Abstract
Because of their sessile lifestyle, plants have evolved adaptations to environmental factors, including UV-B present in solar radiation. To gain a better understanding of the initial events in UV-B acclimation, we have analyzed a 10 min to 1 h time course of transcriptome responses in irradiated and shielded leaves, and immature maize ears to unravel the systemic physiological and developmental responses in exposed and shielded organs. After 10 min of UV-B exposure, 262 transcripts are changed by at least two-fold in irradiated leaves, and this number doubles after 1 h. Indicative of the rapid modulation of transcription, 130 transcripts are only changed after 10 min. This is true not only in irradiated leaves, but also in shielded tissues. After 10 min of exposure, the overlap in transcriptome changes in irradiated and shielded organs is significant; however, after 30 min of UV-B, there are only two transcripts showing similar UV-B regulation between the three organs; 35 are similarly regulated in both IL and SL. Therefore, at longer irradiation times, there is more specificity of responses, and these are organ-specific. We suggest that early signaling in different tissues may be elicited by common signaling pathways, while at longer exposure times responses become more specific. To identify metabolites as possible signaling molecules, we looked for compounds that increased within 5–90 min in both irradiated and shielded leaves, to explain the kinetics of profound transcript changes within 1 h. We found that myoinositol is one such candidate metabolite; and we also demonstrate that if 0.1 mM myoinositol is applied to leaves of greenhouse maize, some metabolites that are changed by UV-B are also changed similarly by the chemical treatment. Therefore, this metabolite can partially mimic UV irradiation.
Keywords: UV-B radiation, Zea mays, microarray analysis, metabolomics, myoinositol, UVR8
Introduction
Solar radiation contains light qualities that are essential for photosynthesis, but certain wavelengths can damage cells. Because of their sessile lifestyle, plants have evolved adaptations to different environmental factors, including solar radiation. Plants contain multiple photoreceptors: phytochromes for perceiving red/far red, cryptochromes, and phototropins for blue/UV-A, and at least one UV-B receptor, UVR8, which was recently identified (Rizzini et al., 2011). In addition to acting as a developmental and physiological signal, low fluence UV-B photons also cause cellular damage by generating photoproducts in DNA and by direct damage to proteins, lipids, and RNA. Elevated UV-B radiation has pleiotropic effects on plant development, morphology, and physiology (Frohnmeyer and Staiger, 2003; Blanding et al., 2007), but the coordination of systemic responses is not well-understood.
In response to the inevitable exposure to damaging UV-B radiation, plants have UV-induced mechanisms of protection and repair, such as the accumulation of UV-absorbing sunscreen compounds (Stapleton and Walbot, 1994; Bieza and Lois, 2001) and the use of UV-A photons by photolyase enzymes to repair most UV-B-induced DNA damage (Britt, 1996). The plant epidermis, by virtue of accumulating numerous phenolic compounds and cuticular waxes, absorbs 90–99% of solar UV-B radiation with minimal absorption of photosynthetically active radiation (PAR; Li et al., 1993; Stapleton and Walbot, 1994; Landry et al., 1997). Maize mutants lacking purple anthocyanin pigments show increased UV-B-mediated DNA damage (Stapleton and Walbot, 1994) and increased sensitivity measured by transcriptome profiling (Casati and Walbot, 2003). Despite the documented protective role of anthocyanins, leaf pigmentation has been bred out of modern maize. We recently discovered that maize landraces adapted to high UV-B in the mountains of Mexico and South America accumulate colorless flavones in seedling, juvenile, and adult leaves. Flavones are excellent UV-B sunscreens, absorbing virtually all harmful radiation, and they are transparent to PAR (Casati and Walbot, 2005). Modern maize lines lack flavone accumulation in leaves and are much more sensitive to UV-B than these high altitude landraces.
Previously, we used transcriptome, proteome, and metabolic profiling to examine responses in the maize canopy and to track changes in shielded leaves and immature ears over 1 –6 h (Casati et al., 2011a). Using a protocol of irradiating canopy leaves in greenhouse grown maize plants, we have sought to identify signals that coordinate systemic responses. Systemic responses can impact yield by modulating ear growth or kernel properties, in addition to the “short-term” and readily repaired damage to DNA and photosynthetic reaction centers in irradiated leaves. Transcript diversity is decreased more than 10% as organs respond to UV-B, indicative of a major reprogramming of gene expression (Casati et al., 2011a). Exposure of just the top leaf substantially alters the transcriptome of both irradiated and shielded organs, with greater changes as additional leaves are irradiated. We found that there is specificity in the responses; for example, some phenylpropanoid pathway genes were expressed only in irradiated leaves and, correspondingly, some phenylpropanoid precursors to sunscreen compounds only accumulated in these leaves (Casati et al., 2011a). Candidates in early steps of signal transduction and possible signal molecules were also identified in the controlled greenhouse conditions in which no UV-B is present until a singular treatment period. Because field-grown maize experiences fluctuating UV-B levels and variation in other environmental conditions, we have also compared the transcriptome, proteome, and metabolome changes after 4 h of supplementary UV-B irradiation in naturally UV-B-acclimated field plants to the same genotype grown in the greenhouse in the absence of UV-B (Casati et al., 2011b). The absolute number of transcript differences was higher in the naïve greenhouse plants than in acclimated field plants. Common elements included transcripts for genes involved in sunscreen biosynthesis and some regulators. Thus, prior acclimation to UV-B results in fewer transcript and protein losses and metabolite changes.
To gain a better understanding of the initial events in UV-B acclimation, we now report a 10 min to 1 h time course of transcriptome responses in irradiated and shielded leaves and in immature maize ears to unravel the systemic physiological and developmental responses in exposed and shielded organs. To identify metabolites as possible signaling molecules, we looked for compounds that increased within 5–90 min in both irradiated and shielded leaves, to explain the kinetics of profound transcript changes within 1 h. We found that myoinositol is one such candidate metabolite, and it also has support from RNA profiling: after 1 h UV-B, transcripts for myoinositol-1-phosphate synthase, the rate-limiting biosynthetic step in plants, are decreased in both irradiated and shielded leaves suggesting down-regulation of biogenesis as is typical for many hormone and small effectors (Casati et al., 2011a). In this paper, we demonstrate that if 0.1 mM myoinositol is applied to leaves of greenhouse maize, some metabolites that are changed by UV-B are also changed similarly by the chemical treatment. Therefore, this metabolite can partially mimic UV irradiation.
Materials and Methods
Samples and treatments
W23 maize was grown for 5 weeks in the greenhouse using the same protocol as previously described (Casati et al., 2011a). The afternoon prior to treatment, 12 plants were moved underneath UV-B lamps (Phillips, TL 20 W/12); the bulbs were covered with cellulose acetate (CA) to exclude wavelengths <280 nm. The topmost two leaves were threaded through slits in polyester plastic (PE); after acclimating overnight, these leaves received 5, 10, 30, 60, or 90 min UV-B exposure (UV-B intensity of 2 W m−2, UV-A: 0.65 W m−2). Exposure times were centered on 11 am, 5 h after sunrise and supplemental greenhouse lighting (33% of summer noon solar fluence from sodium vapor, UV-A blue fluorescent, and metal halide lamps). As a control, untreated plants (C) were used; these correspond to plants that were irradiated with the UV-B lamps covered with the PE filter that absorbs UV-B. A biological replicate consisted of tissue samples pooled from three plants, i.e., samples from the center of the blades and the topmost 2–3 cm ear. Four biological replicates for each treatment (a total of 12 plants) were used in all assays. The three sample types (irradiated leaf, shielded leaf, and immature ear) were immediately flash-frozen in liquid nitrogen after harvesting. Irradiated leaf samples were collected from both irradiated leaves while shielded leaf samples were taken from the two leaves immediately below the PE plastic. Leaves were separated from the midrib before being frozen, and blade segments about 8 cm long were cut out with scissors. Immature ears were cut in half and distributed randomly among the different collection tubes for RNA analysis.
