Abstract
PTEN (phosphatase and tensin homolog deleted on chromosome 10) functions as a major tumor suppressor gene and is frequently deleted in different types of tumors including prostate cancer (PCa). It was hypothesized that germ-line genetic changes of PTEN affect susceptibility to PCa. Both common (with a minor allele frequency ≥5%) and rare (with a minor allele frequency <5%) germ-line variants of PTEN were comprehensively evaluated. A total of 15 germ-line variants were identified by re-sequencing the PTEN gene, including 5′ untranslated region, all nine exons, exon-intron junctions and 3′ untranslated region, in 188 probands of hereditary prostate cancer (HPC) families recruited from Johns Hopkins Hospital. Two microsatellite markers surrounding PTEN were used to test the co-segregation of 10 rare variants, which may give rise to highly penetrant in HPC. Two common single nucleotide polymorphisms (SNPs) were evaluated in the 188 HPC families using a family-based association study approach. To study low penetrant SNPs in PCa susceptibility, 33 SNPs covering PTEN were selected from the whole genome-wide association studies (GWAS) from our available case-control studies in Sweden (Cancer of the Prostate in Sweden (CAPS)) and the publicly available cancer genetic markers of susceptibility (CGEMS) study. Germ-line copy-number variations (CNVs) in PTEN were assessed in CAPS. Co-segregation of germ-line variants and PCa was not observed among HPC families and no significant differences in the allele frequencies were observed in sporadic cases and controls, aggressive and non-aggressive PCa (P >0.05). These results suggest that germ-line variants in PTEN do not have an important role in PCa susceptibility.
Keywords: co-segregation, family-based association, population-based association, prostate cancer, PTEN, sequencing
INTRODUCTION
Located on chromosome 10q23, PTEN is frequently deleted in multiple tumors including prostate cancer (PCa). PTEN has been shown to be involved in many cellular functions including apoptosis, cell-cycle control, cell adhesion and migration; however, the primary function of PTEN is to dephosphorylate and inactivate the second messenger PIP3.1 It exerts its tumor suppressor function by acting as a negative regulator in the PI3K signaling pathway.1 Loss of PTEN has been shown to contribute to PCa development in mouse models,2–5 and mutations or deletions of PTEN have been found to occur frequently in PCa in human, especially in advanced disease.6–10
Deleterious germ-line variants in PTEN were not found to contribute to PCa susceptibility in previous studies, including mutation screening, linkage studies and genome-wide association (GWA) studies in case–control sporadic PCa populations.11–16 Some limitations of these studies were small sample size, low-coverage of the PTEN gene for mutation screening and the lack of evaluation of co-segregation of PTEN germ-line variants and PCa risk. In this report, a comprehensive assessment of germ-line variants in the PTEN gene for PCa susceptibility was carried out, which included mutation screening of a sizeable PTEN region in a large hereditary prostate cancer (HPC) sample, co-segregation analysis of high penetrant variants (rare variants with a minor allele frequency ≥5%) in HPC families, association analysis of low penetrant variants (common single nucleotide polymorphisms (SNPs) with a minor allele frequency <5%) in sporadic PCa and evaluation of copy-number variations (CNVs) in PTEN in sporadic PCa populations.
MATERIALS AND METHODS
Subjects
Hereditary prostate cancer (HPC) families
A detailed description of the HPC study families has been presented in previous work.17 In total, 188 HPC families with at least three first-degree relatives having a diagnosis of PCa were recruited into the study by investigators at the Brady Urology Institute at Johns Hopkins Hospital (JHH, Baltimore, MD, USA). The mean age at diagnosis for these individuals was 64.3 years. Approximately 91.8% of the families were Caucasian and 9.2% were Black. The average number of affected individuals per family was 5.08. The number of families with 3, 4 and 5 or more affected individuals was 28, 47 and 113, respectively. Probands were selected based on the following priorities: affected member with metastatic disease, local invasion, highest Gleason score, highest preoperative prostate-specific antigen (PSA) levels and youngest age at diagnosis.
JHH case–control population
The 1527 cases were from a consecutive series of men who underwent treatment for PCa at JHH. Diagnosis of PCa of the cases was confirmed by pathology reports. Paraffin-embedded tissue and frozen tissue were available for 95 and 35% of cases, respectively. The mean age, PSA, Gleason Score and stage for the cases were 58.5 years, 7.9 ng ml−1, 6.8 and 1.5, respectively. The 482 unaffected controls were recruited from a series of consecutive men that participated in PCa screenings at the Brady Urological Institute at JHH. Mean age and PSA for the controls were 57.5 years and 1.6 ng ml−1. DNA from blood samples was available for all cases and controls.
Cancer of the prostate in Sweden (CAPS) population
The population-based, case–control study in Sweden, named Cancer Prostate in Sweden (CAPS) was used to complete fine-mapping in the study.18 Cases with pathologically diagnosed PCa were recruited from regional cancer registries in Sweden, between July 2001 and October 2003. DNA from blood samples, tumor-node metastasis stage, Gleason grade (biopsy) and PSA levels were available for 2899 cases. Controls were recruited concurrently from the Swedish Population Registry and matched with cases according to age and geographic region. DNA from blood samples was available for 1722 controls. Subsets of the CAPS population were used for two genome-wide association study (GWAS) studies: GASP1 and GASP2. GASP1 included 500 cases and 500 controls and GASP2 included 1500 cases and 500 controls.
Cancer genetic markers of susceptibility (CGEMS) population
The published data from the National Cancer Institute (CGEMS) study was also used.19,20 The study population was the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial and it included 1177 cases and 1105 controls.
