Abstract
Mobile elements account for almost half of the mass of the human genome. Only the retroelements from the non-LTR (long terminal repeat) retrotransposon family, which include the LINE-1 (L1) and its non-autonomous partners, are currently active and contributing to new insertions. Although these elements seem to share the same basic amplification mechanism, the activity and success of the different types of retroelements varies. For example, Alu-induced mutagenesis is responsible for the majority of the documented instances of human disease induced by insertion of retroelements. Using copy number in mammals as an indicator, some SINEs have been vastly more successful than other retroelements, such as the retropseudogenes and even L1, likely due to differences in post-insertion selection and ability to overcome cellular controls. SINE and LINE integration can be differentially influenced by cellular factors, indicating some differences between in their amplification mechanisms. We focus on the known aspects of this group of retroelements and highlight their similarities and differences that may significantly influence their biological impact.
Keywords: LINE, SINE, Alu, Retropseudogene, Non-LTR retrotransposon, SVA, TPRT, Review
2. INTRODUCTION TO THE ACTIVE NON-LTR RETROELEMENTS
Completion of the human genome sequence revealed mobile element numbers to be higher than expected, making up nearly half the mass of the genome (1). Most of these sequences represent fossils of previously active elements. Mobile elements are classified into RNA (Class I) or DNA (Class II) transposons based on the type of intermediate used in their amplification mechanism. Class I RNA transposons, also commonly referred to as retroelements or retrotransposons, are further subdivided into two families based on the presence or absence of long terminal repeats (LTRs). These elements amplify through a process termed retrotransposition (2), during which an RNA intermediate is reverse transcribed to generate a new copy. Although LTR- and non-LTR retrotransposons both use an RNA intermediate, their mechanism of insertion varies greatly (reviewed in 3). Evidence of current mobile element activity in humans is mostly limited to the subgroup of non-LTR retrotransposons: Long Interspersed Element -1 (LINE-1 or L1) and its non-autonomous partners, such as SVA and Alu. Recent publications (4–6) and data from the 1000 human genome project (7) demonstrate that the occurrence of new insertions is much higher than previously estimated dramatically contributing to diversity. Elements or inserts generated by an L1-driven retrotransposition process are identified by the characteristic hallmark features which include flanking target site duplications (TSD) of varying lengths, the usual presence of a 3’ poly adenosine stretch or A-tail and its integration into an A-rich sequence (Figure 1).