Microarray experiments
RNA extraction and microarray hybridization were done as described in Casati and Walbot (2008). Data acquisition, image processing, and spot flagging and removal were performed as described in Skibbe et al. (2009). The median foreground values for each channel from the Agilent Feature Extraction software were first normalized using the lowess method of the limma package in R (Smyth, 2005) within each array, and then using limma's quantile method between all arrays. Probes were classified as “on” if their expression value was more than 3.0 SDs above the average foreground intensity of the Agilent negative controls, providing a 0.13% FDR (∼50 probes). For differential expression, we used the unadjusted p-value generated by limma in the R programming environment for calculation of FDR. Differentially expressed probes were identified using a two-fold cutoff for expression ratios with a limma-assigned p-value < 0.05. Probes were included in the analysis if at least 75% of the replicate expression values (i.e., three of four) were classified as “on.” Microarray data were deposited in GEO under ID GSE30278.
Metabolite profiling
Extraction, liquid partition, and derivation prior to GC–MS analysis were performed as described by Lisec et al. (2006). For analysis, four biological replicates per treatment with a second group of technical replicates were utilized (eight total data points). GC–MS analysis was performed using an autosystem XL Gas Chromatograph and a Turbo Mass Spectrometer (Perkin Elmer) in the Facultad de Ciencias Bioquímicas y Farmacéuticas – UNR facilities. One microliter split injection (split ratio 1:40) was injected at 280°C. The capillary column used was aVF-5 ms column (Varian, Darmstadt, Germany) with the following dimensions: 30 m × 0.25 mm inner diameter and a 0.25-μm film with helium as carrier gas with constant flow at 1 mL/min. The temperature program was 5 min at 70°C, 5 min ramp to 310°C, and final heating for 2 min at 310°C. The transfer line to the MS was set to 280°C. Spectra were monitored in the mass range m/z = 70–600. Tuning and all other settings were according to manufacturer's recommendations.
Chromatograms were acquired with TurboMass 4.1 software (Perkin Elmer). The NIST98mass spectral search program1 (National Institute of Standards and Technology, Gaithersburg, MD, USA) was the software platform. The MS and retention time index were compared with the collection of the Golm Metabolome Database (Kopka et al., 2005; Schauer et al., 2005). MS matching was manually supervised and matches accepted with thresholds of match >650 (with maximum match equal to 1000) and retention index deviation <1.0%. Peak heights were normalized using the amount of the sample fresh weight and ribitol for internal standardization. Relative metabolite contents were determined and statistical analyses were performed using ANOVA tests in Sigma Stat 3.1.
qRT-PCR assays
Primers were designed using the PRIMER3 software (Rozen and Skaletsky, 2000) and are listed in Table 1. Three micrograms of total RNA were used for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR was carried out in a DNA Engine OPTICON2 (MJ Research, division of Bio-Rad, Hercules, CA, USA), as described in Casati and Walbot (2004). Three replicates were performed for each sample, plus template-free samples; a melting curve analysis was performed at the termination of the assay to judge whether a single product was produced. To normalize the data to the NT (no treatment) control, primers for cyanase were used. To confirm the size of the PCR products, and to check that they corresponded to a unique and expected PCR product, the final PCR products were separated on a 2% agarose gel when first used and PCR products were verified by sequencing. Data was processed using PCR Miner2 and analyzed using the average efficiency of each transcript and average Ct of replicate samples to compute relative R0 (Zhao and Fernald, 2005). Differential expression was then calculated according to untreated (NT) controls.
Table 1.
List of primers used for qRT-PCR experiments.
| Primer | Primer sequences |
|---|---|
| BM500597-L | CACAGCTTTGCTTCCCTCTT |
| BM500597-R | GATGCCCATTTATGCTTTGG |
| TC292940-L | GGGAGGGAATGTCTTGTTCA |
| TC292940-R | TTGCAGGCAGCATTGTTTAG |
| AF112150-L | CACCTTATGGCCGAGTCAAT |
| AF112150-R | GTCTGGACCTGGACCTGTGT |
| TC280980-L | TCGGATTCACCACTTTCCTC |
| TC280980-R | GTTGGAAGCTGGAACACTCC |
| AW129897-L | CCCTCGGAACTGTCATCATT |
| AW129897-R | TAGGTGGACAGCGAATAGGG |
| TC308488-L | ATGGCCTACGCAATCTGTTC |
| TC308488-R | CCGAGAACAACCCGATTAGA |
| TC303498-L | GTGAACACAGCCACAGTTGG |
| TC303498-R | ATGTTGCGTTGCGTCTACAG |
| CB278279-L | CTCATCCGGGAGTACGATGT |
| CB278279-R | CCATGGCCTCTTGCTGTAGT |
| TC301764-L | GAGTCGCTGGACAACATCCT |
| TC301764-R | TCTTGCTTGATGGAGACGTG |
Myoinositol treatments
Myoinositol (10–0.01 mM, tissue-culture grade Sigma Chemical Co., St. Louis, MO, USA) was applied in a soaked 8 cm × 12 cm paper towel to the center of the blade of a full expanded canopy leaf, and leaf samples were collected from the treated leaf zone and from the leaf immediately below it (untreated leaf, naturally 180° rotated from the treated leaf to minimize drips); the myoinositol treated leaf is equivalent to UV-B-exposed leaf and the untreated leaf corresponds to a shielded leaf in a UV-B protocol. Leaf samples were collected from blade segments excised on either side of the midrib after 10 and 30 min, and used for metabolite extraction. As controls, similar towels soaked with water or 1 mM glucose were used.
Results
Microarray hybridization design
Previously, we found that shielded organs, such as immature ears and shielded leaves, exhibited significant transcriptome and metabolite changes after 1 h of UV-B irradiation of the two top leaves (Casati et al., 2011a). Given the broad scope of changes evident within 1 h, much shorter UV-B exposure times are required to unravel the systemic physiological and developmental responses in exposed and shielded organs. For this purpose, adult maize plants were covered with a plastic sheath that absorbs UV-B (PE, see Materials and Methods), while two adult leaves per plant were irradiated with UV-B radiation through a plastic that allows UV-B transmittance (CA, Casati et al., 2011a). Plants were irradiated using UV-B lamps for 5, 10, 15, 30, and 90 min. After qRT-PCR analysis with a panel of UV-B-expressed genes (Table 1), many significant changes were seen in the early time points (not shown); therefore, 10 and 30 min were chosen for transcriptome analysis on a microarray along with the 60 min time point from the longer time course UV-B irradiation experiments (Casati et al., 2011a). Using these three time points, the transcriptomes from irradiated and shielded leaves (IL and SL, respectively), and shielded immature ears (IE) were analyzed in order to identify early UV-B responses in these organs.