Sequencing and genotyping of SNP and microsatellite markers
The entire genomic sequence of PTEN was re-sequenced, including the ~2kb region of the promoter, all coding regions, the 3′ and 5′ untranslated regions, and regions that were evolutionarily conserved from multiple species comparison in probands (n=188) of the HPC families. Figure 1 outlines the gene organization of PTEN and Table 1 lists the primers used to amplify the PCR products, the sizes of amplified PCR fragments and the annealing temperature for each pair of primers. All reactions were performed using Taq polymerase (Invitrogen, Carlsbad, CA, USA) at a 10-µl scale consisting of 20 ng genomic DNA, 0.2 °moll−1 of each primer, 0.2 mmoll−1 of each deoxynucleotide triphosphate, 1.5 mmoll−1 MgCl2, 20 mmoll−1 Tris-HCl, 50 mmoll−1 KCl and 0.5 units Taq polymerase (Invitrogen). PCR cycling conditions were as follows: 94 °C for 3 min followed by 33 cycles of 94 °C for 30 s, a specified annealing temperature for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 6 min. The PCR products were purified using the QuickStep PCR Purification kit (Edge BioSystems, Gaithersburg, MD, USA) to remove deoxynucleotide triphosphates and excess primers. All sequencing reactions were carried out using dye terminator chemistry (BigDye, ABI, Foster City, CA, USA) and then precipitated using 75 ± 5% ethanol. Samples were loaded onto an ABI 3700 DNA analyzer after adding 8 µl formamide. SNPs were identified using Sequencher software version 4.0.5 (Gene Codes, Ann Arbor, MI, USA).
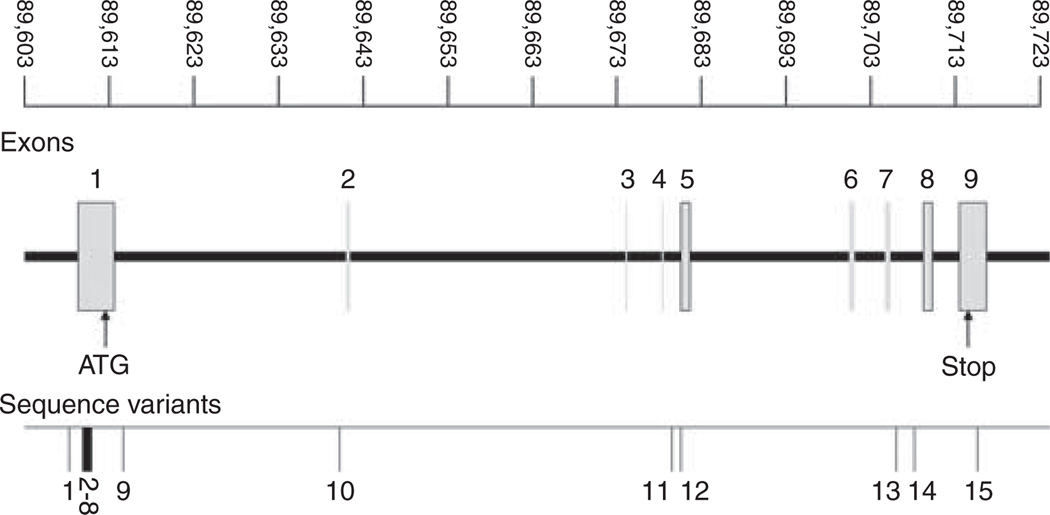
Figure 1.
PTEN gene structure with sequence variants indicated (map distance in kb); 15 sequence variants were found. Physical location: 89613715–89 718 382bp, NCBI Build 35.
Table 1.
Primers used for mutational analysis and SNPs identification in PTEN (89 613 715–89718 382 bp), NCBI Build 35
| PCR ID | Forward primer (5′-3 ′) | Reverse primer (5′-3 ′) | PCR product (bp) | Annealing temperature |
|---|---|---|---|---|
| 1 | AAGGGGGAATCTCTAGGCAA | AGCATGCCCAGTGTAGCTG | 453 | 62 |
| 2 | AGCAAGTGCAGCTGCAGG | AGAGGCTGCACGGTTAGAAA | 579 | 62 |
| 3 | CTCTCAGCGCCTGTGAGC | GGACTTGGCGGTAGCTGAT | 638 | 62 |
| 4 | CCAGTCGCTGCAACCATC | TGCAACCAGGCAAGAGTTC | 473 | 68 |
| 5 | CTGTCCATGTGGAAGTTACCTT | CTGTATCCCCCTGAAGTCCA | 460 | 62 |
| 6 | CCATAGAAGGGGTATTTGTTGG | CATGAATCTGTGCCAACAATG | 420 | 58 |
| 7 | AAAGATTCAGGCAATGTTTGTT | ATGCTGCACTTTAGTCTTCCTG | 350 | 60 |
| 8 | TTTCTCTGGAATCCAGTGTTTC | CAGATCCAGGAAGAGGAAAGG | 413 | 60 |
| 9 | GCTACGACCCAGTTACCATAGC | CCAATGAGTTGAACAACAAAGC | 404 | 60 |
| 10 | TTGCTTGAGATCAAGATTGCA | CTCACCAATGCCAGAGTAAGC | 446 | 60 |
| 11 | TTTTGTTGACTTTTTGCAAATG | GTCAAGCAAGTTCTTCATCAGC | 442 | 62 |
| 12 | AAGAGGGTCATTTAAAAGGCC | TCTGACACAATGTCCTATTGCC | 453 | 60 |
| 13 | CCAGAGAATGAACCTTTTGATG | ACCCTTCGGAAACCTCTCTTAG | 520 | 68 |
| 14 | GTTCTGTCACCAACTGAAGTGG | AGCATCCACAGCAGGTATTATG | 679 | 63 |
Genotyping for the two common SNPs (rs12573787 and rs1903858) was carried out using the MassARRAY system (Sequenom, San Diego, CA, USA) in the 188 HPC families for which DNA samples were available. The SNP rs1903858 was selected to be genotyped in an additional 1527 sporadic PCa cases and 482 unaffected controls from the JHH case–control population. PCR and extension reactions were carried out according to the manufacturer’s instructions and extension product sizes were determined by mass spectrometry.