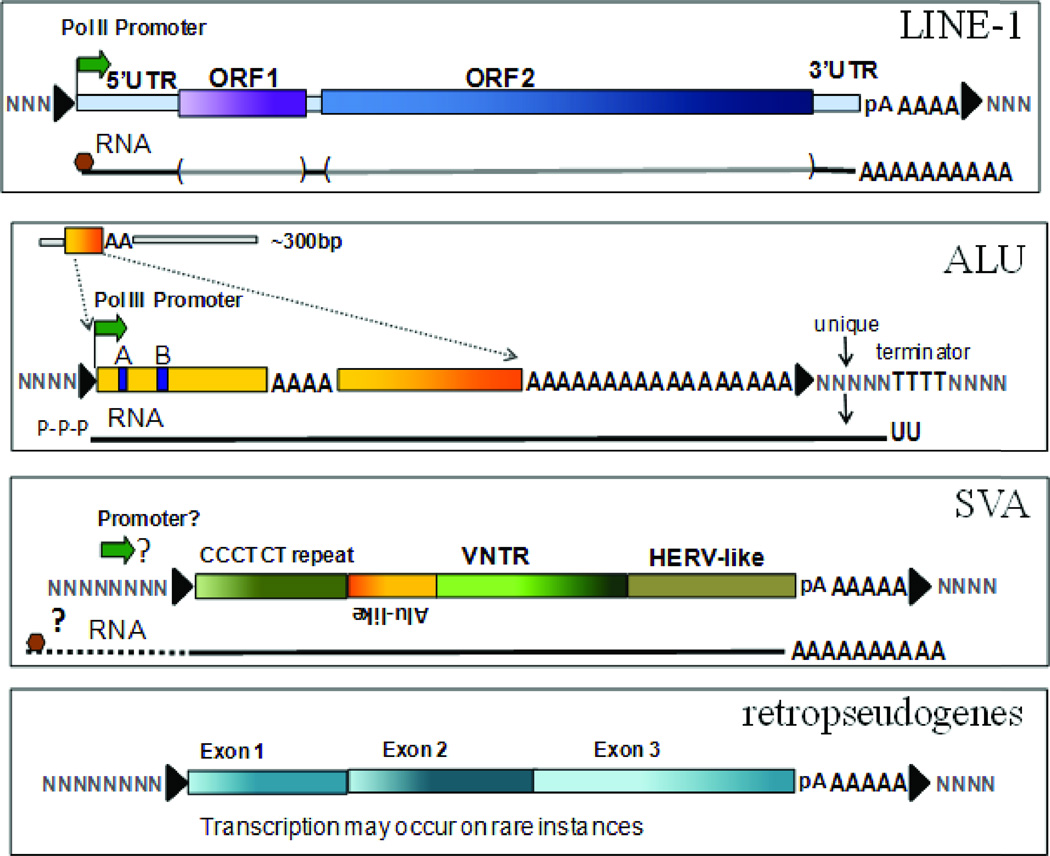
Figure 1.
Schematic representation of the structural organization of active human retroelements and their transcripts. Alu, SVA and LINE-1 belong to the non-LTR group of currently active human retroelements. Each panel shows the schematic of the element with the expected transcript (represented as a line and labeled RNA) shown below it. The boundaries of a retroelement are defined by the presence of tandem site duplications (TSDs) (shown as black arrowheads) generated during the retrotransposition process and which are a hallmark of retrotransposed sequences. Ns represent the flanking genomic sequence. The transcription start sites are represented by a green arrow. LINE-1 (L1) elements are composed of a 5’UTR that contains an internal RNA polymerase II promoter, followed by two open reading frames (ORF1 and ORF2) and a 3’UTR containing a polyadenylation signal (pA) and an A-tail. Full-length L1 transcripts are polyadenylated and capped (represented as a brown hexagon). Alu elements are about 300 bp in length (shown at the top of the panel, with a larger version that includes structural details shown below). Alu elements are composed of two non-identical sequences (monomers shown in yellow) separated by an adenine-rich region and flanked at the 3’ end by a longer A-rich region commonly referred to as an “A-tail”. The Alu left monomer contains the internal bipartite RNA pol III promoter composed of an A and B box (shown in blue). Alu transcripts will usually contain the 3’ flanking unique genomic sequence (shown in gray) past the A-tail due to the lack of a RNA pol III terminator sequence (TTTT) within the Alu sequence. Alu transcripts are thought to have a cap-like structure composed of a gamma-monomethyl phosphate (represented as P-P-P) that has been shown to be present in other pol IIII transcripts (135, 136). SVA elements are composite elements that contain a 5’ region with a variable number of CCCTCT repeats, an antisense Alu-like sequence, a variable number tandem repeat (VNTR) region, and a HERVK-like region previously referred to as SINE-R. A promoter has yet to be identified (represented as “?”), but it is presumed that RNA polymerase II likely drives transcription to generate a capped (brown hexagon) and polyadenylated transcript. Retropseudogenes or processed pseudogenes can originate from the retrotransposition of either RNA pol II or pol III transcripts. Retropseudogenes also are flanked by the hallmark TSD. Although the retrotransposed copies are not expected to contain a promoter sequence with them when they mobilize, sometimes the new flanking genomic sequence into which they insert provides promoter function, and thus some retropseudogenes generate transcripts.
