Introduction
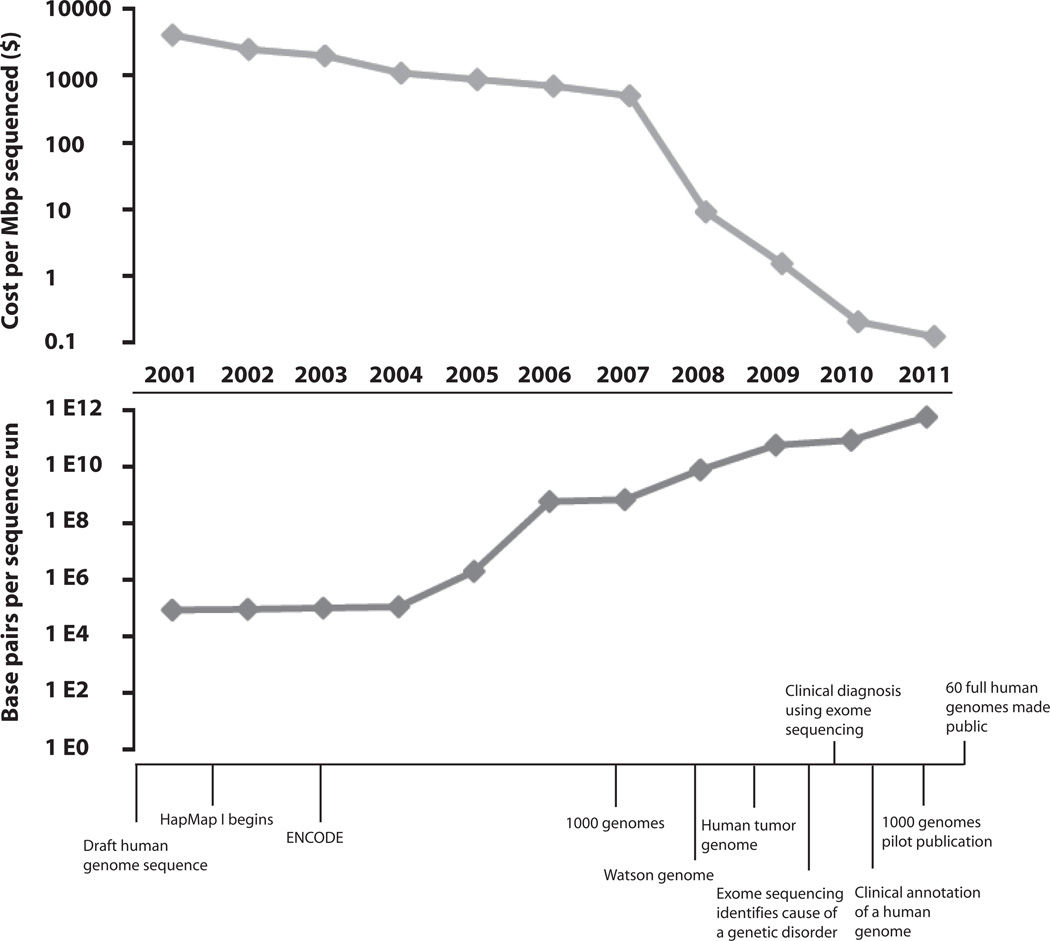
We are in the midst of a time of great change in genetics that may dramatically impact human biology and medicine. The completion of the human genome project,1,2 the development of low cost, high-throughput parallel sequencing technology, and large-scale studies of genetic variation3 have provided a rich set of techniques and data for the study of genetic disease risk, treatment response, population diversity, and human evolution. Newly-developed sequencing instruments now generate hundreds of millions to billions of short sequences per run, allowing for rapid complete sequencing of human genomes. These technological advances have facilitated a precipitous drop (Figure 1) in the cost per base pair of DNA sequenced. To capitalize on the potential of these technologies for research and clinical applications, translational scientists and clinicians must become familiar with a continuously evolving field. In this review we will provide a historical perspective on human genome sequencing, summarize current and future sequencing technologies, highlight issues related to data management and interpretation, and finally consider research and clinical applications of high-throughput sequencing, with specific emphasis on cardiovascular disease.
Figure 1.
Sequencing milestones, costs, and output since completion of the human genome project. Note logarithmic scale for sequencing costs and bases produced per sequence run.
Historical perspective
Genome sequencing has become synonymous with high-throughput sequencing, but it is instructive to revisit historical milestones. Though James Watson and Francis Crick published the first description of the crystallographic double helix DNA structure in 1953,4 it was not until two decades later, with the nearly simultaneous development of Maxam-Gilbert and Sanger sequencing,5,6 that DNA sequencing became widely available to the research community. The Sanger method, which is based on DNA chain termination with a small concentration of radio- or fluorescently-labeled di-deoxy nucleotide triphosphate (dNTPs) molecules followed by size separation by gel electrophoresis, became the research and commercial standard due to technical ease and reliability of results. This was the standard sequencing technology for over three decades and remains the method of choice for sequencing short segments of DNA and confirming genotypes from other technologies. Sanger sequencing, in conjunction with several methods for identifying the approximate genetic locations (“loci”) harboring variations in DNA associated with disease, was the method used to define the basis of many Mendelian, or single gene disorders.
More recently, a modified Sanger approach was the main sequencing engine for the first draft human genome sequence, which was produced by sequencing 500 to 600 base pair segments of DNA in parallel (“shotgun sequencing”) and “assembly” of these sequence fragments into contiguous stretches of DNA (“contigs”) based on sequence overlap.1,2,7 Two sequences were released nearly simultaneously; the first was a product of the decade-long publicly funded Human Genome Project,1 and the second was released by the Celera Corporation, led by Craig Venter and colleagues.7 The accuracy and read lengths generated by this technology were advantageous to sequencing projects in which no template or “reference” sequence was available, but the sequence time (years) and cost (estimated at between 300 million and 3 billion dollars) of these early efforts precluded the use of this technology for large-scale humane genome sequencing. However, the completion of a human genome “reference” sequence allowed for the development of a “next generation” of sequencing instruments that substantially reduced DNA sequencing time and cost.