The microsatellite markers D10S1686 and D10S185, which lie on either side of the PTEN gene in the region of 100.5 ~102.5 cM and span an interval of ~9.6 Mb, were genotyped in HPC families with rare sequence variants to estimate the co-segregation of sequence variants with PCa in HPC families. The genotyping method has been previously described in detail.21
SNPs in the PTEN gene selected from genome-wide association study (GWAS)
To study low penetrant SNPs in PCa susceptibility, 33 SNPs within the PTEN gene were selected from the GWAS of GASP1, GASP2 and CGEMS (see Figure 2). Data from the three existing GWAS studies, which were deposited from Hapmap (http://www.hapmap.org, rel#21, dbSNPb125) was used to test the 33 SNPs spanning the complete PTEN gene (105.3 kb), GeneChip Human Mapping 500k Array, 5.0 Array (Affymetrix, Santa Clara, CA, USA) and HumanHap 300 BeadChips (Illumina, San Diego, CA, USA) were used for GASP1, GASP2 and CGEMS.19,22 The detection of SNPs and CNVs in the CAPS population by means of Affymetrix SNP has been previously described in detail.22,23 Results for a subset of the 33 SNPs spanning PTEN were obtained from the GWAS (results from GASP1, GASP2 and CGEMS) and were used for the imputation of the remaining SNPs.
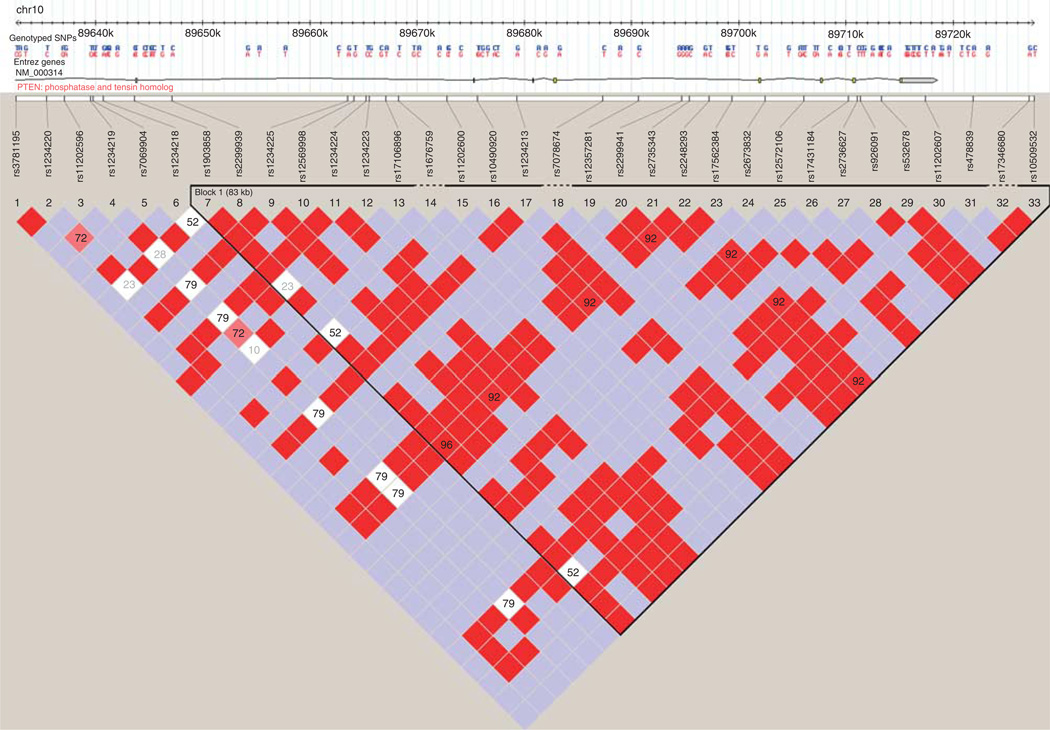
Figure 2.
33 SNPs spanning PTEN (89 623 195–89 728 531 bp, NCBI build 37); Color scheme is based on standard D′/LOD score option in Haploview 4.1.
Statistical methods
Family-based association tests (FBAT) were performed for two SNPs (rs12573787 and rs1903858) in HPC families including 1286 affected and unaffected members, using the software package FBAT.24 Linkage and linkage disequilibrium between PCa and SNPs were assessed by FBAT based on the data from nuclear families, sibships or a combination of the two.
Parametric and non-parametric analyses were carried out to test the co-segregation of the sequence variants in the HPC families using GeneHunter Plus. In the parametric analysis, the frequency of the susceptibility allele was assumed to be 0.003, and penetrance for non-carriers and carriers was 0.001 and 1.0, respectively. In the recessive model, the frequency of the susceptibility allele was set to be 0.15, and the penetrances for heterozygous carriers and homozygous carriers were assumed to be equal.
Linkage heterogeneity among families (HLOD) was estimated using HOMOG (Ott 1999). In non-parametric analysis, LOD scores were calculated using the Kong and Cox exponential allele sharing model score (KCLOD). Multipoint linkage statistics were performed at 0.5-cM intervals across the region covering PTEN gene.
To test associations between the SNPs and CNVs and PCa, allele frequencies between cases and controls for each SNP were compared. LD structure was formed by Haploview (http://www.broadinstitute.org/mpg/haploview). The differences in allele frequencies between cases and controls were tested using standard contingency χ2 tests, and P-values were determined via χ2 approximation. All of the known SNPs in PTEN were imputed based on the genotyped SNP information in the HapMap Phase II data (CEU) using a computer program, IMPUTE.25 Odds ratio and 95% confidence interval (95%) were estimated using an unconditional logistic regression with adjustment for age and geographic region, assuming an additive model of inheritance.
RESULTS
SNP identification
A total of 15 genetic variants were identified in the sequenced region of PTEN in the 188 HPC probands. Five of the variants had already been previously reported (rs1044322, rs12573787, rs1903858, rs34051577 and rs1044487) and none of the 15 was located in exons (http://www.ncbi.nlm.nih.gov/SNP/snp_ref). The location and frequency of each variant are presented in Figure 1 and Table 2. Of the 15 sequence variants, 5 are common variants with a minor allele frequency ≥5% and 10 are rare variants with a minor allele frequency <5%. Of the five common variants, two are SNPs and the three are insertion/deletions.