L1 belongs to the family of non-LTR retrotransposons. There are about half a million L1 copies in the human genome. However, most L1 copies are 5’ truncated (1, 8) or contain inactivating mutations so that only about 100 are thought to be retrotranspositionally active (9). The human L1 is approximately 6000 bp in length with two open reading frames, ORF1 and ORF2 (Figure 1). ORF1 codes for a 40 kDa RNA binding protein thought to function as a nucleic acid chaperone (10). The ORF2p is a 149 kDa multifunctional protein with endonuclease and reverse transcriptase capabilities. Both proteins are essential for L1 retrotransposition (11). The L1 encoded proteins are thought to bind to the RNA to form a cytoplasmic RNP (12–14). Because L1 codes for the proteins required for its amplification, it is referred to as an autonomous element. The L1 5’UTR contains an atypical internal RNA polymerase II (pol II) promoter that initiates transcription upstream of the promoter (15). This helps L1 preserve the ability to self-mobilize since the promoter sequence is present in the transcript and can be passed on to new inserts. Another unusual feature of the L1 5’UTR is the presence of an antisense promoter (ASP), which transcribes the upstream flanking sequence (16, 17).
Alu elements are classified as SINEs (Short Interspersed Elements), due to their short length (~300 bp) and their transcription by RNA polymerase III (pol III). Because Alu has no coding capacity, it depends on L1 to obtain the factors needed for retrotransposition and thus considered a non-autonomous element. With over 1 million copies in the human genome (1), Alu is one of the most successful SINE elements known. However, the vast majority of the Alu copies in the human genome are not retrotranspositionally active. Only a few Alu copies are likely to be active Alu source elements (18). Alu elements are ancestrally derived from the 7SL RNA gene (19), the nucleic acid component of the signal recognition particle (SRP) involved in protein secretion (20). The region of the 7SL gene sharing sequence homology to Alu is defined as the “Alu domain”. An Alu is composed of two different GC-rich monomers separated by an adenine rich region and flanked at the 3’ end by another polyA stretch usually referred to as an “A-tail” (Figure 1). Only the 5’ monomer or “left monomer” of an Alu maintains the internal bipartite RNA polymerase III promoter composed of an A and B box (21). However, the Alu sequence does not contain an RNA pol III terminator (usually four thymidine residues). Thus, transcription of a typical Alu element will continue past the A-tail and into the genomic flank, resulting in Alu transcripts from various loci containing unique 3’ sequences of different lengths (Figure 1).
SVA (SINE/VNTR/Alu) elements are a composite of several sequences (Figure 1). About 2700 SVA elements have been identified in the human genome (22). A “full-length” SVA consists of 5’ tandem array of a hexameric sequence (CCCTCT), a segment with two partial Alu sequences in reverse orientation, followed by variable number tandem repeat (VNTR) segment, and a human endogenous retrovirus-K (HERV-K) like sequences, also referred to as SINE-R, at its 3’end. A promoter has yet to be identified within the SVA sequence. Because SVA displays the characteristic hallmarks of L1-mediated retrotransposition and predicted to have no coding capacity, this element is considered a non-autonomous member of the non-LTR retrotransposon family. However, due to the limited knowledge of its amplification mechanism, further classification of the SVA element as a SINE is still controversial. Not only is SVA uncommonly long (over 4kb) for a SINE which is typically 100–300 bp in length, this element is unlikely to be transcribed by RNA polymerase III due to the presence of multiple internal RNA pol III termination signals.
Retropseudogenes or processed pseudogenes arise from the reverse transcription of mRNA from a large variety of genes. Estimates for the number of retropseudogenes present in the human genome vary from about 8000 (23) up to 33,000 (24) depending on the parameters used for identification. Because they derive from processed transcripts, retropseudogenes typically do not contain introns and many contain a recognizable 3’ polyA tail and flanking TSD (Figure 1). Some genes are more prone to retropseudogene formation, thus the majority of retropseudogene copies being derived from a few genes. Some of the retropseudogenes with higher copy numbers include the highly expressed housekeeping genes GAPDH, cyclophilin, cytochrome c and the ribosomal proteins, which contribute to 20% of the total retropseudogene copies (23). One study suggests a strong association between short length genes and higher numbers of retropseudogene copies (25).