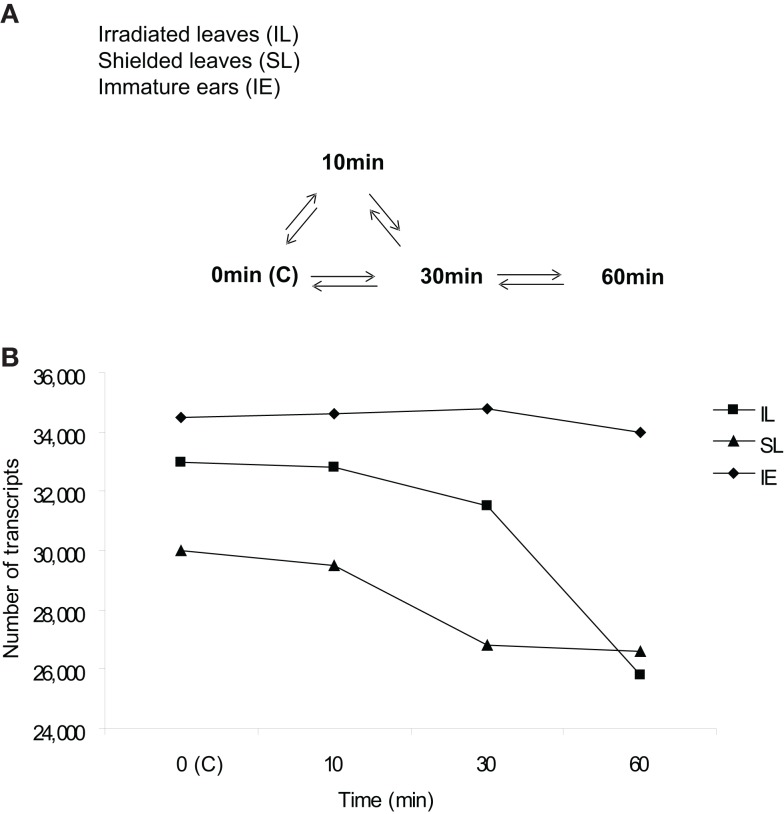
Microarray design is diagrammed in Figure 1A. A highly sensitive, custom-designed Agilent® 4 K × 44 K array with 60-mer probes and internal spike-in control probes quantified transcript abundance and non-specific hybridization for ∼39,000 maize transcripts (Casati and Walbot, 2008). Hybridization signals were scored as present at three-fold above the SD of the average hybridization to the non-specific, non-hybridizing, control probes; with this criterion, the false discovery rate is 0.13%. Transcriptome differences were assessed from leaf or ear samples pooled from three individuals, and four independent biological replicates were performed with symmetrical dye labeling to minimize systematic errors (Kerr and Churchill, 2001). The correlation in quantitative comparisons between datasets among biological replicates was r2 = 0.90–0.99.
Figure 1.
Microarray design and transcriptome size in UV-B-irradiated and shielded leaves and immature ears after different UV-B irradiation times. (A) Microarray hybridization design with direct comparisons to measure UV-B effects on signaling initiation from irradiated (irradiated leaves, IL) to shielded tissues (shielded leaves, SL; and immature ears, IE). All time course treatments had two leaves exposed above the PE. Each arrow represents a pair of samples hybridized on one array; in every case there is one cy3-labeled sample compared to one cy5-labeled sample. For each sample type there were four biological replicates, generating at least four hybridization comparisons, except where noted. No UV-B irradiation or control (0 min or C) plants did not receive any UV-B, see Materials and Methods for details of the tissue sampling protocol. (B) Transcriptome size in IL and SL, and IE from plants that were irradiated with UV-B in the two upper leaves after 10, 30, and 60 min of exposure, and control plants that did not receive any UV-B (0 min, C).
As shown in Figure 1B, maize leaves (L) and immature ears (IE) express a substantial number of genes. Under greenhouse control conditions without any UV-B (0 min, labeled C), immature ears showed the highest number of transcripts expressed, and the total number of transcripts was not significantly affected by a short UV-B treatment up to 1 h. Leaves from control plants showed a slightly lower number of expressed transcripts expressed; however, UV-B exposure decreases transcriptome diversity substantially, both in irradiated (IL) and shielded (SL) leaves (Figure 1B).
Short time course of transcriptome responses
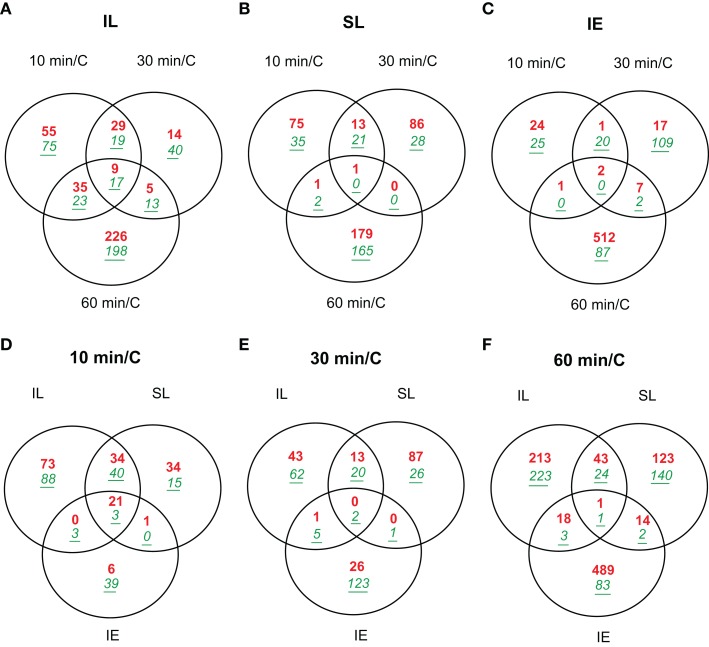
Figure 2A shows that 262 transcripts are changed by at least two-fold (p < 0.05) in irradiated leaves after 10 min of UV-B irradiation dropping to 146 transcripts at 30 min, and then finally rising to 526 transcripts by 1 h (File S1 in Supplementary Material). Thus, transcriptome changes in UV-B-treated leaves are measured as fast as after 10 min of irradiation, and there is rapid modulation over the subsequent 20 and 50 min intervals. For example, 51% of the differentially expressed transcripts at 30 min are similarly changed after only 10 min of irradiation (Figure 2A), while the other half of the 10 min transcript class now show levels similar to control values.
Figure 2.
Venn diagrams comparing transcriptome changes in leaves and immature ears from UV-B-irradiated plants in only two adult leaves compared to non-irradiated control plants (C). Plants were irradiated over a time course of 10, 30, and 60 min. Intersection of genes differentially expressed in irradiated leaves (A); shielded leaves (B); and immature ears (C) during 10, 30, and 60 min. Intersection of genes differentially expressed after 10 min (D); 30 min (E); and 60 min (F) in IL, SL, and IE. Transcripts showing changes higher than two-fold (p < 0.05) were included in the classification. Up-regulated transcripts are in red (bold), down-regulated transcripts are in green (italics, underlined).
Of the transcripts regulated at 10 min, 130 are not significantly altered at longer irradiation times. These probably correspond to mRNAs for proteins that participate in early responses to UV-B in maize. In this group, there are 29 DNA-binding proteins including transcription factors (TFs, 11 up-regulated, 18 down-regulated, Table 2) and seven proteins that participate in signal transduction (two up-regulated, five down-regulated, Table 2) and are candidates to participate in early UV-B-signaling. Moreover, UVR8, a putative UV-B photoreceptor in Arabidopsis (Rizzini et al., 2011), is rapidly increased by UV-B after 10 min in IL and SL. UVR8 is a UV-B-specific signaling component in Arabidopsis that mediates low fluence photomorphogenic responses, and it is required for UV-B-induced expression of the gene encoding the HY5 transcription factor in co-operation with COP1 (Ulm et al., 2004; Brown et al., 2005). UVR8 and COP1 interact directly and rapidly in the nucleus in planta after UV-B exposure (Oravecz et al., 2006), and this very early step in UV-B-signaling is proposed to initiate UV-B acclimation (Favory et al., 2009). In maize, UVR8 is down-regulated by UV-B at exposure times longer than 1 h in irradiated leaves (Casati et al., 2011a). In contrast, after 1 h canopy exposure, shielded maize leaves show up-regulation of UVR8, a step that may be an acclimation to increase subsequent sensitivity to UV-B or a clue that UVR8 may have functions that do not require direct irradiation. Thus, systemic signaling results in up-regulation in shielded organs after a delay, a step that may potentiate subsequent acclimation to UV-B.
Table 2.