Table 2.
Sequence variants of PTEN identified in 188 HPC probands
| Variant ID(position)a | Sequence variants | Position | Frequency |
|---|---|---|---|
| 1 (−1287) | SNP–1287C/G | Promoter | CG: 1.2%; CC: 98.8%; GG: 0 |
| 2 (−1060) | SNP–1060C/G | 5′UTR | CG: 0.7%; CC:99.3%; GG:0 |
| 3 (−976) | SNP–976G/C | 5′UTR | GC:0.7%; GG:99.3%; CC:0 |
| 4 (−904) | SNP–904G/A | 5′UTR | GA: 3.4%; GG:96.6%; AA:0 |
| 5 (−546) | INDEL–546 5GGC/6GGC | 5′UTR | 5/6:2.6%; 5/5:97.4%; 6/6:0 |
| 6 rs12573787 (−511) | SNP–511G/A | 5′UTR | GA:22%; GG:77%; AA:0.7% |
| 7 (−500) | INDEL–500 5GGC/6GGC/7GGC | 5′UTR | 5/6:0.7%; 5/7:0.7%; 5/5:98.6%; 6/6:0; 7/7:0 |
| 8 (−461) | SNP–461G/A | 5′UTR | GA:0.7%; GG:99.3%; AA:0 |
| 9 (210) | SNP 210T/C | Intron 1 | TC:0.6%; TT:99.4%; CC:0 |
| 10 rs1903858 (29460) | SNP 29460C/T | Intron 1 | CT:47.3%; TT:43.2%; CC:9.5% |
| 11 (66729) | INDEL 66729 TCTTA/_ | Intron 4 | TCTTA/_:44%; _/_:45.6%; TCTTA/TCTTA:10.6% |
| 12 (68506) | INDEL 68506 9T/10T | Intron 4 | 9/10:1.8%; 9/9:98.2%; 10/10:0 |
| 13 (93573) | SNP 93573G/A | Intron 7 | GA:1.8%; GG:98.2%; AA:0 |
| 14 (96422) | INDEL 96422 15T/14T | Intron 7 | 15/14:100%; 15/15:0; 14/14:0 |
| 15 (101068) | INDEL 101068 11T/10T | 3′UTR | 11/10:100%; 11/11:0; 10/10:0 |
Abbreviations: HPC, hereditary prostate cancer; SNP, single nucleotide polymorphism; UTR, untranslated region.
Position is relative to ATG translation start.
Co-segregation of rare SNPs and PCa in HPC families
LOD scores for two markers (D10S1686 and D10S185) flanking PTEN located in the region of 100.5 ~102.5 cM are shown in Table 3. HLOD scores were used to present the LOD score for the family group of each SNP. Both dominant and recessive parametric analyses were carried out, and evidence of co-segregation of rare SNPs in the HPC families was not observed. Non-parametric analysis was also carried out and co-segregation of the rare SNPs and PCa was not found (data not shown).
Table 3.
Co-segregation of 10 rare SNPs in PTEN in HPC families
| LOD score (HLOD) | |||||||
|---|---|---|---|---|---|---|---|
| Dominant | Recessive | ||||||
| Variant ID | Family number |
100.5 cM |
101.5 cM |
102.5 cM |
100.5 cM |
101.5 cM |
102.5 cM |
| 1 | 2 | 0.09 | 0.11 | 0.12 | 0.08 | 0.07 | 0.07 |
| 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 2 | 0.03 | 0.03 | 0.03 | 0.04 | 0.04 | 0.03 |
| 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 3 | 0 | 0 | 0.02 | 0 | 0.01 | 0.02 |
| 13 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| All_above_fam | 23 | 0 | 0 | 0 | 0 | 0 | 0 |
Abbreviations: HPC, hereditary prostate cancer; SNP, single nucleotide polymorphism.
Family-based association study (FBAT)
Two common SNPs, rs12573787 and rs1903858, which were in the 5′ untranslated region and intron 1, were further genotyped in the FBAT. An association between the two variants and PCa was not observed, suggesting the variants were not overtransmitted from parents to affected sons in the 188 HPC families including 1286 affected and unaffected family members (Table 4, P >0.05). Subset analysis of FBAT were also carried out by breaking the families down by the number of affected males (number=3, 4 and ≥5) and age at onset (<65 or >65). None of these sub groups showed significant association for these two SNPs in FBAT analysis (all P >0.05).
Table 4.
Results of the FBATs for two common SNPs in PTEN
| SNPs | Chromosome position |
Alleles | Risk allele |
Allele frequency |
P-valuesa |
|---|---|---|---|---|---|
| rs12573787 | 89 613 696 | A/G | A | 0.12 | 0.9 |
| rs1903858 | 89 643 666 | C/T | C | 0.7 | 0.99 |
Abbreviations: FBATs, family-based association tests; SNP, single nucleotide polymorphism.
FBAT test: HPC cases vs controls.
Association test in sporadic PCa cases and controls
The SNP rs1903858 was selected for association analysis among sporadic PCa cases and controls. The frequency of rs1903858 is 0.31 in cases (n=1527) and 0.30 in controls (n=482) from the JHH population. No significant difference in the allele frequencies was found between sporadic PCa cases and controls (P=0.592).
Of the 33 SNPs that spanned the PTEN gene that were selected for association testing with sporadic PCa, there were 28 common SNPs (≥5% frequency) identified with an average spacing of one common SNP every ~3.7 kb. In the PTEN gene, four SNPs were genotyped in GASP1 and GASP2 and six SNPs were genotyped in CGEMS GWAS studies. The remaining SNPs in PTEN were imputed based on the four SNPs genotyped in GASP1 and GASP2 GWAS or six SNPs genotyped in CGEMS GWAS. None of the 33 SNPs (genotyped or imputed) was found to be significantly associated with PCa susceptibility in any these studies, shown as in Table 5 (P >0.05). The 33 SNPs were also tested for their association with PCa aggressiveness in CGEMS and GASP2 populations (all cases in GASP1 population are aggressive). None of them was found to be significantly associated with aggressiveness in these two populations, as shown in Table 6 (all P >0.05).