“Next generation” sequencing technologies
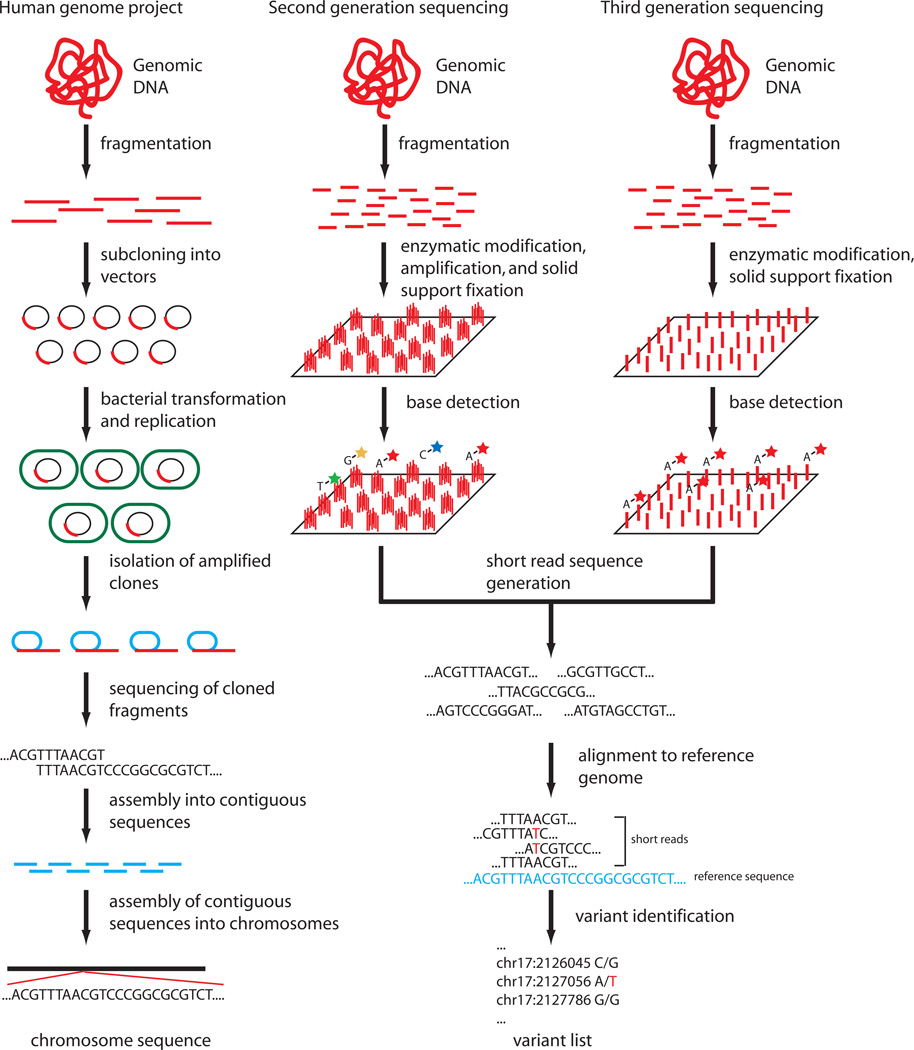
The development of a draft human genome sequence, which has subsequently been revised to constitute a “reference” human genome sequence, facilitated the development of “next generation” sequencing (NGS). NGS is a broad term that refers to a set of methods for: 1) genomic template preparation, or the methodology for processing genomic DNA for downstream sequencing; 2) near simultaneous, or “massively-parallel”, generation of millions to billions of short sequence reads; 3) alignment of sequence reads to a reference sequence; 4) sequence assembly from aligned sequence reads and genetic variant discovery (Figure 2). Most investigators use the output from this final step, a list of genotypes for positions with at least one allele that differs from a reference sequence (“variants”) in all downstream analysis. Thus, “whole genome sequence” data generally refers not to ~3 billion diploid genotypes that cover the known chromosomal positions but the 3–4 million genotypes in each genome that differ from the reference sequence. Several NGS technologies exist that differ primarily in methods for clonal amplification of short fragments of DNA and sequencing the resulting short DNA fragments. Each has specific advantages in terms of read length, accuracy, and throughput (Table 1). All currently forego the time-consuming bacterial cloning step that was used for library preparation in the Human Genome Project. For full details of the technical aspects of each sequencing technology, we refer the reader to recent technological reviews.8,9 We will briefly review each technology here with a focus on advantages, disadvantages and specific sequencing applications for each platform. One issue that deserves specific mention is that of read length. Shorter sequence reads (100 base pairs or shorter) are well suited to the biochemical reactions employed by most of the sequencing technologies. However, the generation of short reads complicates sequence assembly, particularly in repetitive regions of the genome. The generation of longer sequence reads (1000 base pairs or longer) simplifies this task. Furthermore, the use of longer sequence reads spanning several variants aids in resolution of “haplotype phase”, which is the assignment of each allele in a heterozygous genotype to one chromosome of each homologous pair, e.g., the assignment of an “A” allele in a “A/G” genotype to a paternally-derived segment of chromosome 13.
Figure 2.
Three generations of human genome sequencing technology. Three groups of sequencing technology are depicted: sequencing in the human genome project; second generation sequencing as exemplified by the Illumina HiSeq 2000; third generation sequencing as exemplified by the Helicos Heliscope single molecule sequencer.
Table 1.
Sequencing Platform Comparison
| Platform | Amplification | Sequencing | Detection | Read Length | Output per Run |
Run Time |
|---|---|---|---|---|---|---|
| Second-generation sequencing platforms | ||||||
| 454 | Emulsion PCR on beads |
Unlabeled nucleotide incorporation |
Detection of light emitted by release of PPi |
Variable (400 bp for single end sequencing) |
400--600 Mbp |
10 h |
| SOLiD | Emulsion PCR on beads |
Ligation of 2- base encoded fluorescent oligonucleotides |
Fluorescence emission from labeled oligonucleotides |
75+35 bp | 20--30 Gbp | 7 d |
| Illumina | Array-based enzymatic amplification |
Fluorescently labeled end- blocked nucleotide incorporation |
Fluorescence emission from nucleotides |
2[times]100 bp |
100--200 Gbp |
8 d |
| Complete | Rolling-circle replication of short segments of DNA into nanoballs |
Ligation of fluorescently labeled oligonucleotide probes |
Fluorescence emission from oligonucleotide probes |
2[times]35 bp |
20--60 Gbp | 12 d |
| Third-generation sequencing platforms | ||||||
| Helicos | NA | Single dye- labeled nucleotides are added sequentially and incorporated by polymerases by use of single DNA molecular templates |
Microscopy of fluorescently labeled nucleotides |
2[times]25– 55 bp |
21--35 Gbp | 8 d |
| Pacific Biosciences |
NA | Incorporation of fluorescently labeled nucleotides by polymerases on solid support |
Zero-mode waveguide imaging of fluorescent nucleotide incorporation by individual polymerases |
2[times]1000 bp |
75--100 Mbp (projected); 5--10 Mbp (actual usable sequence) |
30 min |
| Oxford nanopore |
NA | Processive endo- or exonuclease activity feeds individual bases or whole DNA strands through protein or solid- state nanopores |
Current disruption across nanopore corresponds to nucleotide structure |
Variable | Variable | Variable |
| Ion Torrent | Variable | DNA polymerase incorporation of unlabeled nucleotides added sequentially to solid-state microwells |
Solid-state detection of hydrogen ions released by nucleotide incorporation |
200 bp | 10 Mbp to 1 Gbp |
2 h |
bp indicates base pair; Gbp, one billion base pairs; Mbp, one million base pairs; PPi, pyrophosphate; and PCR, polymerase chain reaction.