Other retrotransposed sequences from non-coding genes can be found in the human genome attesting to the promiscuity of the L1 retrotransposition machinery. These retrotransposed sequences will be flanked by the hallmark TSD. There are multiple examples of RNA polymerase III-generated transcripts that have been retrotransposed. For example, the human genome contains several hundred retropseudogene copies of 7SL (26) and multiple examples of U6 copies. Interestingly, many of the retrotransposed U6 sequences are found as part of a chimera with a 5’ truncated L1 sequence. This chimeric element is postulated to form in vivo by a “template switch” mechanism, where the ORF2p switches between the L1 RNA to the U6 transcript during reverse transcription (27). Another relatively successful family of RNA pol-III retrotransposition events is derived from the Y (hY) RNA genes associated with the Ro60 autoantigen with almost 1000 copies (28). Among the shorter retrotransposed sequences found in mammals are the “tailless” inserts derived from portions of tRNA or pre-tRNA sequences (29). Interestingly, retrotransposed copies from another mobile element, the endogenous retrovirus HERV-W, an LTR-retrotransposon, have also been reported (30).
3. EXPRESSION OF NON-LTR RETROELEMENTS
3.1. Expression of L1: the driving force
Expression of RNA is a requisite for amplification of retroelements. The vertical transmission of retroelements provides evidence of the expression somewhere in the germline. Because the non-autonomous elements depend on L1 products, understanding the distribution and extent of L1 expression is of great importance. L1 activity requires the L1 transcript as a template for the new copy as well as the expression of both ORF1 and ORF2 proteins (11). ORF1 protein (ORF1p) appears to be more abundant and easier to detect, making the evaluation of its endogenous expression more common in the literature. Endogenous expression of ORF1 has been reported in several human cell lines including teratocarcinoma and choriocarcinoma cells (31). Different studies using a variety of tumor samples detected ORF1p in breast and testicular cancers, pediatric germ cell tumors, ileal carcinoids, bladder and pancreatic neuroendocrine tumors including some samples of prostate and colorectal tumors (32–37). Although most examples detected ORF1p expression in the cytoplasm, some cancers displayed a nuclear localization of ORF1p. In these cases, nuclear detection of ORF1p correlated with poor prognosis (32). A detailed analysis of ORF1p expression in mice demonstrated its temporal regulation in germ line and steroidogenic tissue (38). ORF1p has also been detected in somatic cells (syncytiotrophoblasts from placenta) of adult mice (39) and in different regions of the brain of L1-transgenic mice (40). At the time of publication, data on endogenous ORF2p expression in human tissues are scarce. One study detects ORF2p in a variety of tissues including male gonads, prespermatogonia of fetal testis and germ cells of adult testis, Leydig, Sertoli and microvascular endothelial cells (41). As expected, ORF1p expression was also observed in the same cell types.
Detection of L1 proteins in a cell is not a reliable indicator of L1 retrotransposition activity as both ORF1p and ORF2p may derive from defective L1 copies and thus be non-functional. Analysis of L1 RNA expression is complex due to extensive processing by splicing (42, 43) and/or premature polyadenylation (44) of L1 transcripts. Northern blot analysis of L1 transcripts presents the advantage allowing distinction between full-length and other L1 products (Figure 2A), but may be limited in sensitivity. In contrast, RT-PCR approaches can detect very small amounts of L1 RNA. However, it is difficult to envision an RT-PCR approach that distinguishes between full-length and processed L1 products making RT-PCR data unreliable as an indicator of L1 activity. The application of some of the newer technologies such as paired end RNAseq may prove valuable for evaluating L1 transcripts. However, due to the large L1 copy number, even small amounts of DNA contamination will skew the data by particularly enriching for sequence reads matching the 3’ regions of L1, as the 5’ truncated inserts are more abundant than full length L1 elements. Another limitation of using methods based on short sequence reads derives from the inability to distinguish between reads that derive from L1 fragments present within other non-L1 mRNAs and bona-fide L1 transcripts. In addition, most of these methodologies lack information on the orientation of the obtained sequence (sense vs. antisense), making it difficult to discern those reads derived from RNA products generated by the antisense activity of the L1 promoter or other flanking promoters. Published data demonstrate that expression of full-length as well as processed L1 transcripts is widespread in human somatic tissues, including human adult stem cells, and that the amount of L1 RNA and proportion of processed products varies extensively between tissues (45).