Table 5.
Frequency of 33 SNPs spanning PTEN in three GWAS studies
| CGEMS (1177 cases/1105 controls) | GASP1 (500 cases/500 controls) | GASP2 (1500 cases/500 controls) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Minor allele |
F_A▲ | F_U▲ | Pa | ORa | Status | F_A▲ | F_U▲ | Pa | ORa | Status | F_A▲ | F_U▲ | Pa | ORa | Status |
| rs3781195 | 89 632 589 | G | 0.03 | 0.02 | 0.81 | 1.05 | Imputed | 0.03 | 0.01 | 0.011 | 2.29 | Imputed | 0.02 | 0.02 | 0.79 | 1.07 | Imputed |
| rs1234220 | 89 635 453 | G | 0.10 | 0.10 | 0.63 | 1.05 | Original | 0.11 | 0.09 | 0.19 | 1.22 | Imputed | 0.10 | 0.12 | 0.17 | 0.85 | Imputed |
| rs11202596 | 89 637 134 | A | 0.09 | 0.08 | 0.70 | 1.04 | Original | 0.04 | 0.04 | 0.57 | 0.86 | Imputed | 0.05 | 0.05 | 0.95 | 0.99 | Imputed |
| rs1234219 | 89 639 557 | G | 0.08 | 0.07 | 0.60 | 1.06 | Imputed | 0.06 | 0.06 | 0.96 | 1.01 | Imputed | 0.06 | 0.08 | 0.24 | 0.84 | Imputed |
| rs7069904 | 89 639 818 | A | 0.02 | 0.02 | 0.16 | 1.34 | Imputed | 0.02 | 0.01 | 0.046 | 2.72 | Imputed | 0.01 | 0.00 | 0.61 | 1.33 | Imputed |
| rs1234218 | 89 640 739 | T | 0.07 | 0.07 | 0.84 | 1.02 | Imputed | 0.05 | 0.06 | 0.74 | 0.94 | Imputed | 0.06 | 0.07 | 0.24 | 0.84 | Imputed |
| rs1903858 | 89 643 666 | G | 0.31 | 0.28 | 0.03 | 1.16 | Imputed | 0.29 | 0.30 | 0.78 | 0.97 | Imputed | 0.27 | 0.28 | 0.95 | 0.99 | Imputed |
| rs2299939 | 89 647 130 | A | 0.19 | 0.19 | 0.63 | 1.04 | Original | 0.08 | 0.10 | 0.24 | 0.79 | Imputed | 0.09 | 0.08 | 0.45 | 1.14 | Imputed |
| rs1234225 | 89 663 549 | T | 0.35 | 0.32 | 0.08 | 1.12 | Imputed | 0.34 | 0.35 | 0.67 | 0.96 | Imputed | 0.33 | 0.35 | 0.39 | 0.93 | Imputed |
| rs12569998 | 89 664 137 | G | 0.14 | 0.13 | 0.29 | 1.10 | Imputed | 0.14 | 0.14 | 0.85 | 0.98 | Original | 0.13 | 0.13 | 0.71 | 1.04 | Original |
| rs1234224 | 89 665 276 | G | 0.35 | 0.32 | 0.08 | 1.12 | Imputed | 0.34 | 0.35 | 0.67 | 0.96 | Imputed | 0.33 | 0.35 | 0.39 | 0.93 | Imputed |
| rs1234223 | 89 665 570 | C | 0.10 | 0.10 | 0.70 | 1.04 | Imputed | 0.11 | 0.09 | 0.21 | 1.21 | Imputed | 0.10 | 0.12 | 0.17 | 0.85 | Imputed |
| rs17106896 | 89 667 087 | T | 0.05 | 0.04 | 0.23 | 1.19 | Imputed | 0.02 | 0.01 | 0.049 | 2.69 | Imputed | 0.01 | 0.00 | 0.40 | 1.59 | Imputed |
| rs1676759 | 89 668 276 | G | 0.03 | 0.02 | 0.72 | 1.07 | Imputed | 0.01 | 0.01 | 0.58 | 1.35 | Imputed | 0.01 | 0.00 | 0.34 | 1.68 | Imputed |
| rs11202600 | 89 672 813 | C | 0.07 | 0.07 | 0.84 | 1.02 | Imputed | 0.07 | 0.07 | 0.98 | 1.00 | Original | 0.08 | 0.10 | 0.053 | 0.79 | Original |
| rs10490920 | 89 675 623 | C | 0.14 | 0.13 | 0.29 | 1.10 | Imputed | 0.14 | 0.14 | 0.82 | 0.97 | Imputed | 0.13 | 0.13 | 0.75 | 1.04 | Imputed |
| rs1234213 | 89 679 301 | A | 0.35 | 0.32 | 0.07 | 1.12 | Imputed | 0.34 | 0.35 | 0.66 | 0.95 | Imputed | 0.33 | 0.35 | 0.39 | 0.93 | Imputed |
| rs7078674 | 89 687 333 | T | 0.00 | 0.00 | NA | NA | Imputed | 0.00 | 0.00 | NA | NA | Imputed | 0.00 | 0.00 | NA | NA | Imputed |
| rs12357281 | 89 690 651 | C | 0.09 | 0.08 | 0.75 | 1.04 | Imputed | 0.04 | 0.04 | 0.65 | 0.89 | Imputed | 0.05 | 0.05 | 0.88 | 0.97 | Imputed |
| rs2299941 | 89 694 699 | G | 0.14 | 0.13 | 0.23 | 1.11 | Imputed | 0.14 | 0.14 | 0.99 | 1.00 | Imputed | 0.13 | 0.12 | 0.87 | 1.02 | Imputed |
| rs2735343 | 89 695 409 | C | 0.35 | 0.32 | 0.09 | 1.11 | Imputed | 0.34 | 0.35 | 0.65 | 0.95 | Imputed | 0.33 | 0.35 | 0.37 | 0.93 | Imputed |
| rs2248293 | 89 697 245 | C | 0.35 | 0.32 | 0.07 | 1.12 | Original | 0.31 | 0.31 | 0.98 | 1.00 | Imputed | 0.30 | 0.33 | 0.16 | 0.88 | Imputed |
| rs17562384 | 89 699 396 | C | 0.19 | 0.19 | 0.59 | 1.04 | Imputed | 0.08 | 0.10 | 0.25 | 0.79 | Imputed | 0.09 | 0.08 | 0.44 | 1.14 | Imputed |
| rs2673832 | 89 702 453 | G | 0.11 | 0.11 | 0.92 | 0.99 | Original | 0.01 | 0.02 | 0.22 | 0.56 | Imputed | 0.02 | 0.01 | 0.38 | 1.35 | Imputed |
| rs12572106 | 89 706 087 | C | 0.14 | 0.13 | 0.27 | 1.10 | Imputed | 0.14 | 0.14 | 0.82 | 0.97 | Imputed | 0.13 | 0.13 | 0.75 | 1.04 | Imputed |
