Abstract
ArsD is a metallochaperone that delivers As(III) to the ArsA ATPase, the catalytic subunit of the ArsAB pump encoded by the arsRDABC operon of Escherichia coli plasmid R773. Conserved ArsD cysteine residues (Cys12, Cys13 and Cys18) construct the As(III) binding site of the protein, however a global structural understanding of this arsenic binding remains unclear. We have obtained NMR assignments for ArsD as a starting point for probing structural changes on the protein that occur in response to metalloid binding and upon formation of a complex with ArsA. The predicted solution structure of ArsD is in agreement with recently published crystallographic structural results.
Keywords: NMR assignments, Arsenic, ARS operon, Metallochaperone
Biological context
Arsenic is one of the most toxic elements found in the environment and ranks first on the Environmental Protection Agency Superfund list of Hazardous Substances (http://www.atsdr.cdc.gov/cercla/07list.html). Since arsenic is toxic to most life forms, it is no surprise that nearly every organism, from bacteria to archaea to eukaryotes, including humans, have evolved detoxification systems for the metalloid (Bhattacharjee and Rosen 2007). In Escherichia coli, the arsRDABC operon of plasmid R773 confers resistance to arsenite and antimonite, where ArsAB is an ATPases that pumps trivalent metalloids (As(III) and Sb(III)) out of the cell. ArsD has recently been demonstrated to be a metallochaperone for As(III), ferrying the metalloid through the cytosol to the catalytic subunit of the efflux pump, the ArsA ATPase (Lin et al. 2006). In various ars operons, the arsD gene is almost always adjacent to the arsA gene, suggesting a related function. Interaction with ArsD increases the affinity of ArsA for As(III), which in turn increases the detoxification efficiency for the system (Lin et al. 2007). Direct transfer of As(III) from ArsD to ArsA occurs in the presence of chelators, consistent with the channeling of the metalloid between protein partners; transfer is dependent on the presence of ATP (Yang et al. 2010). These studies indicate a direct interaction between ArsD and ArsA during transfer of As(III) from one protein to the other.
In this study, we have undertaken NMR characterization of ArsD to provide a basis for understanding the structural aspects of how the protein functions. ArsD is a homodimer of two 120-residue subunits in which residues Cys12, Cys13 and Cys18 form the As(III) binding site required for arsenic metallochaperone activity (Lin et al. 2007). Each subunit has eight cysteine residues, including three vicinal pairs, Cys12–13, Cys112–113 and Cys119–120. Although all three vicinal pairs bind As(III), only Cys12, Cys13 and Cys18 are conserved, and these three are required for chaperone activity. The crystal structure of apo ArsD was recently reported (Ye et al. 2010). The overall structure of the ArsD monomer has a thioredoxin fold, with a core of four beta-strands flanked by four α-helices (PDB code 3MWH). The region of ArsD with the cysteines that form the arsenic binding site was not visible in that structure, but a structure of an oxidized form of an ArsD homologue, showing the location of the cysteines in disulfide bonds, has been reported (PDB code 3KTB). In this report, we have initiated determination of the solution structure of ArsD for comparison with the static structure and to allow identification of amino acid-specific chemical environmental changes in ArsD coupled with binding of arsenite and antimonite or during the interaction with ArsA. Therefore, the work outlined in this report provides the basis for future experiments directed at understanding the mechanism by which ArsD transfers As(III) to ArsA and hence mediates arsenic toxicity in E. coli.
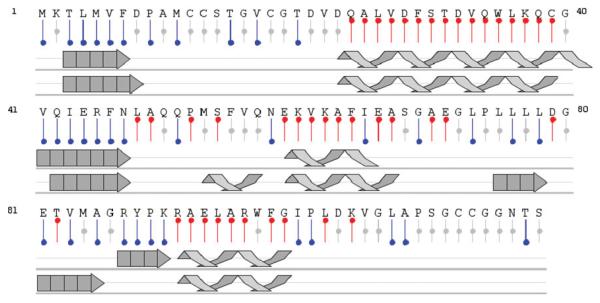
Dispersion in the high-resolution 15N TROSY HSQC spectrum (Pervushin et al. 2000) indicates the protein is well folded, with the number of peaks consistent with the protein existing as a symmetric homodimer (Fig. 1). Assignments for the 118 amino acid ArsD construct were completed for the majority of native protein residues. In the absence of As(III), amide chemical shifts for the three conserved cysteines (Cys12, Cys13 and Cys18) occur in the unfolded region of the spectrum, indicating these residues exist in an unstructured environment. Chemical shift analysis of the backbone atom resonances (Wishart et al. 1991) was applied to predict the secondary structure for the protein in solution (Fig. 2). Prediction analysis suggests the protein is flanked by a 7 residue N-terminal β-strand. The active site residues Cys12, Cys13 and Cys18 all exist in the unstructured region between β-strand 1 and α-helix 1. In addition, the protein contains an extensive C-terminal unstructured tail ranging from residues 100 to 118.