List of transcripts encoding transcription factors, sensing, and signal transduction proteins that are UV-B-regulated in IL after 10 min of exposure.
| Category | Transcript | Description | Match | Log2 UV-B/C |
|---|---|---|---|---|
| Transcription/DNA-binding | TC307568 | *Heat-shock factor RHSF5 | GRMZM2G125969 | 1.08 |
| TC302156 | MYB-like protein E1 | GRMZM2G145041 | 1.16 | |
| TC315780 | *Regulator of chromosome condensation-like | GRMZM2G337819 | 1.63 | |
| TC304508 | *Transcription factor MADS57 | GRMZM2G044251 | 1.38 | |
| TC312622 | Homeobox-leucine zipper protein HAT14 | GRMZM2G127537 | 1.06 | |
| TC289946 | *Heat-shock factor RHSF6 | GRMZM2G010871 | 1.75 | |
| BG841242 | *TGACG-motif binding factor | GRMZM2G137046 | 1.44 | |
| TC280929 | *MADS box protein | GRMZM2G148693 | 1.30 | |
| TC294340 | *CCAAT-binding transcription factor | GRMZM2G104396 | 1.41 | |
| TC308958 | *Heat-shock factor RHSF7 | GRMZM2G165972 | 2.33 | |
| AF112150 | *MADS box protein 3 | GRMZM2G072582 | 2.39 | |
| TC300388 | WRKY transcription factor | GRMZM2G120320 | −1.18 | |
| TC310737 | *WRKY transcription factor 53-like | GRMZM2G449681 | −1.43 | |
| TC280931 | BTH-induced ERF transcriptional factor 1 | GRMZM2G052667 | −1.13 | |
| TC297607 | WRKY transcription factor 13-like | GRMZM2G141299 | −1.06 | |
| TC300180 | Ethylene-responsive element binding factor | GRMZM2G020150 | −1.05 | |
| TC295419 | *LigA | GRMZM2G131340 | −1.33 | |
| TC293183 | Wound inducive mRNA | GRMZM2G006468 | −1.16 | |
| TC298370 | *Cyclin T2-like protein | GRMZM2G081580 | −1.28 | |
| TC301163 | Homeobox protein 1-like | GRMZM2G445634 | −1.83 | |
| TC288985 | Ethylene-responsive transcription factor 7 | GRMZM2G307665 | −1.13 | |
| TC293926 | *Transcription factor WRKY32 | GRMZM2G324999 | −1.07 | |
| TC300365 | HAT dimerisation domain-containing protein-like | −1.06 | ||
| TC294981 | *Light-induced protein CPRF-2 | GRMZM2G073427 | −2.01 | |
| TC307533 | *Ethylene-responsive transcription factor 3 | GRMZM2G020054 | −2.95 | |
| TC285041 | *DNA-binding protein RAV1-like | GRMZM2G059939 | −1.54 | |
| TC313485 | *NAC domain transcription factor | GRMZM2G127379 | −1.20 | |
| TC310581 | *CCR4-associated factor-like protein | GRMZM2G177340 | −1.04 | |
| BM333902 | *Zinc finger protein LSD1 | GRMZM2G089106 | −1.74 | |
| Signal transduction | TC312631 | Ser–thr protein kinase | GRMZM2G135359 | 1.00 |
| TC314581 | Serine:threonine-specific receptor protein kinase-like | GRMZM2G092776 | 1.26 | |
| CO528246 | CBL-interacting protein kinase | GRMZM2G052067 | −1.01 | |
| TC294050 | EF-hand Ca2+-binding protein CCD1 | −1.84 | ||
| TC312158 | *EF-hand calcium binding protein-like | GRMZM2G357595 | −1.61 | |
| DN205713 | SNF1-related kinase regulatory gamma subunit 1 | GRMZM2G173536 | −1.17 | |
| TC282953 | 1-phosphatidylinositol-4-phosphate 5-kinase-like | GRMZM2G476448 | −1.08 | |
| UV-B sensing | TC305402 | *UV-B-resistance protein UVR8-like | GRMZM2G337819 | 1.55 |
| TC305401 | *UV-B-resistance protein UVR8-like | GRMZM2G302245 | 1.61 |
*Transcripts that are also UV-B-regulated in SL after 10 min.
Previously, we found that at longer UV-B exposure times (for example 4 h), maize UVR8 is down-regulated both in irradiated and shielded leaves (Casati et al., 2011a). Thus, UVR8 in maize is only transiently up-regulated in leaves then down-regulated. If UVR8 is a UV-B sensor in maize, reducing receptor concentration could be important for acclimation to reduce the amplitude of responses.
In shielded leaves after 10 min canopy irradiation, the number of UV-B-regulated transcripts is about 1.8-fold lower than in irradiated leaves (148 transcripts); however, two-thirds of these (98) are also increased in IL at the same time point (Figure 2D; File S4 in Supplementary Material). Nineteen of the overlapping transcripts correspond to transcription factors/DNA-binding proteins; therefore, the signal that induces transcription must be transmitted quickly to the shielded leaves. Although there is some overlap between UV-B-regulated transcripts in SL after 10 and 30 min of treatment (35 transcripts, 24% of the transcripts changed at 10 min; File S2 in Supplementary Material), most transcripts that are changed after 10 min quickly return to their basal expression levels in the absence of UV-B. After 1 h of UV-B, almost all UV-B-regulated transcripts in SL are specific for this time point (348 total transcripts, only four shared with the other time points; Figure 2B; File S2 in Supplementary Material).
In immature ears, fewer transcripts are altered compared to leaves; however, even after just 10 min UV-B exposure, 73 mRNAs are differentially regulated compared to controls (Figure 2C; File S3 in Supplementary Material). Twenty-three of them remain changed after 30 min of irradiation, but as measured for leaf transcripts, most of the very early regulated transcripts return to their basal levels of expression by 30 min. The 2 to 3-cm immature maize ears are wrapped within a whorl of husk leaves behind a leaf sheath, and they are unlikely to receive any UV-B radiation at this stage. Nonetheless, the immature ears are UV-B responsive: two of the up-regulated TFs in Table 2 (heat-shock factor RHSF6 and an ethylene-responsive transcriptional coactivator-like protein) and one down-regulated TF (an ethylene-responsive transcription factor 3) are shared with irradiated and shielded leaves. Table 3 lists all 24 transcripts common among the three organs after 10 min UV-B. It is interesting to note that seven (29%) correspond to heat-shock proteins (HSPs). We previously found that a number of HSPs were down-regulated by UV-B after longer exposure times in IE (Casati et al., 2011a). Together, our results suggest that changes in the expression levels of this group of proteins may have an important role in UV-B responses, both in irradiated and shielded tissues, and transient changes in their levels would be important for acclimation to this radiation. Finally, after 1 h of exposure, the number of UV-B-regulated transcripts increased in IE with most of them specific to this time point (611 transcripts; Figure 2C; File S3 in Supplementary Material). This is similar to the shielded leaves measurements.
Table 3.
List of transcripts that are only UV-B-regulated in all irradiated and shielded leaves and immature ears after 10 min of exposure.