Of the NGS platforms that are currently commercially available, the 454 (454 Life Sciences/Roche) instrument was developed first. This platform is based on “pyrosequencing” which detects light emitted by secondary reactions initiated by the release of pyrophosphate during nucleotide incorporation.10 Advantages include long reads and facile “mate-pair” sequencing, a method that sequences both ends of a previously circularized DNA molecule. Pairing reads that span tens of kilobases of genomic template sequence further facilitates haplotype phasing and the identification of structural genetic variation such as deletions and insertions of large segments of DNA. Disadvantages include systematic errors in reading frame (“frame shift errors”) in certain circumstances and lower throughput and higher sequencing costs than other commercial technologies.
SOLiD (Applied Biosystems by Life Technologies) sequencing utilizes sequencing-by-ligation in which the sequence of a DNA template is read by competitive ligation of 2-base probes to the nascent DNA strand.11 Advantages include throughput (~20–30 Gbp per run), and base-level error information encoded in the 2-base sequences, both of which make the platform suitable for human whole genome and exome variant discovery. The main disadvantage is the necessity to work with unconventional data formats for sequence reads and the reference genome.
The Illumina/Solexa (Illumina, Inc.) platform is widely used for a variety of applications, including human whole genome and exome variant discovery and transcriptome sequencing (“RNAseq”), by virtue of easily prepared paired-end sequencing libraries, high throughput, and ease of analysis of its short read information. After genomic DNA isolation, fragmentation, and several enzymatic modification steps, sequencing libraries are amplified from single DNA strands on glass surfaces. The resultant templates are sequenced using an approach in which fluorescently labeled “end-blocked nucleotides,” which do not allow further DNA polymerization, are incorporated by DNA polymerase, the base-specific fluorescent color is detected via fluorescence imaging, the end block and fluorescent tag is enzymatically cleaved, and the process is repeated following image storage, yielding image-encoded nucleotide sequences.12 Drawbacks include comparatively short sequence reads (≤100 bp) and practical limits to insert sizes for paired end sequencing.
Complete Genomics, Inc. provides a sequencing service, in contrast to other companies that have primarily focused on providing sequencing instruments, that is targeted solely towards human whole genomes. The instrument uses sequencing-by-ligation of hundreds of “DNA nano-balls,” or chained-replicates of 70-base-pair sequences of sheared genomic DNA modified by adaptor inserts.13 Theoretical throughput exceeds that of any of the NGS technologies described thus far.
“Third” generation sequencing technologies
A “third” generation of sequencing instruments has been developed that is defined by the lack of DNA or RNA amplification in template library preparation (“single molecule sequencing”, Figure 2). By foregoing this step, these technologies require less genomic DNA, avoid PCR-introduced error and amplification bias, and may be superior for high-throughput sequencing applications, such as transcriptome sequencing (“RNAseq”), that depend on accurate quantification of relative DNA or RNA fragment abundance.
The first of these single-molecule sequencing technologies is the Helicos Heliscope (Helicos BioSciences). The specific Helicos chemistry is based on single-molecule sequencing by cyclic reversible terminator nucleotide incorporation.14 A single dye molecule is used to label the dNTPs and fluorescence microscopy is used to image the dye in sequencing reactions carried out on single molecule templates on solid support. The order in which each fluorescently labeled dNTP is added to the sequencing reactor determines the base sequence at that position. Notably, the instrument is also suitable for direct RNA sequencing without conversion to complementary DNA (cDNA), thus avoiding error and copy number bias associated with reverse transcription.15
Pacific Biosciences have recently developed a method for imaging individual DNA polymerase molecules as they synthesize a nascent DNA molecule covalently attached to solid support.16 Advantages include read information that is theoretically as long as 1 kb or longer and real-time sequencing kinetics that reflect nucleotide methylation state and DNA secondary structure.17
Life Technology’s Ion Torrent device is targeted towards individual laboratories interested in a small footprint, medium throughput sequencing platform. This sequencing engine is based on detection of hydrogen ions released from nucleotides incorporated into the growing DNA strand.18 This signal is detected in a solid-state semiconductor akin to a miniaturized pH meter, and the technology is theoretically suitable to single-molecule sequencing. Throughput is currently low (< 1 Gbp per run), but the release of higher density chips has made sequencing of transcriptomes and exomes feasible.
“Nanopore” sequencing technologies detect base-specific changes in ionic flux as DNA traverses small pores in solid surfaces that are placed in an electric field.19 Advantages to this method include theoretically unparalleled sequencing speed and minimal template preparation. At this point, however, detection speed and accuracy remain significant technological hurdles, as the transit speed of nucleic acids through nanopores in even minimal electric fields is several orders or magnitude higher than the highest detection frequency. Several enzymatic methods have been developed to slow transit time and facilitate detection of changes in ionic flux.20,21
Processing high-throughput sequence data
Data generation from high-throughput sequencing is becoming less expensive and time consuming. Generating sequence data, however, is only the first step in extracting usable information from high-throughput sequencing sequencing. For output from most currently available sequencing platforms, several tasks must be performed prior to downstream analysis: 1) short read mapping, or alignment of each sequence read to a reference genome to identify the genomic sequence represented by the short read; 2) base calling at every genomic position covered by aligned short reads; 3) identification of sequence variation from the reference genome. The percentage of base positions that are read by properly aligned short reads is described by “coverage.” The number of times that a single base position is read by short read sequences is termed “depth of coverage” and most investigators currently consider 30-fold (“30×”) average depth of coverage as a benchmark for high-quality genome sequence data. Prior to discussing these data management issues, it is worth highlighting some of the limitations of the current approach that utilizes a haploid reference sequence, that is, a sequence that has only one base for every genomic position.
The human reference genome and its limitations
The human reference genome currently used for short read alignment and variant calling (NCBI reference genome22) is derived from a collection of DNA samples from a small number of anonymous donors. It is currently the only “finished-grade” human genome in that it was assembled de novo from long sequence reads and covers ~ 99% of known chromosomal positions with high fidelity. However, it represents a very small sampling of human genetic variation. Analysis in our lab using the 1000 genomes population variation data demonstrated that at ~1.6 million genomic positions, the NCBI reference sequence differed from the major, or most frequent, allele in each of the three HapMap populations, including ~ 800,000 positions at which all three population groups have major alleles that differ from the NCBI reference allele.23 Additionally, the reference sequence contains thousands of common and rare disease risk alleles, including more than twenty rare disease susceptibility alleles such as the Factor V Leiden allele associated with hereditary thrombophilia.23,24 Various approaches to addressing these issue have been suggested, including the use of a “major allele” reference sequence. We have recently used this approach to identify the putative genetic basis for familial thrombophilia in a family quartet using whole genome sequencing.23 Notably, the multi-genic risk for this trait we identified included the Factor V allele conferring activated protein C resistance, which would not have been identified in homozygous state using the NCBI reference genome for variant identification.