| rs17431184 | 89 710 231 | C | 0.19 | 0.19 | 0.60 | 1.04 | Imputed | 0.08 | 0.10 | 0.25 | 0.79 | Imputed | 0.09 | 0.08 | 0.43 | 1.15 | Imputed |
| rs2736627 | 89 711 074 | C | 0.09 | 0.10 | 0.43 | 0.92 | Imputed | 0.01 | 0.02 | 0.22 | 0.56 | Imputed | 0.02 | 0.01 | 0.38 | 1.35 | Imputed |
| rs926091 | 89 711 392 | T | 0.14 | 0.13 | 0.27 | 1.10 | Imputed | 0.14 | 0.14 | 0.82 | 0.97 | Imputed | 0.13 | 0.13 | 0.75 | 1.04 | Imputed |
| rs532678 | 89 713 322 | T | 0.35 | 0.32 | 0.08 | 1.12 | Imputed | 0.34 | 0.35 | 0.65 | 0.95 | Imputed | 0.33 | 0.35 | 0.37 | 0.93 | Imputed |
| rs11202607 | 89 717 394 | T | 0.07 | 0.07 | 0.99 | 1.00 | Original | 0.05 | 0.06 | 0.74 | 0.94 | Imputed | 0.06 | 0.07 | 0.24 | 0.84 | Imputed |
| rs478839 | 89 721 850 | G | 0.40 | 0.42 | 0.41 | 0.95 | Original | 0.40 | 0.38 | 0.39 | 1.11 | Imputed | 0.38 | 0.37 | 0.89 | 1.01 | Imputed |
| rs17346680 | 89 727 081 | A | 0.04 | 0.03 | 0.29 | 1.19 | Imputed | 0.04 | 0.02 | 0.003 | 2.24 | Original | 0.07 | 0.08 | 0.11 | 0.80 | Original |
| rs10509532 | 89 727 534 | T | 0.14 | 0.13 | 0.26 | 1.10 | Original | 0.14 | 0.14 | 0.78 | 0.96 | Original | 0.13 | 0.13 | 0.75 | 1.04 | Original |
Abbreviations: CGEMS, cancer genetic markers of susceptibility; GWAS, whole genome-wide association study; OR, odds ratio; SNPs, single nucleotide polymorphisms.
P and OR are calculated based on an unconditional logistic regression with adjustment for age and geographic region, assuming an additive model of inheritance.
F_A, frequency in affected cases; F_U, frequency in controls.
Table 6.
Frequency of 33 SNPs spanning PTEN in 2 GWAS studies
| CGEMS (650 aggressive/527 non-aggressive) | GASP2 (637 aggressive/863 non-aggressive) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Minor allele |
F_A▲ | F_U▲ | Pa | ORa | Status | F_A▲ | F_U▲ | Pa | ORa | Status |
| rs3781195 | 89 632 589 | G | 0.03 | 0.03 | 0.82 | 1.06 | Imputed | 0.02 | 0.02 | 0.84 | 0.95 | Imputed |
| rs1234220 | 89 635 453 | G | 0.11 | 0.10 | 0.70 | 1.06 | Original | 0.11 | 0.09 | 0.15 | 1.19 | Imputed |
| rs11202596 | 89 637 134 | A | 0.09 | 0.08 | 0.58 | 1.09 | Original | 0.04 | 0.05 | 0.44 | 0.85 | Imputed |
| rs1234219 | 89 639 557 | G | 0.08 | 0.07 | 0.47 | 1.13 | Imputed | 0.07 | 0.06 | 0.26 | 1.20 | Imputed |
| rs7069904 | 89 639 818 | A | 0.03 | 0.02 | 0.41 | 1.27 | Imputed | 0.01 | 0.00 | 0.21 | 1.91 | Imputed |
| rs1234218 | 89 640 739 | T | 0.07 | 0.07 | 0.69 | 1.07 | Imputed | 0.07 | 0.06 | 0.30 | 1.17 | Imputed |
| rs1903858 | 89 643 666 | G | 0.32 | 0.30 | 0.26 | 1.11 | Imputed | 0.27 | 0.28 | 0.66 | 0.96 | Imputed |
| rs2299939 | 89 647 130 | A | 0.19 | 0.20 | 0.49 | 0.93 | Original | 0.08 | 0.09 | 0.19 | 0.80 | Imputed |
| rs1234225 | 89 663 549 | T | 0.36 | 0.33 | 0.10 | 1.16 | Imputed | 0.33 | 0.33 | 0.72 | 1.03 | Imputed |
| rs12569998 | 89 664 137 | G | 0.14 | 0.14 | 0.95 | 0.99 | Imputed | 0.13 | 0.13 | 0.56 | 0.94 | Original |
| rs1234224 | 89 665 276 | G | 0.36 | 0.33 | 0.10 | 1.16 | Imputed | 0.33 | 0.33 | 0.72 | 1.03 | Imputed |
| rs1234223 | 89 665 570 | C | 0.10 | 0.10 | 0.68 | 1.06 | Imputed | 0.11 | 0.09 | 0.15 | 1.19 | Imputed |
| rs17106896 | 89 667 087 | T | 0.05 | 0.04 | 0.19 | 1.30 | Imputed | 0.01 | 0.01 | 0.32 | 1.58 | Imputed |
| rs1676759 | 89 668 276 | G | 0.03 | 0.03 | 0.98 | 1.01 | Imputed | 0.01 | 0.01 | 0.96 | 1.02 | Imputed |
| rs11202600 | 89 672 813 | C | 0.07 | 0.07 | 0.69 | 1.07 | Imputed | 0.09 | 0.07 | 0.06 | 1.29 | Original |
| rs10490920 | 89 675 623 | C | 0.14 | 0.14 | 0.95 | 0.99 | Imputed | 0.12 | 0.13 | 0.48 | 0.93 | Imputed |
| rs1234213 | 89 679 301 | A | 0.36 | 0.33 | 0.09 | 1.16 | Imputed | 0.33 | 0.33 | 0.75 | 1.03 | Imputed |
| rs7078674 | 89 687 333 | 0 | 0.00 | 0.00 | NA | NA | Imputed | 0.00 | 0.00 | NA | NA | Imputed |
| rs12357281 | 89 690 651 | C | 0.09 | 0.08 | 0.64 | 1.07 | Imputed | 0.04 | 0.05 | 0.38 | 0.84 | Imputed |