| Transcript | Description | Match | Log2_10 min/C | ||
|---|---|---|---|---|---|
| IL | SL | IE | |||
| TC305560 | F5 family protein | 3.96 | 4.57 | 3.18 | |
| TC305561 | Unknown | 4.26 | 4.30 | 2.94 | |
| TC288590 | Extensin class I | GRMZM2G097135 | 3.36 | 3.62 | 2.74 |
| TC302397 | Ethylene-responsive transcriptional coactivator-like protein | GRMZM2G051135 | 4.55 | 4.33 | 2.47 |
| TC305946 | Unknown | GRMZM2G021816 | 2.54 | 3.05 | 2.25 |
| TC312940 | Small heat-shock protein | GRMZM2G080724 | 2.69 | 3.37 | 2.24 |
| TC293599 | DNAJ-like protein | GRMZM2G039886 | 1.92 | 2.39 | 1.89 |
| TC289946 | Heat-shock factor RHSF6 | GRMZM2G010871 | 1.75 | 2.13 | 1.88 |
| CB179674 | GDNF family receptor alpha 4 precursor | 1.83 | 2.21 | 1.80 | |
| TC307526 | Unknown | GRMZM2G165272 | 2.14 | 2.68 | 1.79 |
| TC307608 | Unknown | GRMZM2G098696 | 1.77 | 1.88 | 1.67 |
| TC313835 | Blr6628-like protein | GRMZM2G168261 | 3.16 | 3.07 | 1.64 |
| TC300184 | Mitochondrial small heat-shock protein 22 | GRMZM2G007729 | 2.83 | 3.57 | 1.50 |
| TC306900 | Unknown | GRMZM2G044251 | 2.16 | 2.24 | 1.44 |
| TC289461 | Unknown | GRMZM2G140994 | 1.34 | 1.55 | 1.41 |
| TC293598 | DNAJ-like protein | GRMZM2G119316 | 1.54 | 1.73 | 1.31 |
| DT644608 | DNAJ heat-shock protein | GRMZM2G098058 | 1.50 | 2.26 | 1.27 |
| TC313845 | Heat-shock protein | GRMZM2G361605 | 1.13 | 1.56 | 1.27 |
| TC282722 | RNA recognition motif (RRM)-containing protein | GRMZM2G125529 | 1.31 | 1.66 | 1.24 |
| TC290709 | Outward-rectifying potassium channel | GRMZM2G351342 | 1.85 | 2.00 | 1.11 |
| TC279806 | HSP70 | GRMZM2G351416 | 1.59 | 2.01 | 1.00 |
| TC288130 | HP8 peptide | GRMZM2G037015 | −3.21 | −2.60 | −2.93 |
| TC307533 | Ethylene-responsive transcription factor 3 | GRMZM2G020054 | −2.95 | −2.64 | −2.13 |
| TC283547 | Cytokinesis regulating protein-like | GRMZM2G404126 | −1.06 | −1.55 | −1.37 |
In terms of dissecting the network of maize responses to UV-B, it is important to note that although the total number of UV-B-regulated transcripts after 10 min of irradiation is fewer than after 60 min in all tissues analyzed (Figure 2), there is a higher proportion of overlap in transcripts between the different organs changed after 10 min than after longer exposure times (Figures 2D,E; Files S4–S6 in Supplementary Material). We hypothesize that early signaling in different organs is elicited by common signaling pathways, while at longer exposure times, responses and their coordination becomes more organ-specific.
Originally, a panel of genes showing differential regulation after 1 h of UV-B irradiation in a long time course experiment (Casati et al., 2011a) was selected; and primers were designed for qRT-PCR assays to pick shorter time points for the current microarray experiment presented in this work. Thus, to validate the microarray results, we compared the expression patterns of a subset of these transcripts representing types that are on, off, up- or down-regulated by the short UV-B treatments in microarray experiments and qRT-PCR assays (Table 4). For most transcripts, there is a very good correlation between the results obtained using both techniques. However, in some cases (for example, Xylanase Inhibitor I at time 30 min) there are some differences. We think that differences may be due to the use of different techniques, as microarray hybridization may be detecting more than one transcript from the same gene family, so the expression pattern measured can be an average of the expression of two or more genes, while qRT-PCR was done using primers that are specific for only one probe. Despite this, most values from qRT-PCR correspond closely in magnitude to the microarray results for these transcripts, demonstrating that the microarray data are highly reproducible and the qRT-PCR assay was effective in picking significant time points for microarray analysis. Additionally, where a value from one of the channels is missing, we were still able to confirm directional patterns of expression from the microarray. For instance, for 70 kDa peptidylprolyl isomerase, the pattern on the microarray is “off” at 0 min and “on” at 10 min. The qPCR for 10 min UV-B confirms this result by being up-regulated and thus turned on with an expression ratio of 1.86 relative to the no treatment control.
Table 4.
Confirmation of microarray data by qRT-PCR assays.
| Microarray pattern after UV | Accession number | Description | log2 (10 min UV-B/C) |
log2 (30 min UV-B/C) |
||
|---|---|---|---|---|---|---|
| qRT-PCR | array | qRT-PCR | Array | |||
| Up, Off/On or increased | BM500597 | Phosphoethanolamine N-methyltransferase | 0.53 | N.D. | 1.26 | 0.73 |
| TC292940 | 70 kDa peptidylprolyl isomerase | 1.86 | OffOn | 1.76 | OffOn | |
| AF112150 | MADS box protein 3 | 2.23 | 1.52 | 2.43 | 2.07 | |
| TC280980 | Xylanase Inhibitor I | 0.50 | N.D. | 1.86 | 0.70 | |
| Down, On/Off or Unchanged | AW129897 | CLN3 protein | −0.81 | −0.69 | 0.14 | N.D. |
| TC308488 | NADP-dependent leukotriene B4 12-hydroxydehydrogenase | −2.13 | OnOff | 0.74 | N.D. | |
| TC303498 | LigA | −3.47 | OnOff | −0.53 | N.D. | |
| CB278279 | TFIIF-alpha family protein | −1.22 | OnOff | −0.55 | N.D. | |
| TC301764 | Male sterility MS5 family protein | −1.35 | N.D. | −0.99 | −1.01 | |
(1) Log2 ratio for both qPCR and microarray (p-values < 0.05): all of these are same “direction” and reasonably concordant, as explained above. (2) Log2 ratio for qPCR and pattern for microarray (OffOn or OnOff): we are missing a reading for either the control or 10/30 m timepoint, but the qPCR still shows whether or not we are “on” or “off” at 10 or 30 m (all of this category match). (3) ND (no data): a p-value > 0.05, too high to have confidence and thus not included in the final result.
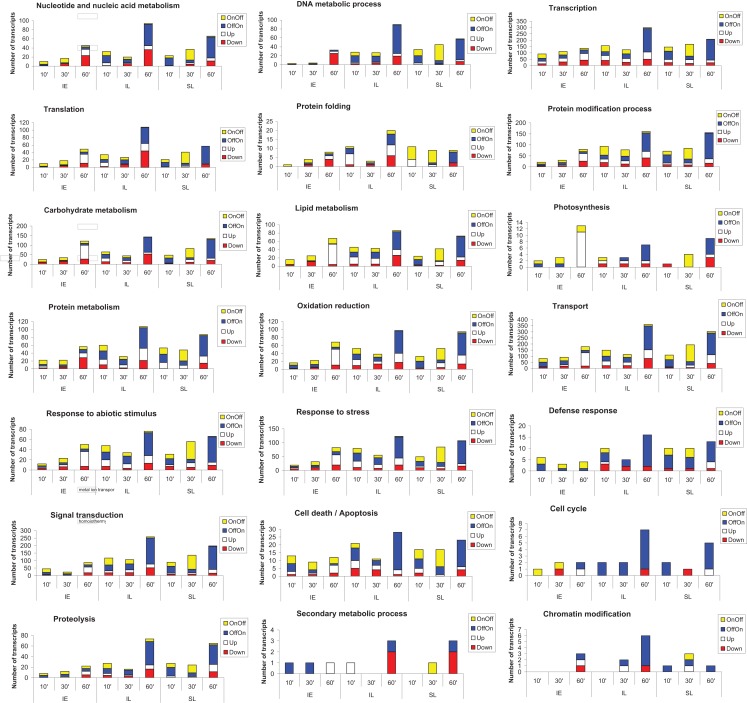
Go classification of transcripts
The impact of UV-B on 18 major cellular processes was assessed by GO classification of transcripts from four expression categories (those that were turned on or off, or that were up- or down-regulated versus the non-irradiated control) for each of the three organs analyzed (IL, SL, and IE) in the 1 h time point (Figure 3). UV-B perception and systemic signaling to shielded organs has a major impact in all 18 GO categories. The proportion of transcripts in each of the four expression types is distinctive for irradiated leaf, shielded leaf, and immature ears. The transcription/transcriptional regulation category has the highest representation (Figure 3). As mentioned above, there are three common TFs that are UV-B-regulated at 10 min in all three organs, and at 30 and 60 min differential expression of more tissue-specific transcription factors is evident. In the transcription category, the number of mRNAs is significantly increased after 1 h UV-B in all tissues, prerequisite to the major reprogramming of gene expression that occurs over the following hours in continuous UV-B (Casati et al., 2011a). For the signal transduction category, after 10 min irradiation, the number of transcripts changed is higher in leaves than in ears, with more transcripts changed in IL than in SL. We conclude that UV-B-signaling is more prominent in tissues that are directly exposed to UV-B than in shielded ones.