Aligning sequence reads to the human reference genome
There are several programs for mapping short reads to a reference genome; for an in-depth comparison of alignment programs, we direct the reader to a recent work by Li and Homer.25 Historically, mapping alignment with quality (“MAQ”) was the most widely used alignment algorithm,26 but this algorithm has been supplanted by other open-source solutions that are superior for longer (>35 bp) sequence reads. Though several alignment algorithms can be run on high-memory multiple core desktops and even laptops, parallel computing architecture, which utilizes multiple processors to perform alignment tasks simultaneously, reduces the time required for alignment several fold. Unfortunately, few individual labs currently are able to provide this computing power. One solution is on-demand distributed or parallel computing architecture, i.e., “cloud” computing. This approach is economical in the sense that elastic parallel computing environments allow users to select and utilize only processing and storage capacity necessary for current tasks.
Identifying single nucleotide variants and small insertions/deletions
Following alignment to the reference genome, sequence reads are compared at every genomic position, producing a base call for each chromosomal position. For in-depth discussion of genotype calling from next generation sequence data, including the use of linkage disequilibrium for genotype determination and probabilistic genotypes for low- and intermediate coverage sequencing, such as that employed in the 1000 genomes project, we direct the reader to a recent work by Nielsen, et al.27 A variety of different algorithms incorporate base quality, which specifies the confidence of each base call within the individual short reads, mapping quality, or confidence of accurate mapping of each short read to the specified genomic locus, and the number of bases contributing to each of the possible 16 genotypes at a position, into a probabilistic score for genotypes at every chromosomal location. The most likely genotype is compared to the reference sequence, and, typically, only positions containing at least one base differing from the reference sequence are retained for downstream analysis. This fact has several important implications. First, the reference base is crucial to the identification of genetic variation: if the haploid reference base harbors the same allele predisposing to disease as the subject being sequenced, it will not appear in the variant list, potentially leading to underestimation of the burden of certain disease-associated alleles. Second, comparison between individuals, e.g., in co-segregation and linkage studies, can be complicated by the degree of overlap between genetic variant sets such that the assumption of homozygous reference allele calls can bias exploratory studies for causative variants. Several variant calling solutions, notably, SAMtools28 and the Genome Analysis Toolkit (GATK)29 have base calling algorithms that facilitate cohort-wide variant identification, which addresses this problem. Third, the reference sequence represents a small sampling of human genetic variation, and as large scale sequencing efforts are undertaken, ethnicity-specific major allele differences may impact alignment of short reads against the current reference genome and subsequent variant identification.
Identifying large structural variants
Large structural rearrangements > 1kb, termed structural variants (SVs), encompass large deletions, duplications, insertions, and inversions, and transposons. Largely ignored in many early sequencing efforts, emerging evidence suggests that these structural variants are strongly associated with several Mendelian and complex diseases, including familial dilated cardiomyopathy, autism spectrum disorders, idiopathic mental retardation, schizophrenia, and Crohn’s disease.30–35 In some cases these large genetic variants underly > 15% of disease diagnoses.36 Several methods have been developed for identification of SVs, but three main methods have generally been accepted and are used for identification of specific types of SVs. A complementary, hybridization-based method for identifying SVs, comparative genomic hybridization, will not be discussed further here. Notably, however, due to high false positive rates for SV detection using high-throughput sequencing, this and other PCR-based methods are often used to confirm candidate SVs.
The first method for identification of structural variants is mate pair sequencing,37 which is based on sequencing two ends of a DNA molecule following circularization, providing paired short read sequence information separated by hundreds to thousands of base pairs. A related technique, paired end sequencing, is used routinely in most commercial sequencing technologies to provide paired short sequence reads from each end of an amplified linear DNA molecule. Comparison of median insert size and orientation from paired end reads to homologous chromosomal segments in the reference genome is used to identify structural rearrangements.38 Though sensitive for inversions and other “copy neutral” SVs, or SVs that do not change the copy number of the affected chromosomal region, and somewhat well suited to identifying start and end points of SVs (“breakpoints”), detection scope is limited by the size of the insert, in that only structural rearrangements spanned by the insert can be detected.
A second method for identification of structural rearrangements is based on regional variation in read depth, which is in turn dependent on copy number of the genomic region interrogated. Several methods have been developed for identification of significant differences in read depth in genomic regions relative to median read depth.39–43 This method for identification of SVs is ideally suited for identification of large insertions and deletions, but has limited capability to resolve breakpoints, and cannot distinguish copy neutral SVs from normal sequence.
The third method for identification of large SVs is split-read mapping, which is based on mapping elements with inserts in the reference genome or the sample genome to contiguous short read sequences by using one end of the read as an anchor and the other end to search for possible breakpoints, yielding single-nucleotide level breakpoint resolution and novel sequence discovery in some cases.44 Finally, candidate structural variants are often compared to known structural variants identified using population-scale sequencing or genotyping to provide probabilities of false discovery and improved breakpoint resolution.45
Variant quality control and genotype validation
Validation of sequence data has become a particularly difficult problem in interpretation of genetic variants discovered via high-throughput sequencing. Per genotype error rates for commercially available high-throughput sequencing technologies achieving an average depth of coverage of >30× are currently between one in every 1000 to one in every 100,000 bases. By comparison, per-genotype error rates for Sanger sequencing, the current standard for clinical applications, is between one in 100,000 and one in 1,000,000 base pairs. Filtering variants via a combination of quality score metrics for individual short reads and final genotypes can minimize errors. Roach, et al, and our group have demonstrated that leveraging family genotype information can also be useful for error identification, in that pedigree-based allele inheritance analysis can be used to identify not only inconsistencies with Mendel’s laws of inheritance, but regions in which short reads have been incorrectly mapped or genotyped.46 We have recently demonstrated a >90% reduction in the error rate by sequestering variants identified in these regions.23
Despite these and other advances in error reduction, however, high-throughput sequencing platforms do not yet provide the level of confidence about individual variants that would be required for routine incorporation into clinical care. To date, clinically important variants have mostly been re-sequenced using Sanger-based chemistry or confirmed with oligonucleotide genotyping arrays. Both approaches are time- and resource-intensive. Alternative capture-based approaches, in which either a standard commercial or custom oligonucleotide set is used to select genomic regions of interest for high-coverage high-throughput resequencing, are also costly and time-consuming. Validation of small structural variants such as insertions and deletions is even more difficult, often requiring bacterial cloning of single strands prior to re-sequencing. Until the accuracy of high-throughput sequencing improves such that primary data does not require orthogonal confirmation, data validation will continue to be a major barrier to widespread incorporation of high-throughput sequence data into clinical applications.