Figure 2.
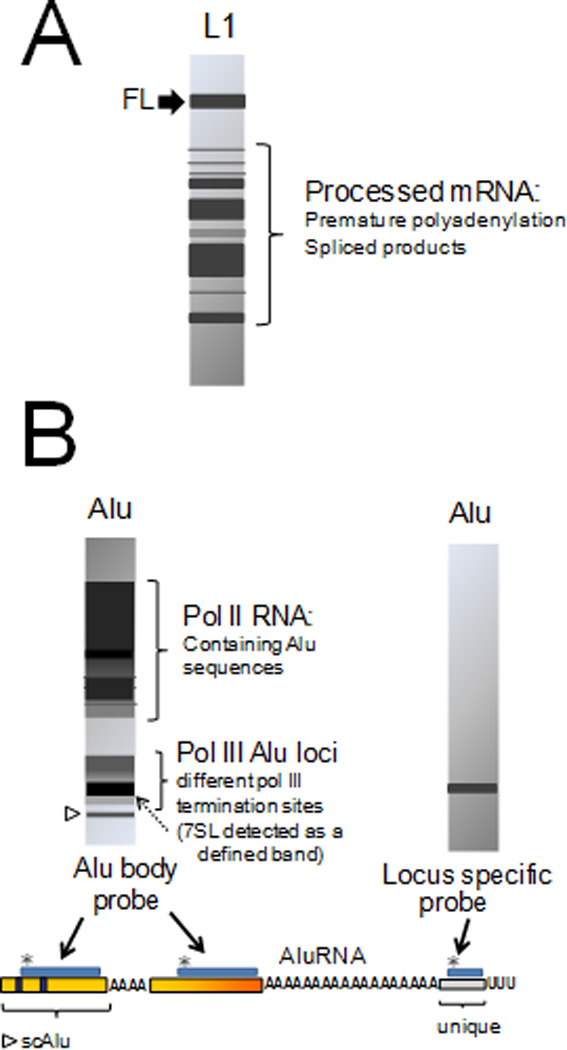
RNA profiles of L1 and Alu elements. Schematic representations of northern blot analysis of L1 (A) and Alu (B) are shown. L1 transcripts are extensively processed both by premature polyadenylation and splicing (42, 44). An L1 RNA profile may show full-length (FL) transcripts (indicated by a black arrow), in addition to a variety of processed products depending on the human tissue analyzed (45). Alu RNA profiles will vary depending on the probe utilized. The structure of a typical Alu transcript is shown at the bottom. Probes (shown as blue boxes marked with asterisks *) that hybridize with the Alu body sequence will detect both large (over 1 kb) and small (100–600 bp) RNA products (schematic shown on the left). The large products observed include RNA pol II transcripts that contain Alu sequences, while the smaller products observed include the RNA pol III transcribed Alu RNAs of various lengths, the left monomer, a degradation intermediate known as small cytoplasmic Alu (scAlu (54), indicated by an open arrowhead) and the 7SL transcript (56), indicated by a dotted arrow) due to shared sequence homology. The use of a probe that hybridizes to the unique sequence (derived from the genomic flank) will only detect those RNA pol III Alu transcripts generated from that specific locus (schematic shown on the right).
The subcellular localization of L1 components is of importance for two reasons: 1) after transcription, the L1 RNA needs to reach the cytoplasm for translation and 2) because of the cis-preference exerted by L1 transcripts (46), the L1 RNA undergoing translation and its two proteins, ORF1p and ORF2p [likely as an RNP (12, 47)], must reach the nucleus to generate a new insert. Analyses using living and fixed cells showed the colocalization of the L1 RNA and both the ORF1 and ORF2 proteins in the cytoplasm (47, 48). The L1 components are often observed associated with markers of cytoplasmic stress granules (49). Previous studies by the same group, using a GFP tagged L1 ORF2p showed that the deletion of its carboxy-terminus caused the protein to also localize to nucleoli (50). However, the importance of the association of L1 components with stress granules and the nucleolus is unclear, as it is unknown if they represent functional intermediate steps of the retrotransposition cycle or the accumulation of byproducts.