Figure 3.
GO classification of transcripts into categories. Transcripts that were turned on (OffOn) or off (OnOff), or that were up- or down-regulated over the 1 h time course experiment were used. Transcripts that belonged to 18 major cellular processes were used for the classification.
Transport is another category showing a significant number of UV-B-regulated transcripts at short times (Figure 3). This category comprises proteins that participate in various types of transport, for example protein (GO:0006886), ion (GO:0006813), vesicle-mediated (GO:0016192), or dicarboxylic acid transport (GO:0006835); signaling components of the UV-B cascade are probably transported by at least some of these proteins.
It is interesting that transcripts that encode genes in secondary metabolism are not significantly represented in our classification for 10, 30, and 60 min exposure times. Many secondary metabolism-associated transcripts, including mRNAs for enzymes in flavonoid metabolism, are induced after longer exposure times (Casati et al., 2011a). A similar result is observed for other categories, such as DNA metabolism including DNA repair. It is clear that in completely shielded IE, that never receive any direct UV-B, there are only a few transcripts in this group (Figure 3). In SL, although there are more mRNAs represented in this category than in IE, this number is lower than in IL, especially when comparing the 60 min time point. Transcripts involved in DNA repair metabolism are induced at longer times of exposure, when damaged DNA is accumulated, and this mostly occurs in organs that are directly irradiated with UV-B (Casati et al., 2011a).
Identification of UV-B-induced metabolomic changes
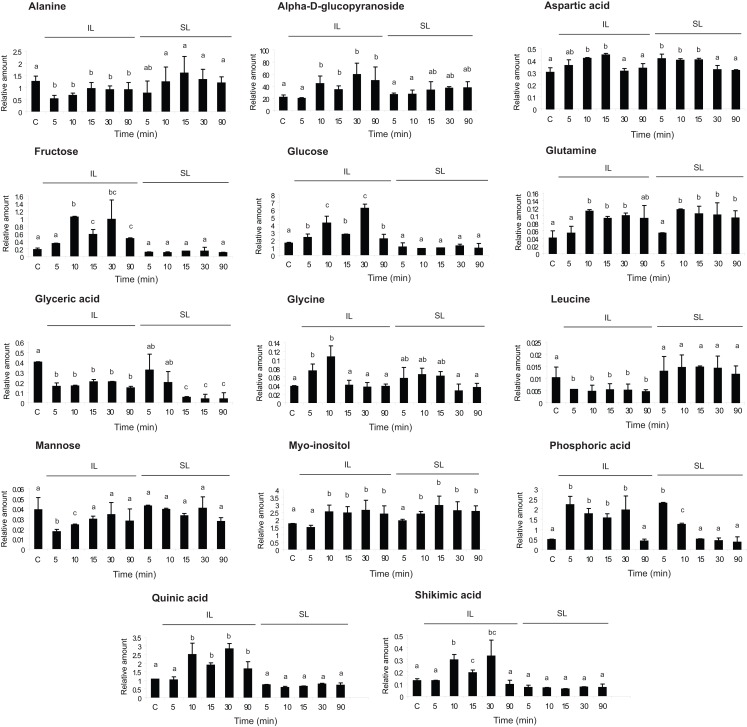
To identify potential signal molecules that move quickly from irradiated leaves to shielded organs, we conducted metabolic profiling using GC–MS (see Materials and Methods). Because transcriptome analysis identified changes within 10 min, metabolite samples were analyzed after 5, 10, 15, 30, and 90 min of UV-B irradiation for comparison to untreated control plants (C). We identified 84 compounds, and 14 of these had a statistically significant change by UV-B in at least one time point (Figure 4, one way ANOVA).
Figure 4.
Metabolic profiling from UV-B-irradiated leaves. Metabolites from maize leaves from irradiated (IL) and shielded (SL) leaves from maize plants that were covered with a plastic sheath that absorbs UV-B, and only two adult leaves per plant were irradiated with UV-B radiation during 5, 10, 15, 30, and 90 min. As a control, samples from non-irradiated leaves (C) were included. Statistical analysis was done using one way ANOVA; statistically significant differences are labeled with letters a, b, c, and d (α = 0.05).
Five metabolites were increased in both IL and SL (aspartic, phosphoric, and glyceric acids, glutamine, and myoinositol, Figure 4) while nine metabolites were restricted to irradiated leaves: alanine, an alpha-d-glucopyranoside, fructose, glucose, glycine, leucine, mannose, shikimic acid, and quinic acid (Figure 4). The last two compounds are in the phenylpropanoid pathway, suggesting that the synthesis of some phenylpropanoid sunscreens initiates only in exposed tissues. Metabolites restricted to irradiated leaves are not translocated to shielded tissues nor do mobile signals induce them in shielded organs, in accordance with our previous results (Casati et al., 2011a). In contrast, metabolites modulated by UV-B in both irradiated and shielded leaves are potential signal molecules synthesized in exposed leaves and translocated to shielded organs; or alternatively, an unknown signal could be transmitted to shielded tissues, and this signal could induce the synthesis of these compounds in shielded tissues. Myoinositol is of particular interest in light of our previous microarray results (Casati et al., 2011a). We reported that transcripts for myoinositol-1-phosphate synthase were down-regulated by UV-B in both IL and SL after 4 h of UV-B irradiation (Casati et al., 2011a), which would be predicted to increase the levels of the precursor myoinositol. Either lowered levels of myoinositol-1-phosphate or elevated myoinositol could be signaling molecules coordinating UV-B responses. Figure 4 shows that myoinositol levels are rapidly increased after 10 min of UV-B both in IL and SL, confirming it as a UV-B-signaling candidate. There are additional metabolites that show changes in both IL and SL; these are intermediates of primary metabolism. We hypothesize that these are unlikely to be specific signals but probably reflect global metabolic changes that are induced by UV-B.
Putative role of myoinositol or a myoinositol derivative in UV-B-signaling in maize
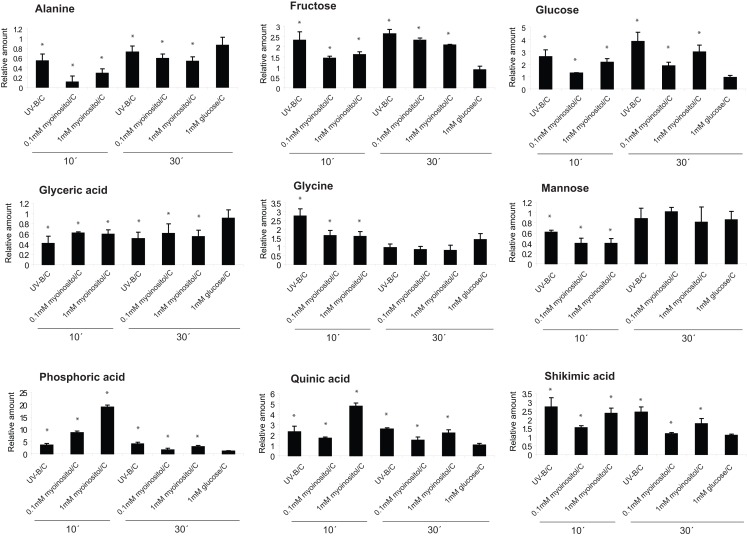
To evaluate the role of myoinositol as a candidate mobile signal, we applied different concentrations of myoinositol (10–0.01 mM) using a soaked paper towel resting on a canopy leaf (see Materials and Methods); then metabolome changes were analyzed in the treated leaf zone, and in the neighboring more mature untreated leaf from the same plant after 10 and 30 min. The control was a plant in which a water-soaked towel was applied. Metabolomic changes after 10 and 30 min of a 0.1 and 1-mM myoinositol treatment were assayed in treated and untreated leaves; and they were compared to those elicited by UV-B. Figure 5 shows that, of the 14 metabolites with altered levels after 10 min of UV-B in Figure 4, nine are similarly altered after the myoinositol treatment. A parallel treatment with 1 mM glucose elicited none of these changes (Figure 5). Therefore, myoinositol can elicit a subset of the metabolic effects of UV-B; however, it is probable that additional compounds contribute to systemic signaling.