Haplotype phasing using high throughput sequence data
Resolution of haplotype phase is important to understanding shared disease-associate chromosomal segments containing variants that tend to be inherited en bloc, compound heterozygous (two or more risk alleles in one gene) and oligogenic (two or more risk alleles in multiple genes) genotype-phenotype associations, regulatory effects of genetic variation, and differential parent of origin effects in disease assocation studies.47 Furthermore, large databases of phased sequence data will be important resources for genome-wide association studies that utilize imputation, or estimation of genotypes not assayed by other technologies such as chip-based genotyping. This practice has become commonplace as investigators combine datasets to improve power to detect disease associations of small magnitude, and will be important for investigating rare variant effects. Short-read high throughput sequence data alone does not provide information about haplotype phase. However, several statistical algorithms based on pedigree information, common population haplotypes, and paired short reads have been developed that are applicable to high-throughput sequence data.47–50 Moreover, several investigators have developed experimental methods for haplotype phasing based on sorting individual metaphase chromosomes and subsequent sequencing,51 or from a combination of long-insert cloning and next-generation sequencing.52 Further development of these methods will be critical to the use of this tool for investigating disease biology.
High-throughput sequencing and Mendelian disease genetics
The utility of high-throughput sequencing for investigation of disease genetics is great. The application of NGS for the identification of cardiovascular disease-associated loci has resulted in several notable successes, including the identification of BAG3 mutations as a cause of dilated cardiomyopathy, mutations in SMAD3 associated with familial aortic aneurysms, and AARS2 and ACAD9 in familial mitochondrial cardiomyopathy35,53–55 (Table 2). These studies have provided intriguing hypotheses for follow-up work characterizing novel pathways in human cardiovascular disease. The genetic basis for several non-cardiovascular diseases has similarly been explored using exome and whole-genome sequencing (Table 3). Notably, two studies have demonstrated the promise of NGS in aiding clinical diagnosis and management. Choi, et al used exome sequencing to identify a mutation in SLC26A3 in a patient with the suspected renal salt-wasting Bartter syndrome; this finding allowed them to make the unanticipated diagnosis of congenital chloride diarrhea and modify clinical care accordingly.61 Worthey, et al used exome sequencing to identify a missense mutation in the gene XIAP in a patient with intractable Crohn’s-like inflammatory bowel disease, establishing a diagnosis of X-linked inhibitor of apoptosis (XIAP) deficiency. Subsequent allogeneic stem cell transplant resulted in dramatic improvement in the patient’s gastrointestinal disease.66
Table 2.
Exome and whole genome sequencing for cardiovascular disease gene identification.
| Disease | Inheritance model |
Sequenced subjects | Putative loci identified |
Validation | Reference |
|---|---|---|---|---|---|
| Complex I deficiency with hypertrophic cardiomyopathy |
Autosomal recessive |
One proband | ACAD9 | Wild-type cDNA complementation in fibroblasts; compound heterozygous or homozygous mutations found in ACAD9 in 120 index cases with complex I deficiency |
54 |
| Dilated cardiomyopathy |
Autosomal dominant |
Four affected family members |
BAG3 | 7 structural and single- nucleotide variants found in BAG3 in 311 unrelated probands; knockdown of bag3 in zebrafish recapitulated the phenotype |
35 |
| Familial thoracic aortic aneurysm |
Autosomal dominant |
Two distantly- related affected individuals in one family |
SMAD3 | Cosegregation in family; sequencing of 181 additional probands identified three additional SMAD3 mutations in four families |
53 |
| Infantile mitochondrial cardiomyopathy |
Autosomal recessive |
One proband | AARS2 | Co-segregation of mutation in a separate family; metabolomic analysis of post-mortem heart and skeletal muscle of proband demonstrating increased alanine levels |
55 |
| Kabuki syndrome | Autosomal dominant |
Ten unrelated cases | MLL2 | Sanger sequencing confirmation in 26 of 43 additional cases |
56 |
| Moyamoya disease | Complex | Index case from each of eight families |
RNF213 | Genome wide linkage analysis; combination of linkage analysis and sequencing of RNF213 in 42 index cases; case-control study in 958 subjects of East Asian ancestry |
57 |
| Sick sinus syndrome (SSS) |
Complex | 7 individuals with SSS and the rs28730774[T] variant associated with SSS; 80 individuals without SSS |
MYH6 | Genotyping in 469 SSS cases and 1185 controls |
58 |
Table 3.
Selected studies using exome and whole genome sequencing for non-cardiovascular disease gene identification.
| Disease | Inheritance model | Sequenced subjects | Putative loci identified |
Validation | Reference |
|---|---|---|---|---|---|
| Exome sequencing projects | |||||
| Carnevale, Malpuech, Michels, and oculo-skeletal- abdominal syndromes |
Autosomal recessive |
One proband | MASP1 | Co-segregation in two additional families |
59 |
| Charcot-Marie-Tooth neuropathy |
Autosomal recessive |
Two affected family members |
GJB1 | Co-segregation in the family | 60 |
| Congenital chloride losing diarrhea* |
Autosomal recessive |
One proband with suspected Bartter syndrome |
SLC26A3 | Sanger sequencing identification of homozygous variants in SLC26A3 in 5 of 39 unrelated patients with suspected Bartter syndrome; clinical followup demonstrating evidence of chloride losing diarrhea |
61 |
| FADD deficiency | Autosomal recessive |
One proband with biological features of ALPS |
FADD | Co-segregation in the family; In vitro assay of FADD protein levels and apoptotic activity |
62 |
| Familial amyotrophic lateral sclerosis |
Autosomal dominant |
Two affected individuals |
VCP | Sanger sequencing of a cohort of 210 familial ALS cases |
63 |
| Familial combined hypolipidemia |
Autosomal recessive |
Two affected siblings | ANGPTL3 | Co-segregation in family and frameshift mutations at same locus associated with phenotype in previous work |
64 |
| Fowler syndrome | Autosomal recessive |
Two unrelated cases | FLVCR2 | Previous reports of co- segregation in one unrelated family |
65 |
| Intractable inflammatory bowel disease; X-linked inhibitor of apoptosis deficiency* |
X-linked recessive | One proband | XIAP | Functional assay of PBMCs recapitulating XIAP deficiency and impaired immune reactivity. Bone marrow transplant of affected child improved disease |
66 |
| Joubert syndrome 2 | Autosomal recessive |
Proband and mother | TMEM216 | Parallel linkage mapping and candidate re-sequencing in 13 cases from 8 kindreds |
67 |
| Mental retardation | Autosomal recessive |
Two unaffected parents of five affected siblings |
TECR | Co-segregation in family | 68 |
| Mental retardation | Sporadic | Ten unrelated cases and their unaffected parents |
Several | Previous functional evidence suggests a role for the gene in mental retardation |
69 |
| Miller syndrome | Autosomal recessive |
Four affected individuals in three kindreds |
DHODH | Co-segregation by Sanger sequencing in three separate families |
70 |
| Neonatal diabetes mellitus | Autosomal dominant |
One proband | ABCC8 | Absence of mutation in healthy controls |
71 |
| Non-syndromic hearing loss (DFNB82) |
Autosomal recessive |
Single proband | GPSM2 | Co-segregation in family, Not found in 192 controls and 192 unrelated cases |
72 |
| Primary Lymphedema | Autosomal dominant |
One proband | GJC2 | Co-segregation by Sanger sequencing in four additional families |
73 |
| Schinzel-Giedion syndrome |
Sporadic | Four unrelated cases | SETBP1 | Sanger sequencing of 9 unrelated cases and 188 controls |
74 |
| Seckel syndrome | Autosomal recessive |
Single proband | CEP152 | Parallel linkage analysis and candidate region sequencing in a separate family; morphological analysis of CEP152-deficient mitotic cells |
75 |
| Spinocerebellar ataxia | Autosomal dominant |
Four affected family members |
TGM6 | Co-segregation of mutations in the same gene in a second family |
76 |
| Genome sequencing projects | |||||
| Charcot-Marie-Tooth Neuropathy |
Autosomal recessive |
One proband | SH3TC2 | Co-segregation with the phenotype by Sanger sequencing in family |
77 |
| Metachondromatosis | Autosomal dominant |
One proband and partial linkage analysis in family |
PTPN11 | Co-segregation by Sanger sequencing in a second family |
78 |
| Miller syndrome | Autosomal recessive |
Two affected offspring and both parents |
DHODH, DNAH5, and KIAA0556 | Previously published exome sequencing (reference70) |
46 |
| Sitosterolemia* | Autosomal recessive |
One proband | ABCG5 | Presence of sitosterolemia in blood sample after weaning; previous association with sitosterolemia |
79 |
Sequence information used to make clinical diagnosis.