| rs2299941 | 89 694 699 | G | 0.14 | 0.14 | 0.98 | 1.00 | Imputed | 0.12 | 0.13 | 0.41 | 0.91 | Imputed |
| rs2735343 | 89 695 409 | C | 0.36 | 0.33 | 0.11 | 1.15 | Imputed | 0.33 | 0.33 | 0.79 | 1.02 | Imputed |
| rs2248293 | 89 697 245 | C | 0.36 | 0.33 | 0.11 | 1.15 | Original | 0.30 | 0.30 | 0.75 | 0.97 | Imputed |
| rs17562384 | 89 699 396 | C | 0.19 | 0.20 | 0.47 | 0.93 | Imputed | 0.08 | 0.09 | 0.17 | 0.79 | Imputed |
| rs2673832 | 89 702 453 | G | 0.11 | 0.10 | 0.44 | 1.11 | Original | 0.02 | 0.02 | 0.38 | 0.76 | Imputed |
| rs12572106 | 89 706 087 | C | 0.14 | 0.14 | 0.98 | 1.00 | Imputed | 0.12 | 0.13 | 0.48 | 0.93 | Imputed |
| rs17431184 | 89 710 231 | C | 0.19 | 0.20 | 0.47 | 0.93 | Imputed | 0.08 | 0.10 | 0.15 | 0.78 | Imputed |
| rs2736627 | 89 711 074 | C | 0.09 | 0.09 | 0.64 | 1.07 | Imputed | 0.02 | 0.02 | 0.38 | 0.76 | Imputed |
| rs926091 | 89 711 392 | T | 0.14 | 0.14 | 0.98 | 1.00 | Imputed | 0.12 | 0.13 | 0.48 | 0.93 | Imputed |
| rs532678 | 89 713 322 | T | 0.36 | 0.33 | 0.11 | 1.15 | Imputed | 0.33 | 0.33 | 0.80 | 1.02 | Imputed |
| rs11202607 | 89 717 394 | T | 0.07 | 0.07 | 0.67 | 1.07 | Original | 0.07 | 0.06 | 0.31 | 1.17 | Imputed |
| rs478839 | 89 721 850 | G | 0.39 | 0.42 | 0.25 | 0.91 | Original | 0.38 | 0.38 | 0.92 | 0.99 | Imputed |
| rs17346680 | 89 727 081 | A | 0.04 | 0.04 | 0.98 | 1.00 | Imputed | 0.07 | 0.06 | 0.20 | 1.22 | Original |
| rs10509532 | 89 727 534 | T | 0.14 | 0.14 | 0.91 | 0.99 | Original | 0.12 | 0.13 | 0.48 | 0.93 | Original |
Abbreviations: CGEMS, cancer genetic markers of susceptibility; GWAS, whole genome-wide association study; OR, odds ratio; SNPs, single nucleotide polymorphisms.
P and OR are calculated based on an unconditional logistic regression with adjustment for age and geographic region, assuming an additive model of inheritance.
F_A, frequency in aggressive cases; F_U, frequency in non-aggressive cases.
Copy-number variations (CNVs) analysis
In addition to testing the association of germ-line SNPs and PCa susceptibility, germ-line CNVs in PTEN and the association with sporadic PCa was also evaluated with three SNP probes (rs12569998, rs11202600 and rs17107001) at PTEN region on Gene-Chip Human Mapping 5.0 Array (Affymetrix). The CNV defined by the three probes in PTEN was not associated with PCa risk from GWAS in CAPS population (data not shown).
DISCUSSION
In this report, germ-line sequence variants of PTEN in both HPC and sporadic prostate caner were comprehensively evaluated. The PTEN gene was screened, including the 5′ untranslated region, all nine exons, exon–intron junctions and 3′ untranslated regions, in HPC probands. The co-segregation of PTEN mutations/sequence variants in HPC families and association in a sporadic PCa population were also evaluated. To summarize, 10 rare and 5 common variants were identified by sequencing, however, were unable to identify any potentially important mutations, such as mutations in exons from germ-line DNA samples of 188 HPC probands. There was no statistically significant difference in allele frequencies of the two common SNPs, rs12573787 and rs1903858, between cases and controls after genotyping 1286 HPC members and no statistical difference in the allele frequency of one common SNP after genotyping in 1527 PCa cases and 482 controls. Additionally, there was no evidence of co-segregation of variants in PTEN in the HPC families. The 33 SNPs that spanned the PTEN gene showed no association between PTEN and PCa susceptibility in the CAPS and CGEMS population. None of the 33 SNPs was shown to be associated with aggressiveness of PCa. Finally, in the results of evaluating CNVs in PTEN from the available data in CAPS, there were no detectable CNV changes associated with PCa (data not shown). The results suggest that PTEN gene variants do not have an effect on PCa susceptibility and aggressiveness.