Figure 5.
Effect of myoinositol in UV-B-regulated metabolites. Metabolites changed from maize leaves treated with 0.1 and 1 mM myoinositol compared with a control leaf that was soaked with water; and also after UV-B irradiation for 10 and 30 min. As a control, samples from leaves that were treated with 1 mM glucose were used. Statistical analysis was done using Student T-test; statistically significant differences are labeled with * (p < 0.05).
Discussion
Under normal solar fluence, UV-B damages macromolecules but it also elicits physiological and developmental changes in plants. Previously, we found that leaves exposed to UV-B ∼three-fold higher than ambient solar noon for brief periods (1–2 h) generate signals that result in transcriptome changes in completely shielded distant organs such as immature ears and leaves wrapped in UV-B filters (Casati and Walbot, 2004). In addition, as a follow up experiment, we used transcriptome, proteome, and metabolic profiling to quantify maize canopy responses and changes in shielded leaves and immature ears, and track the kinetics of alterations in exposed and shielded organs (Casati et al., 2011a). Our results demonstrated that exposure of just the top leaf substantially altered the transcriptome of both irradiated and shielded organs, with greater changes as additional leaves were irradiated (Casati et al., 2011a). We found that some phenylpropanoid pathway genes were expressed only in irradiated leaves, reflected by the accumulation of some phenylpropanoid precursors only in these leaves. Moreover, UV-B-regulated transcriptome, proteome, and metabolome changes occurred in shielded organs within 1 h; candidates in early steps of signal transduction and possible signal molecules were identified utilizing a time course experiment. To define the most rapid canopy and shielded organ responses that occur before 60 min, we now report a transcriptome and metabolome study over a shorter time course from 5 to 90 min of UV-B irradiation. Tracking response kinetics to elevated UV-B in irradiated leaves and identifying the signals produced there that subsequently elicit systemic changes in reproductive organs should elucidate how UV-B decreases plant yield beyond what is predicted from the modest impact on photosynthesis.
The first question in our experiments was to identify transcripts that encode candidates in early UV-B-signaling in irradiated and shielded tissues. After 10 min of UV-B exposure, 262 transcripts are changed by at least two-fold (p < 0.05) in irradiated leaves, and this number doubles after 1 h (Figure 2A). Indicative of the rapid modulation of transcription, 130 transcripts in this list are only changed after 10 min. This is true not only in IL, but also in shielded tissues such as leaves and immature ears, where there are 110 and 49 mRNAs, respectively, with significant changes after 10 min, but not at longer UV-B exposure times (Figures 2B,C). Twenty-four of these rapid responses are shared in the three organs studied (Figure 2D; Table 3), and seven of them correspond to HSP. We previously found that a number of HSPs were down-regulated by UV-B after longer exposure times in IE (Casati et al., 2011a). Together, our results suggest that changes in the expression levels of this group of proteins may have an important role in UV-B responses, both in irradiated and shielded tissues, and transient changes in their levels would be important for acclimation to this radiation.
After 10 min of exposure, the overlap in transcriptome changes in irradiated and shielded leaves is significant: 98 transcripts show similar UV-B regulation (Figure 2D); however, in SL, the number of UV-B-regulated transcripts after this short irradiation time is about 1.8-fold lower than in IL. In the list of overlapping UV-B-regulated transcripts between IL and SL, there are a number of transcription factors that probably participate in early UV-B-signaling in leaves and UVR8 which has been recently proposed as a putative UV-B photoreceptor in Arabidopsis (Table 2, Rizzini et al., 2011). UVR8 is a UV-B-specific signaling component that mediates low fluence photomorphogenic responses, and it is required for UV-B-induced expression of the gene encoding the HY5 transcription factor (Ulm et al., 2004; Brown et al., 2005) in co-operation with COP1. UVR8 and COP1 interact directly and rapidly in the nucleus after UV-B exposure (Oravecz et al., 2006); this is a very early step in UV-B-signaling responses, ensuring UV-B acclimation and protection (Favory et al., 2009). In our previous microarray experiments at longer UV-B exposure times, maize UVR8 is down-regulated by UV-B both in IL and SL (Casati et al., 2011a). If UVR8 is a UV-B sensor in maize, turning down responses appears to be important for successful acclimation. Nonetheless, at shorter irradiation times (less than an hour), maize UVR8 is up-regulated by UV-B in IL and SL (Casati et al., 2011a, this work). We hypothesize that this may be an acclimation to increase subsequent sensitivity to UV-B or an indication that UVR8 has functions that do not require direct UV-B perception.
After 30 min of UV-B treatment, there are only two transcripts showing similar UV-B regulation between the three organs; 35 are similarly regulated in both IL and SL. Therefore, at longer irradiation times, there is an increasing proportion of organ-specific responses. An important result of our experiments is that, even though the total number of UV-B-regulated transcripts after 10 min of UV-B is lower than after 60 min in all samples analyzed (Figure 2), the proportion of overlap in transcriptome changes between organs is more prominent after 10 min than after longer exposure times (Figures 2D,E). We suggest that early signaling in different tissues may be elicited by common signaling pathways, while at longer exposure times responses become more specific.
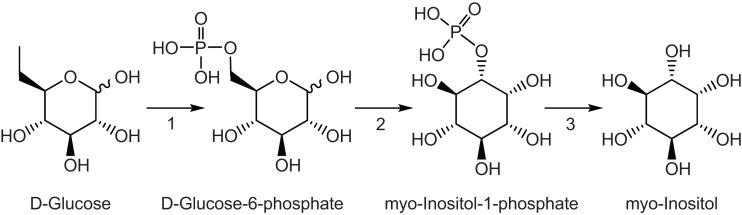
The second aim of this work was to identify potential molecules that could transmit the UV-B signals from irradiated leaves to shielded organs. Because a signaling metabolite(s) must increase quickly in irradiated leaves to trigger transcriptome changes in shielded organs, we predicted that such molecules would show high concentrations relative to untreated plants and increase in shielded organs. By GC–MS we identified 14 metabolites that showed differential levels by UV-B at exposure times shorter than 90 min (Figure 4). Of these, five metabolites were increased both in IL and SL (aspartic, phosphoric and glyceric acid, glutamine, and myoinositol, Figure 4). These are potential UV-B signal molecules that could be translocated from exposed leaves or an unknown mobile signal triggers in situ synthesis in shielded leaves. Previously, we found that levels of myoinositol were increased after a 4 h UV-B treatment in irradiated and shielded leaves and proposed it as a candidate UV-B-signaling compound (Casati et al., 2011a). Myoinositol, ubiquitous in most organisms, is already known to participate in stress responses. For example, in salt- or cold-tolerant plant species, myoinositol biosynthesis plays a role in protection (Bohnert et al., 1995). This molecule is ubiquitous in organisms. This sugar alcohol is synthesized from glucose in three steps: first glucose is phosphorylated by hexokinase, then, glucose-6-P is converted to myoinositol-1-P by the myoinositol-1-phosphate synthase, and this intermediate is finally dephosphorylated by a phosphatase to produce myoinositol (Figure 6). The step catalyzed by myoinositol-1-phosphate synthase is the rate-limiting step for myoinositol biosynthesis in plants (Loewus and Murthy, 2000). In our previous experiments, transcripts for a myoinositol-1-phosphate synthase were found to be decreased by UV-B (Casati et al., 2011a). Thus, if myoinositol acts as a UV-B-signaling molecule, rapid synthesis at shorter times of exposure is expected, while longer times of irradiation would provoke a down-regulation of the gene to moderate the amount of this signaling molecule.