Thus far, these studies have focused on well-characterized diseases with extreme phenotypic manifestations and co-segregation analysis of single gene loci. However, filtering variants by co-segregation with the disease phenotype, as well as by comparison with population controls, e.g., the dbSNP and 1000 genomes genetic variation databases, has not always yielded a definitive answer. This difficulty is further compounded by the inclusion of non-validated SNPs in recent iterations of these databases. Consequently, these repositories now contain a small but definite subset of putative variants that are actually sequencing errors. Filtering of variants in co-segregation studies by their presence in these databases may thus lead to the misidentification of damaging mutations as benign polymorphisms. Further annotation of variants by identity-by-descent status, which seeks to identify common ancestral disease-associated haplotypes, represents an evolution of the co-segregation approach.80,81
High throughput sequencing and complex disease genetics
More recently there has been increasing interest in the use of high-throughput sequencing for association analysis with complex disease. Much of the focus has been on discovering the source of “missing heritability” of common complex diseases. Six years has elapsed since the first publication of a genome wide association study of common genetic variants and common disease.82 Since then, hundreds of highly statistically significant, replicated associations with common disease have been found. However, most alleles identified via this technique confer modest risk, and the heritability of common disease explained by these alleles in isolation or aggregate is low.83 One of the hypotheses for this relative paucity of high-effect associations is that rare variants of large effect contribute in aggregate to common disease. By virtue of their rarity, these variants have not been included on current genotyping arrays and therefore previous GWAS studies have thus been unable to assess their association with disease. Furthermore, several investigators have hypothesized that some of the modest associations between common variants and common disease are mediated via weak linkage disequilibrium between common marker variants and rare, causative variants of large effect.84 Recently, Stefansson, et al used a combination of large-scale chip-based genotyping and intermediate-depth (10×) whole genome sequencing of a smaller cohort of cases and controls to identify a rare variant in a novel locus strongly associated with sick sinus syndrome.58 Importantly, this is the first demonstration of the use of whole genome sequencing to identify an association between a rare variant and complex disease. Though it is not yet cost effective to perform deep whole genome sequencing of large cohorts of individuals with common disease, as sequencing costs drop, genotype-phenotype association studies using whole genome sequencing may become feasible. Meanwhile, several efforts are currently underway to identify coding variants using exome sequencing associated with complex phenotypes.
An important advantage of whole genome over whole exome association studies is the ability to interrogate the noncoding genome. Despite systematic over-representation of protein coding regions on many genotyping arrays, 88% of significant GWAS associations are located in intronic or intergenic regions.85 Thus, exome-targeted sequencing approaches are likely to miss the majority of significant genome wide associations with common disease. For Mendelian disorders, the majority of underlying allelic variants identified thus far disrupt coding regions, and thus many early sequencing efforts have focused on the exome. However, it is likely that more comprehensive variant discovery will be required for discovery of many genotype-phenotype associations.
Genome sequencing and the clinic
Applying the bulk of genetic predictive information to whole genome sequence data from individuals is one of the most difficult tasks in NGS data interpretation. We previously developed and applied a methodology for interpretation of genetic and environmental risk in a single subject using a combination of traditional clinical assessment, whole genome sequencing, and integration of genetic and environmental risk factors,86 and have recently done so for a family quartet.23 A similar approach has been applied to carrier testing for severe recessive childhood disease risk using NGS and for detection of fetal aneuploidy via sequencing of maternal blood samples.87,88 One of the main challenges to the widespread application of these analytical schemes is incomplete and inconsistent status of publicly-available genome annotation databases. Several annotation sources exist for gene regions, including the consensus coding sequence (CCDS) database,89 RefSeq,22 the UCSC KnownGenes database,90 and the GENCODE91 and ENSEMBL92 databases. Each has advantages in terms of coverage and accuracy; however, the inconsistent use of these data in the literature is an issue for replicating research findings. Similarly, several variant databases exist for associations with Mendelian disorders, including the Human Gene Mutation Database,93,94 the Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim), and many disease-specific databases. None are well suited to variant-level annotation of whole genome sequence data and many contain annotation errors and common polymorphisms, by some estimates comprising approximately > 25% of the entries. Furthermore, these databases are contaminated by descriptions of susceptibility loci of questionable impact,87 and mutation annotations are often based on differing builds of the reference genome or outdated gene and protein sequences. Several prediction algorithms exist for predicting variant pathogenicity that are based on different combinations of evolutionary conservation, structural prediction, and physical properties of amino acid substitutions.95–99 However, they are limited in specificity and sensitivity, and concordance between predictions from the various algorithms is low.100 Databases for common variant – common disease associations85,86 and pharmacogenomic associations101 are more complete, but there is a great need for comprehensive, easily searchable, and accurate variant-level association databases as whole genome sequence data becomes more widely available.
