOI] [Google Scholar]
- Peng M., Bi Y. M., Zhu T., Rothstein S. J. (2007a). Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 65, 775–797 10.1007/s11103-007-9241-0 [DOI] [PubMed] [Google Scholar]
- Peng M., Hannam C., Gu H., Bi Y. M., Rothstein S. J. (2007b). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50, 320–337 10.1111/j.1365-313X.2007.03050.x [DOI] [PubMed] [Google Scholar]
- Peng M., Hudson D., Schofield A., Tsao R., Yang R., Gu H., Bi Y. M., Rothstein S. J. (2008). Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 59, 2933–2944 10.1093/jxb/ern148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed A. J., Hageman R. H. (1980). Relationship between nitrate uptake, flux, and reduction and the accumulation of reduced nitrogen in maize (Zea mays L.): II. Effect of nutrient nitrate concentration. Plant Physiol. 66, 1184–1189 10.1104/pp.66.6.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi E., Balconi C., Manusardi C., Gentinetta E., Motto M. (1991). Genetic variation for traits relating to nitrogen content of maize stalks. Euphytica 52, 91–98 [Google Scholar]
- Rosen H. (1957). A modified ninhydrin colorimetric analysis of amino acids. Arch. Biochem. Biophys. 67, 10–15 10.1016/0003-9861(57)90255-2 [DOI] [PubMed] [Google Scholar]
- Scheible W. R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rojas N., Schindelasch D., Thimm O., Udvardi M. K., Stitt M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499 10.1104/pp.104.047019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestak K., Catsky J., Jarvis P. G. (1971). Plant Photosynthetic Production. The Hague: W. Junk [Google Scholar]
- Steenbjerg F., Jakobsen S. T. (1963). Plant nutrition and yield curves. Soil Sci. 95, 69–90 10.1097/00010694-196301000-00012 [DOI] [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186 10.1016/S1369-5266(99)80033-8 [DOI] [PubMed] [Google Scholar]
- Taylor L., Nunes-Nesi A., Parsley K., Leiss A., Leach G., Coates S., Wingler A., Fernie A. R., Hibberd J. M. (2010). Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilisation during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 62, 641–652 10.1111/j.1365-313X.2010.04179.x [DOI] [PubMed] [Google Scholar]
- Tollenaar M., Migus W. (1984). Dry matter accumulation of maize grown hydroponically under controlled-environment and field conditions. Can. J. Plant Sci. 64, 475–485 10.4141/cjps84-070 [DOI] [Google Scholar]
- Vincentz M., Moureaux T., Leydecker M. T., Vaucheret H., Caboche M. (1993). Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 3, 315–324 10.1111/j.1365-313X.1993.tb00183.x [DOI] [PubMed] [Google Scholar]
- Vos J., Putten P. E. L., Birch C. J. (2005). Effect of nitrogen supply on leaf appearance, leaf growth, leaf nitrogen economy and photosynthetic capacity in maize (Zea mays L.). Field Crops Res. 93, 64–73 [Google Scholar]
- Wang R., Guegler K., LaBrie S. T., Crawford N. M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509 10.2307/3871145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Crawford N. (2007). Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 145, 1735–1745 10.1104/pp.107.108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A., Purdy S., MacLean J. A., Pourtau N. (2006). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57, 391–399 10.1093/jxb/eri279 [DOI] [PubMed] [Google Scholar]
- Ying J., Lee E. A., Tollenaar M. (2000). Response of maize leaf photosynthesis to low temperature during the grain-filling period. Field Crops Res. 68, 87–96 10.1016/S0378-4290(00)00107-6 [DOI] [Google Scholar]