Figure 6.
Myoinositol pathway in plants. 1, hexokinase; 2, myoinositol-1-phosphate synthase; 3, phosphatase.
Consequently, to further investigate the role of myoinositol in UV-B-signaling in maize, we tested whether it could mimic the effect of UV-B by applying different myoinositol concentrations on maize leaves, and we compared metabolite changes elicited by this compound with those induced by UV-B radiation. We found that myoinositol induces similar changes in the levels of nine metabolites, paralleling UV-B in both treated (like a UV-B canopy leaf) and untreated (like a shielded organ) leaves (Figure 5). Therefore, myoinositol can partially mimic UV-B irradiation. We hypothesize that additional compounds are required to reconstitute full systemic signaling that shows all the hallmarks and specificity of UV-B-induced acclimations. Identifying and testing candidate signal molecules are major goals for future research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this artcle can be found online at http://www.frontiersin.org/Plant_Genetics_and_Genomics/10.3389/fpls.2011.00033/abstract
Excel file containing list of transcripts that are UV-B-regulated in irradiated leaves from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to non-irradiated leaves by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in shielded leaves from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to non-irradiated leaves by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in immature ears from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to ears from non-irradiated plants by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 10 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 30 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0.05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 60 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Acknowledgments
We thank Mónica Hourcade for GC/MS technical support, Tato Rodriguez for helping with Figures preparation, and Scott Loh for setting up the UV-B apparatus and moving the plants into it. Research supported by USDA National Research Initiative Grant 2008-35100-04578 to Virginia Walbot and by FONCyT grants PICT-2006-00957 and PICT-2007-00711 to Paula Casati. Paula Casati is a member of the Researcher Career of CONICET.
Footnotes
References
- Bieza K., Lois R. (2001). An Arabidopsis mutant tolerant to lethal ultraviolet- B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 126, 1105–1115 10.1104/pp.126.3.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanding C. R., Simmons S. J., Casati P., Walbot V., Stapleton A. E. (2007). Coordinated regulation of maize genes during increasing exposure to ultraviolet radiation: identification of ultraviolet-responsive genes, functional processes and associated potential promoter motifs. Plant Biotechnol. J. 5, 677–695 10.1111/j.1467-7652.2007.00282.x [DOI] [PubMed] [Google Scholar]
- Bohnert H. J., Nelson D. E., Jensen R. G. (1995). Adaptations to environmental stresses. Plant Cell 7, 1099–1111 10.2307/3870060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A. B. (1996). DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 4, 75–100 10.1146/annurev.arplant.47.1.75 [DOI] [PubMed] [Google Scholar]
- Brown B. A., Cloix C., Jiang G. H., Kaiserli E., Herzyk P., Kliebenstein and D. J., Jenkins G. I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230 10.1073/pnas.0504954102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Campi M., Morrow D. J., Fernandes J. F., Walbot V. (2011a). Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12, 10.1186/1471-2164-12-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Campi M., Morrow D. J., Fernandes J., Walbot V. (2011b). Transcriptomic, proteomic and metabolomic analysis of maize responses to UV-B: comparison of greenhouse and field growth conditions. Plant Signal. Behav. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2003). Gene expression profiling in response to ultraviolet radiation in Zea mays genotypes with varying flavonoid content. Plant Physiol. 132, 1739–1754 10.1104/pp.103.022871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2004). Rapid transcriptome responses of maize (Zea mays) to UV-B in irradiated and shielded tissues. Genome Biol. 5, R16. 10.1186/gb-2004-5-3-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2005). Differential accumulation of maysin and rhamnosylisorientin in leaves of high altitude landraces of maize after UV-B exposure. Plant Cell Environ. 28, 788–799 10.1111/j.1365-3040.2005.01329.x [DOI] [Google Scholar]
- Casati P., Walbot V. (2008). Maize lines expressing RNAi to chromatin remodeling factors are similarly hypersensitive to UV-B radiation but exhibit distinct transcriptome responses. Epigenetics 3, 216–229 10.4161/epi.3.4.6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory J.-J., Stec A., Gruber H., Rizzini L., Oravecz A., Funk M., Albert A., Cloix C., Jenkins G. I., Oakeley E. J., Seidlitz H. K., Nagy F., Ulm R. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601 10.1038/emboj.2009.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133, 1420–1428 10.1104/pp.103.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr K. M., Churchill G. A. (2001). Statistical design and the analysis of gene expression microarray data. Genet. Res. 77, 123–128 [DOI] [PubMed] [Google Scholar]
- Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmuller E., Dormann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A. R., Steinhauser D. (2005). GMD@CSB. DB: the golm metabolome database. Bioinformatics 21, 1635–1638 10.1093/bioinformatics/bti236 [DOI] [PubMed] [Google Scholar]
- Landry L. G., Stapleton A. E., Lim J., Hoffman P., Hays J. B., Walbot V., Last R. L. (1997). An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl. Acad. Sci. U.S.A. 94, 328–332 10.1073/pnas.94.1.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ou-Lee T.-M., Raba R., Amundson R. G., Last R. L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B radiation. Plant Cell 5, 171–179 10.1105/tpc.5.2.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A. (2006). Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 1, 387–396 10.1038/nprot.2006.59 [DOI] [PubMed] [Google Scholar]
- Loewus F. A., Murthy P. P. (2000). Myo-inositol metabolism in plants. Plant Sci. 150, 1–19 10.1016/S0168-9452(99)00150-8 [DOI] [Google Scholar]
- Oravecz A., Baumann A., Maté Z., Brzezinska A., Molinier J., Oakeley E. J., Adam E., Schafer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975–1990 10.1105/tpc.105.040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.-J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G. I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 10.1126/science.1200660 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. J. (2000). “Primer3 on the WWW for general users and for biologist programmers,” in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds Krawetz S., Misener S., Totowa N. J. (Totowa, NJ: Humana Press; ), 365–386 [DOI] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M. G., Willmitzer L., Fernie A. R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 10.1016/j.febslet.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Skibbe D. S., Fernandes J. F., Medzihradszky K. F., Burlingame A. L., Walbot V. (2009). Mutator transposon activity reprograms the transcriptomes and proteomes of developing maize anthers. Plant J. 59, 622–633 10.1111/j.1365-313X.2009.03901.x [DOI] [PubMed] [Google Scholar]
- Smyth G. K. (2005). “Limma: linear models for microarray data,” in Bioinformatics and Computational Biology Solutions using R and Bioconductor, eds Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W. (New York: Springer; ), 397–420 [Google Scholar]
- Stapleton A. E., Walbot V. (1994). Flavonoids can protect maize DNA from the induction of ultraviolet-radiation damage. Plant Physiol. 105, 881–889 10.1104/pp.105.3.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Maté Z., Adam E., Oakeley E. J., Schafer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1397–1402 10.1073/pnas.0308044100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fernald R. (2005). Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12, 1045–1062 10.1089/cmb.2005.12.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file containing list of transcripts that are UV-B-regulated in irradiated leaves from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to non-irradiated leaves by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in shielded leaves from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to non-irradiated leaves by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in immature ears from plants that were irradiated during 10, 30, and 60 min in 2 leaves in comparison to ears from non-irradiated plants by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 10 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 30 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0.05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.
Excel file containing list of transcripts that are UV-B-regulated in irradiated and shielded leaves, and immature ears from plants that were irradiated during 60 min in 2 leaves in comparison to non-irradiated samples from the same organs by two-fold (p < 0. 05). Up-regulated transcripts by two-fold are in red, while down-regulated transcripts by two-fold are in green.