This is the first time that germ-line variants of PTEN, including both rare and common variants and CNVs, were comprehensively evaluated in hereditary and sporadic PCa populations. The PTEN region was re-sequenced in a large sample size (n=188) of HPC probands. No deleterious variants were found to contribute to PCa susceptibility, which was consistent with previous studies.13,14 Co-segregation of 10 rare variants, which were believed to give rise to high-penetrant HPC, was further evaluated by testing two microsatellite markers flanking PTEN in the HPC families. However, co-segregation was not observed in the HPC families. Common variants, which were believed to contribute low-penetrant PCa, were explored in sporadic PCa populations and no association was found between the SNPs and PCa risk, consistent with previous studies.15,16 It was also the first time that germ-line CNVs in PTEN were explored for PCa susceptibility. In our study, there were no CNV probes on PTEN region, and three SNP probes were used to define CNVs for PTEN. No CNV defined by the three SNPs at PTEN was found to be associated with sporadic PCa. Based on the available CNV information from Hapmap,26 the nearest CNVs on the 5′ and 3′ flank region of PTEN (89 613 715–89 718 382) are 87 791 026–87 798 355 (~1.8 Mbp on 5′ region) and 90 932 947–90 938 070 (~1.2 Mbp on 3′ region), respectively. It is possible that CNVs at PTEN can be detected by microarrays with higher resolution.
To date, there have not been any germ-line variants in PTEN found to contribute to PCa susceptibility. However, the possibility can not be eliminated that germ-line and/or somatic sequence variants in PTEN may be associated with PCa aggressiveness. In the literature the presence of TMPRSS2: ERG fusion and a PTEN deletion was reported to be associated with a more aggressive form of PCa.27 In our studies, however, germ-line sequence variants of PTEN did not serve as indicator for the aggressiveness of PCa from the GWAS studies in both CGEM and GASP2 population. Therefore, germ-line sequence variants of PTEN could not serve as an indicator for the aggressiveness of Pca.28–31
In conclusion, this study suggests that PTEN germ-line SNPs are unlikely to have an important role in hereditary and non-hereditary PCa susceptibility.
References
- 1.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 2.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. PTEN is essential for embryonic development and tumor suppression. Nature Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine PTEN tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 4.Abate-Shen C, Banach-Petrosky WA, Sun X, Economides KD, Desai N, Gregg JP, et al. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63:3886–3890. [PubMed] [Google Scholar]
- 5.Halvorsen O, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin. Cancer Res. 2003;9:1474–1479. [PubMed] [Google Scholar]
- 6.Dong JT, Li CL, Sipe TW, Frierson HF., Jr Mutations of PTEN/MMAC1 in primary prostate cancers from Chinese patients. Clin. Cancer Res. 2001;7:304–308. [PubMed] [Google Scholar]
- 7.Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp. Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz M, Grignard G, Margue C, Dippel W, Capesius C, Mossong J, et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int. J. Cancer. 2006;120:1284–1292. doi: 10.1002/ijc.22359. [DOI] [PubMed] [Google Scholar]
- 9.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 10.Verhagen PC, Duijin PW, Hermans KG, Looijenga LH, Van Gurp RJ, Stoop H, et al. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J. Pathol. 2006;208:699–707. doi: 10.1002/path.1929. [DOI] [PubMed] [Google Scholar]
- 11.Witte JS, Goddard KA, Conti DV, Elston RC, Lin J, Suarez BK, et al. Genomewide scan for prostate cancer-aggressiveness loci. Am. J. Hum. Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs M, Stanford JL, Jarvik GP, Janer M, Badzioch M, Peters MA, et al. A genomic scan of families with prostate cancer identifies multiple regions of interest. Am. J. Hum. Genet. 2000;67:100–109. doi: 10.1086/302969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooney KA, Tsou HC, Petty EM, Miesfeldt S, Ping XL, Gruener AC, et al. Absence of PTEN germ-line mutations in men with a potential inherited predisposition to prostate cancer. Clin. Cancer Res. 1999;5:1387–1391. [PubMed] [Google Scholar]
- 14.Forrest MS, Edwards SM, Hamoudi RA, Dearnaley DP, Arden-Jones A, Dowe A, et al. No evidence of germline PTEN mutations in familial prostate cancer. J. Med. Genet. 2000;3:210–212. doi: 10.1136/jmg.37.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Shira A, Matarasso N, Rosner S, Bercovich D, Matzkin H, Orr-Urtreger A. Mutation screening and association study of the Candidate prostate cancer susceptibility genes MSR1, PTEN, and KLF6. Prostate. 2006;66:1052–1060. doi: 10.1002/pros.20425. [DOI] [PubMed] [Google Scholar]
- 16.Haiman CA, Stram DO, Cheng I, Giorgi EE, Pooler L, Penney K, et al. Common genetic variation at PTEN and risk of sporadic breast and prostate cancer. Cancer Epidemiol. Biomarkers. Prev. 2006;15:1021–1025. doi: 10.1158/1055-9965.EPI-05-0896. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Gillanders EM, Isaacs SD, Chang BL, Wiley KE, Zheng SL, et al. Genome-wide scan for prostate cance rsusceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate. 2003;57:320–325. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- 18.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 19.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 20.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 21.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 22.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, et al. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69:2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19:S26–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 26.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cnacer is associated with a favorable outcome. Mod. Pathol. 2008;23:1–10. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 28.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 29.Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 30.Nelen MR, van Staveren WC, Peeters EA, Hassel MB, Gorlin RJ, Hamm H, et al. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum. Mol. Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 31.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Association; 1996. [Google Scholar]