3.2. Expression of Alu and other non-autonomous elements
Evaluation of Alu expression is also complex, but for different reasons. First, being particularly enriched in genic regions, Alu sequences are commonly found within RNA pol II transcribed mRNA (51, 52). However, only the RNA pol III Alu transcripts are efficiently retrotransposed (53) making it critical to distinguish between the two types of transcripts. Secondly, Alu elements share sequence homology to the ubiquitously and highly expressed 7SL RNA. Third, Alu transcripts vary in length. Because Alu elements do not contain an RNA pol III terminator within their sequence, transcription will continue past the A-tail into the genomic flanking region (unique sequence) until it reaches an RNA pol III terminator, usually four thymidine residues, somewhere within the downstream genomic DNA (Figure 1). Alu transcripts from different loci will contain a unique 3’ sequence characteristic of the individual element generating the RNA. Due to the diverse location of Alu elements, pol-III generated Alu transcripts can vary in length from 300 bp to over 600 bp. Finally, the pol-III generated Alu transcript can be processed to a smaller product (left monomer) also known as small cytoplasmic Alu (scAlu) (54), which is prevalent due to its higher stability than the Alu dimer RNA (55). Thus, a hybridization analysis which uses a probe that anneals to the Alu sequence (Alu body) will likely detect all of these transcripts generating a variety of smears rather than a distinct band in a Northern blot (see Figure 2B for a schematic example). Due to shared sequence homology, usually the 7SL RNA will appear as an intense discrete band within the smear containing the faster migrating products. Previous work on Alu expression in HeLa cells estimates that 7SL RNA is at least 500 fold more abundant than RNA pol III Alu transcripts (56), making it vastly easier to detect 7SL RNA than Alu RNA. In contrast, a probe to the unique region of the Alu RNA (Figure 2B) should only detect transcripts generated from that specific locus. Size fractionation of transcripts (57) and the use of poly-A selected RNA (58) will enrich for RNA pol III Alu transcripts and reduce the presence of 7SL RNA in the samples. Alu sequences are commonly found in mRNAs; it has been estimated that about 4% of human mRNAs harbor Alu elements (59). One of the better approaches for evaluating Alu expression, is the use of primer extension with reverse transcriptase which gives the advantage of identifying the RNA pol III Alu transcripts by size (60). In contrast, the use of the standard RT-PCR, 3’ RACE using an oligo dT primer and microarray approaches will also encounter the same problem of trying to distinguish between bona fide RNA pol III Alu transcripts and other RNAs containing Alu and Alu-like sequences. Evaluations using these types of approaches, not only will not adequately reflect Alu expression, but in addition, due to favorable oligo annealing, PCR based assays will selectively amplify the younger less mutated Alu elements introducing a bias in the distribution of Alu subfamily expression.
Due to the complexity of Alu expression analysis, reliable data are scarce. Except for the predicted germ line expression, currently there are minimal data available on tissue or developmental expression of Alu elements. Most of the data are derived from a few studies on cell lines. One of these studies demonstrates that transcripts from different Alu subfamilies can be detected in NTera2D1 cells (61). Recently, several laboratories performed genome wide chromatin immunoprecipitation (ChIP) analyses which identified multiple Alu loci (mostly older elements) bound by RNA polymerase III factors in both cancerous and normal cell lines (62–64). Stringent analyses of their data support the active transcription of several of the identified Alu elements of which different loci express in different cell lines. Detection of transcripts from the older non-active Alu subfamilies suggests that expression alone is not sufficient for retrotransposition of Alu elements, but that other factors determine the retrotranspositional capability of individual Alu elements (discussed in section 4.2). Table 1 illustrates the discrepancy between the distribution of Alu subfamily transcripts in relation to their current activity as estimated by reports of diseases caused by de novo insertion of an Alu.