Other applications of high throughput sequencing
Though high-throughput sequencing has become synonymous with whole genome and exome sequencing, there are many other emerging applications for the technology. The first of these is whole transcriptome sequencing, which uses massively parallel sequencing to sequence RNA transcripts in various physiological conditions. This unique application allows for determination of allele-specific expression, information about alternative splicing, RNA editing events, and, via read depth, accurate quantification of messenger RNA (mRNA) copy number, and, therefore, gene expression. Compared with oligonucleotide expression arrays, RNAseq is able to quantify transcript abundance with a greater dynamic range and accuracy at extremes of transcript abundance, allowing for more accurate quantification of gene expression and rich functional genomics information. Matkovich, et al, recently used a unique combination of RNAseq and a new technology, RNA-induced silencing complexes (RISC)-sequencing, to characterize cardiac mRNA regulation by microRNAs, small noncoding RNAs that regulate diverse cellular functions by facilitating mRNA degradation or inhibiting translation.102 Technologies that do not require generation and amplification of a cDNA library, such as the Helicos platform, are particularly well suited to this application because they require no prior knowledge of the transcriptome and avoid biases in gene expression measurements and sequencing errors that are related to reverse transcription.
Secondly, subsets of exomes can be queried in a high-throughput manner in the next generation of candidate gene studies using custom oligonucleotide based capture techniques coupled with high throughput sequencing.103–106 Combined with a pooled case-control approach, these study designs may prove to be valuable to gene finding or comprehensive sequence interrogation (“fine mapping”) of genomic regions that have been linked with inherited disease by other technologies such as array-based genotyping or repetitive element mapping.
Third, there is increasing focus on the use of high-throughput sequencing in clinical diagnosis via the rapid identification of cell-free DNA. Specific to cardiovascular medicine, is the recent demonstration of the use of cell-free sequencing of blood samples for the identification of an organ-specific “transplant DNA” signature correlating with acute cellular rejection in a pilot study of heart transplant recipients.107 With confirmation in larger cohorts, technologies such as these may be combined with other functional assays of the genome such as gene expression arrays108 to obviate the need for endomyocardial biopsy surveillance in select patients.
Lastly, while we have focused in much of this review on inherited genetic information, there is an entirely separate dimension of heritable information that researchers are just beginning to explore on a genome-wide scale. Epigenetic traits, or heritable traits that do not involve DNA sequence changes, are often due to chemical modifications of the DNA molecule such as cytosine methylation in CpG regions.109,110 To date, bisulfite sequencing, in which 5-methyl-cytosine bases are converted to uracil by bisulfite and subsequently sequenced, identifying CpG regions with high uracil content that correspond to methyl-cytosine bases, has been the standard technique. However, single molecule sequencers yield polymerase kinetics information that correlates with methylation status and other structural information such as DNA polymerase footprint and RNA and DNA secondary structure.
Conclusions
The whole genome sequencing era is here. Challenges remain to widespread sequencing of individuals. However, advances in high-throughput sequencing technologies have made possible initial strides in understanding the fundamental genetic basis for inherited disease, and sequencing personal genomes may someday allow for individualization of health care to genetics. Ten years out from the completion of the human genome sequence, we are about to enter an era in which a vast amount of sequencing information will be available to medical researchers and, ultimately, health care professionals. It is incumbent upon physicians and scientists as stewards of this technology to ensure that high quality sequence data is incorporated appropriately into research and clinical endeavors.
Acknowledgments
Sources of funding
FED was supported by NIH/NHLBI training grant T32 HL094274-01A2 and the Stanford Dean’s Postdoctoral Research Fellowship. MTW was supported by NIH National Research Service Award fellowship F32 HL097462. EAA was supported by NIH/NHLBI KO8 HL083914, NIH New Investigator DP2 Award OD004613, and a grant from the Breetwor Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
SRQ is a founder, consultant and equity holder in Helicos BioSciences and a founder, consultant and shareholder of Fluidigm corp. EAA is a founder and stockholder in Personalis.
References
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 5.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 8.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 9.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 11.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling NgB, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 14.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 16.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 17.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 19.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 20.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J Am Chem Soc. 2010;132:17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewey FE, Chen R, Cordero SP, Ormond KE, Caleshu C, Karczewski KJ, Carrillo MW, Wheeler MT, Dudley JT, Byrnes JK, Corenejo OE, Knowles JW, Woon M, Sangkuhl K, Gong L, Thorn CF, Hebert JM, Capriotti E, David SP, Pavlovic A, West A, Thakuria J, Ball MP, Zaranek AW, Rehm HL, Church GM, West JS, Bustamante CD, Snyder M, Altman RB, Klein RJ, Butte AJ, Ashley EA. Phased whole genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002280. e1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, Butte AJ. The reference human genome demonstrates high risk of type 1 diabetes and other disorders. Pac Symp Biocomput. 2011:231–242. doi: 10.1142/9789814335058_0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nature reviews. Genetics. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 31.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, Pakula AT, Eisenhauer NJ, Uhas K, Weik L, Guy L, Care ME, Morel CF, Boni C, Salbert BA, Chandrareddy A, Demmer LA, Chow EW, Surti U, Aradhya S, Pickering DL, Golden DM, Sanger WG, Aston E, Brothman AR, Gliem TJ, Thorland EC, Ackley T, Iyer R, Huang S, Barber JC, Crolla JA, Warren ST, Martin CL, Ledbetter DH. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Zuchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, Reiser J, Martin E, Nickerson DA, Hershberger RE. Genome-wide Studies of Copy Number Variation and Exome Sequencing Identify Rare Variants in BAG3 as a Cause of Dilated Cardiomyopathy. Am J Hum Genet. 2011;88:273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang LY, Abyzov A, Korbel JO, Snyder M, Gerstein M. MSB: a mean-shift-based approach for the analysis of structural variation in the genome. Genome Res. 2009;19:106–117. doi: 10.1101/gr.080069.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang ZD, Gerstein MB. Detection of copy number variation from array intensity and sequencing read depth using a stepwise Bayesian model. BMC Bioinformatics. 2010;11:539. doi: 10.1186/1471-2105-11-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abyzov A, Gerstein M. AGE: defining breakpoints of genomic structural variants at single-nucleotide resolution, through optimal alignments with gap excision. Bioinformatics. 