Fig. 1.
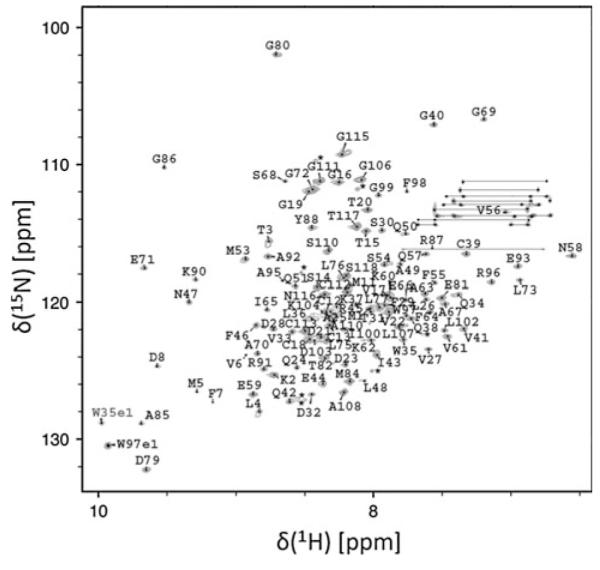
Assigned 1H,15N TROSY HSQC spectrum of ArsD. Data were collected on the National Magnet Laboratory’s Varian INOVA 720 MHz spectrometer at 293 K
Fig. 2.
Summary of proposed secondary structural elements for ArsD in solution (middle) compared with structural characterization obtained from crystallographic analysis (bottom) from CSI analysis using NMRView (Johnson 2004). Single letter residue designations are provided (top) with predicted secondary structural elements displayed in red (helix), blue (strand) or grey (unfolded)
A comparison of predicted secondary structural elements for ArsD in solution with the crystal structure results indicate a reasonable agreement between the structures (Fig. 2), with a few differences. The solution structure is less ordered than the crystal structure, especially in the length and location of α-helix 2 and β-strand 3. By both methods, the conserved cysteines (Cys12, Cys13 and Cys18) exist in an unstructured region between β-strand 1 and α-helix 1, as earlier indicated for the predicted solution structure. It is only when the cysteine residues are arti-factually oxidized to form disulfide bonds that they could be resolved by crystallography. These results suggest this region of apo ArsD is normally very flexible and folds around As(III), allowing the three cysteine residues to form the binding site. In a previous study, ArsD with As(III) bound was modeled and this metallated protein was docked in silico with ArsA (Ye et al. 2010). These modeling experiments offer testable predictions for which the solution characterization of ArsD outlined above will be informative. Therefore, these NMR data provide the basis for future experiments directed at monitoring structural changes coupled with binding of As(III) and/or ArsA. Using the crystal structure as the global structural template, we will be able to map binding surfaces between proteins and monitor structural changes during As(III) binding and delivery in the future using NMR spectroscopy. These results will provide significant insight in the molecular mechanism of ArsD metallochaperone function.
Methods and experiments
Labeling, expression and purification of ArsD
An ArsD construct containing residues 1–118 with the C-terminal cysteine pair (Cys119,Cys120) replaced by a six histidine tag (sequence: MKTLMVFDPA MCCSTGVCGT DVDQALVDFS TDVQWLKQCG VQIERFNLAQ QPM SFVQNEK VKAFIEASGA EGLPLLLLDG ETVMA GRYPK RAELARWFGI PLDKVGLAPS GCCGG NTSHH HHHH) for a total size of 124 residues and a mass of 13,543 Daltons was expressed in E. coli strain BL21(DE3). Uniformly labeled 15N, 15N/13C and 2H/15N/13C protein samples were prepared by growing cells on M9 medium containing 15NH4Cl, 13C glucose and/or 100% D2O (Cambridge Isotope Labs, Andover, MA). Expression and purification of the His6-tagged ArsD was as described previously (Lin et al. 2007). Eluted proteins were concentrated, quickly frozen and stored at −80°C until use. Protein concentrations were between 0.8 and 1.3 mM as determined by UV–Vis spectroscopy at 280 nm (using A280 of 1.0 cm−1 = 75 μM). For NMR experiments, proteins were exchanged into a buffer containing 20 mM sodium phosphate, 100 mM NaCl, 5 mM DTT, 90% H2O/10% D2O (pH 6.0) using Micro Bio-Spin columns (Bio-Rad, Hercules, CA); under similar buffer conditions the protein is a stable homodimer (Lin et al. 2007). NMR samples were cycled through multiple vacuum/Ar purge cycles using a Schlenk line. Samples were then sealed under argon directly within the NMR tube.
Nuclear magnetic resonance spectroscopy (NMR)
NMR spectra were acquired at 293 K on a Varian INOVA 720 MHz spectrometer (National High Field Magnet Laboratory, Tallahassee FL) and on an in house Varian INOVA 600 MHz spectrometer. The 720 MHz spectrometer was equipped with a triple resonance gradient room temperature probe while the 600 MHz spectrometer utilized a Varian triple resonance gradient cold probe. Backbone assignments were made using the following experiments: 15N-HSQC, HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB and CBCA(CO)NH (Cavanagh et al. 1996). Partial side chain atom assignments were made using a 15N TOCSY and a CCONH calibrated to account for the deuterium labeled aliphatic carbons. Spectra were analyzed according to established protocols on a Macintosh G5 PC using the processing programs NMRPipe (Delaglio et al. 1995) and Sparky (Goddard and Kneller 2001). Chemical shift analysis for secondary structure prediction from the NMR data was performed using CSI provided by the Sykes laboratory utilizing backbone and Cb residues only (Wishart et al. 1991).
Assignments and data deposition
Residue assignments for the 248 residue symmetric ArsD homodimer are provided in the 15N HSQC spectrum (Fig. 1). Amide backbone assignments were completed for 109 of the 112 non-proline residues, minus the protein’s N-terminus, 6 prolines and the histidine tag residues. Side chain assignments were achieved for 106 of the 118 residues in the ArsD monomeric primary structure. Chemical shift assignments are deposited in the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) under the accession code BMRB-17231.
Acknowledgments
We thank the National High Magnetic Field Laboratory (NHMFL) in Tallahassee, FL. for 720 MHz NMR time as part of their user’s program. The INOVA 600 MHz spectrometer with cold probe was purchased through NIH support (1S10RR16626-01). The work was supported by NIH grants AI043428 (BPR) and DK06 8139 (TLS).
Contributor Information
Jun Ye, Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, Detroit, MI 48201, USA.
Yanan He, Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, Detroit, MI 48201, USA.
Jack Skalicky, Department of Biochemistry, University of Utah, Salt Lake City, UT 84132, USA.
Barry P. Rosen, Department of Cellular Biology and Pharmacology, Florida International University, Herbert Wertheim College of Medicine, Miami, FL 33199, USA
Timothy L. Stemmler, Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, Detroit, MI 48201, USA
References
- Bhattacharjee H, Rosen BP. Arsenic metabolism in prokaryotic and eukaryotic microbes. In: Nies DH, Simon S, editors. Molecular microbiology of heavy metals. vol 6. Springer-Verlag; Heidelberg: 2007. pp. 371–406. [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR spectroscopy: principles and practice. Academic Press; San Diego: 1996. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2001. [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci USA. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Yang J, Rosen BP. ArsD residues Cys12, Cys13, and Cys18 form an As(III)-binding site required for arsenic metallochaperone activity. J Biol Chem. 2007;282:16783–16791. doi: 10.1074/jbc.M700886200. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Braun D, Fernandez C, Wuthrich K. [15 N, 1H]/[13C, 1H]-TROSY for simultaneous detection of backbone 15 N–1H, aromatic 13C–1H and side-chain 15 N–1H2 correlations in large proteins. J Biomol NMR. 2000;17:195–202. doi: 10.1023/a:1008399320576. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Bio. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Yang J, Rawat S, Stemmler TL, Rosen BP. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry. 2010 doi: 10.1021/bi100026a. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ajees AA, Yang J, Rosen BP. The 1.4 a crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry. 2010;49:5206–5212. doi: 10.1021/bi100571r. [DOI] [PMC free article] [PubMed] [Google Scholar]