2011;27:595–603. doi: 10.1093/bioinformatics/btq713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam HY, Mu XJ, Stutz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nature reviews. Genetics. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 50.Williams AL, Housman DE, Rinard MC, Gifford DK. Rapid haplotype inference for nuclear families. Genome Biol. 2010;11:R108. doi: 10.1186/gb-2010-11-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, Shendure J. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regalado ES, Guo DC, Villamizar C, Avidan N, Gilchrist D, McGillivray B, Clarke L, Bernier F, Santos-Cortez RL, Leal SM, Bertoli-Avella AM, Shendure J, Rieder MJ, Nickerson DA, Milewicz DM. Exome Sequencing Identifies SMAD3 Mutations as a Cause of Familial Thoracic Aortic Aneurysm and Dissection With Intracranial and Other Arterial Aneurysms. Circ Res. 2011;109:680–686. doi: 10.1161/CIRCRESAHA.111.248161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, Uziel G, Lamantea E, Invernizzi F, Poulton J, Rolinski B, Iuso A, Biskup S, Schmidt T, Mewes HW, Wittig I, Meitinger T, Zeviani M, Prokisch H. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 55.Gotz A, Tyynismaa H, Euro L, Ellonen P, Hyotylainen T, Ojala T, Hamalainen RH, Tommiska J, Raivio T, Oresic M, Karikoski R, Tammela O, Simola KO, Paetau A, Tyni T, Suomalainen A. Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. American journal of human genetics. 2011;88:635–642. doi: 10.1016/j.ajhg.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda A, Kikuta K, Takagi Y, Harada KH, Fujiyama A, Herzig R, Krischek B, Zou L, Kim JE, Kitakaze M, Miyamoto S, Nagata K, Hashimoto N, Koizumi A. Identification of RNF213 as a Susceptibility Gene for Moyamoya Disease and Its Possible Role in Vascular Development. PLoS One. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, Stefansdottir H, Gretarsdottir S, Matthiasson SE, Thorgeirsson GM, Jonasdottir A, Sigurdsson A, Stefansson H, Werge T, Rafnar T, Kiemeney LA, Parvez B, Muhammad R, Roden DM, Darbar D, Thorleifsson G, Walters GB, Kong A, Thorsteinsdottir U, Arnar DO, Stefansson K. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirmaci A, Walsh T, Akay H, Spiliopoulos M, Sakalar YB, Hasanefendioglu-Bayrak A, Duman D, Farooq A, King MC, Tekin M. MASP1 mutations in patients with facial, umbilical, coccygeal, and auditory findings of Carnevale, Malpuech, OSA, and Michels syndromes. Am J Hum Genet. 2010;87:679–686. doi: 10.1016/j.ajhg.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montenegro G, Powell E, Huang J, Speziani F, Edwards YJ, Beecham G, Hulme W, Siskind C, Vance J, Shy M, Zuchner S. Exome sequencing allows for rapid gene identification in a Charcot-Marie-Tooth family. Ann Neurol. 2011;69:464–470. doi: 10.1002/ana.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, Puel A, Bacon CM, Rieux-Laucat F, Pang K, Britland A, Abel L, Cant A, Maher ER, Riedl SJ, Hambleton S, Casanova JL. Whole-exome-sequencing-based discovery of human FADD deficiency. Am J Hum Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lalonde E, Albrecht S, Ha KC, Jacob K, Bolduc N, Polychronakos C, Dechelotte P, Majewski J, Jabado N. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum Mutat. 2010;31:918–923. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- 66.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 67.Edvardson S, Shaag A, Zenvirt S, Erlich Y, Hannon GJ, Shanske AL, Gomori JM, Ekstein J, Elpeleg O. Joubert syndrome 2 (JBTS2) in Ashkenazi Jews is associated with a TMEM216 mutation. Am J Hum Genet. 2010;86:93–97. doi: 10.1016/j.ajhg.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caliskan M, Chong JX, Uricchio L, Anderson R, Chen P, Sougnez C, Garimella K, Gabriel SB, Depristo MA, Shakir K, Matern D, Das S, Waggoner D, Nicolae DL, Ober C. Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Hum Mol Genet. 2011;20:1285–1289. doi: 10.1093/hmg/ddq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 70.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, Lobbens S, Simon A, Bellanne-Chantelot C, Letourneau L, Scharfmann R, Delplanque J, Sladek R, Polak M, Vaxillaire M, Froguel P. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, Kanaan M. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ostergaard P, Simpson MA, Brice G, Mansour S, Connell FC, Onoufriadis A, Child AH, Hwang J, Kalidas K, Mortimer PS, Trembath R, Jeffery S. Rapid identification of mutations in GJC2 in primary lymphoedema using whole exome sequencing combined with linkage analysis with delineation of the phenotype. J Med Genet. 2011;48:251–255. doi: 10.1136/jmg.2010.085563. [DOI] [PubMed] [Google Scholar]
- 74.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, Devriendt K, Amorim MZ, Revencu N, Kidd A, Barbosa M, Turner A, Smith J, Oley C, Henderson A, Hayes IM, Thompson EM, Brunner HG, de Vries BB, Veltman JA. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 75.Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nurnberg P, Karaguzel A, Wollnik B. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, Jiang H, Zhang P, Shen L, Guo JF, Li N, Li YR, Lei LF, Zhou J, Du J, Zhou YF, Pan Q, Wang J, Li RQ, Tang BS. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 77.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, Gumbs CE, Pevsner J, Thomas G, Valle D, Hoover-Fong JE, Goldstein DB. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000991. e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Human molecular genetics. 2010;19:4313–4318. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, Albrechtsen A, Andersen G, Cao H, Korneliussen T, Grarup N, Guo Y, Hellman I, Jin X, Li Q, Liu J, Liu X, Sparso T, Tang M, Wu H, Wu R, Yu C, Zheng H, Astrup A, Bolund L, Holmkvist J, Jorgensen T, Kristiansen K, Schmitz O, Schwartz TW, Zhang X, Li R, Yang H, Wang J, Hansen T, Pedersen O, Nielsen R. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 81.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 82.Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang K, Dickson SP, Stolle CA, Krantz ID, Goldstein DB, Hakonarson H. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86:730–742. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000294. e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra64. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S41–S49. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cooper DN, Ball EV, Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, Nickerson DA. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 98.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005;311:179–191. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- 102.Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Friedman TB. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berg JS, Evans JP, Leigh MW, Omran H, Bizon C, Mane K, Knowles MR, Weck KE, Zariwala MA. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: Implications for application to clinical testing. Genet Med. 2011;13:218–229. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, Tilanus MA, Bouwhuis S, Mukhopadhyay A, Scheffer H, Hoefsloot LH, Veltman JA, Cremers FP, Collin RW. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vermeer S, Hoischen A, Meijer RP, Gilissen C, Neveling K, Wieskamp N, de Brouwer A, Koenig M, Anheim M, Assoum M, Drouot N, Todorovic S, Milic-Rasic V, Lochmuller H, Stevanin G, Goizet C, David A, Durr A, Brice A, Kremer B, van de Warrenburg BP, Schijvenaars MM, Heister A, Kwint M, Arts P, van der Wijst J, Veltman J, Kamsteeg EJ, Scheffer H, Knoers N. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am J Hum Genet. 2010;87:813–819. doi: 10.1016/j.ajhg.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 108.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 109.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001316. e1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]