Table 1.
Alu subfamilies genomic distribution, activity and relative expression
| Alu subfamily | % total genomic Alu |
# diseases caused by de novo insertion (%) (107) |
% Alu transcripts (61)$ |
|---|---|---|---|
| S + J | 82 | 0 (0%) | 66 |
| Y | 17 | 11 (27%) | 33 |
| Ya5* | 0.3 | 18 (44%) | 0.8 |
| Yb8* | 0.2 | 12 (29%) | 0.5 |
| Total | 100 | 41 (100%) | 100 |
Includes all subfamily variants,
Only includes data from validated bona fide RNA pol III transcribed Alu elements
Evidence indicates that Alu expression appears to be limited. Because the internal Alu RNA pol III promoter alone is insufficient to drive expression (65, 66), it is likely that even Alu elements with intact A and B boxes are transcriptionally inactive (56). In addition, Alu expression is thought to be downregulated through methylation (58, 67, 68). However, RNA pol III driven Alu expression levels can be transiently increased under various stress conditions that alter chromatin structure (69, 70), such as heat shock (69) and viral infection (71–73). Studies indicate that a few loci are the main contributors to this transient increase in Alu expression in response to stress (74), rather than a global response from all Alu elements in the genome. Considering that each of the one million plus Alu elements is flanked upstream by different sequences, it is not surprising that any individual Alu locus may respond differently to changes in chromatin structure or external stimuli such as viral infection. Early studies detected both full length Alu transcripts and the processed scAlu form primarily in the cytoplasm (54, 58, 60). A recent study using in situ hybridization and Alu-MS2 tagged RNA detected Alu transcripts mostly in the nucleus (47). However, it is unknown if either the nuclear or cytoplasmic localization of Alu transcripts favors its retrotransposition. It is possible that only those Alu transcripts that reach the cytoplasm are capable of sequestering the L1 ORF2p required to retrotranspose.
Data on expression of other retrotransposed sequences vary considerably. Details on SVA expression remain elusive, largely due to the lack of identification of a well-characterized internal promoter. However, a recent discovery shows that some of these elements have acquired promoter sequences through 5’ transduction events (75). This occurs when transcription from an upstream genomic site continues into the SVA sequence and, through splicing, a promoter sequence is juxtaposed with the SVA sequence. Retrotransposition of this transcript generates an insert of an SVA with its own promoter that can subsequently generate new copies creating new SVA subfamilies. Bioinformatic analyses have led to the identification and study of two SVA families that were created through this mechanism: the CpG-SVA family (75) and the SVA_F1 family (76). Evaluations using EST data and RT-PCR demonstrated the expression of the SVA_F1 family in germline, stem cells, tumor cells and different types of somatic cells (76). Interestingly, although L1, Alu and SVA use the L1 retrotransposition machinery, the subcellular localization of their transcripts differs, which possibly contributes to the observed variations in their amplification mechanism (47). Expression of retropseudogenes is unexpected due to the lack of mobilization of the promoter sequence. However, if the copies insert downstream of another promoter, transcription of retropseudogenes is possible. An analysis of pseudogenes identified in ENCODE regions demonstrated that several are transcribed in cell lines or tissues (77). It is still unknown, whether any of the expressed retropseudogenes are capable of furthering their own amplification.
4. INSERTION MECHANISM
4.1. Retrotransposition process and Target Primed Reverse Transcription (TPRT)
The L1 retrotransposition amplification cycle initiates with transcription followed by the localization of the L1 transcript to the cytoplasm for translation. The retrocompetent L1 mRNA is bicistronic, generating both ORF1 and ORF2 proteins required for L1 retrotransposition (11). As mentioned earlier, ORF1p possesses nucleic acid chaperone activity (78, 79), an essential property for L1 retrotransposition (79, 80). L1 RNA shows a strong cis-preference for its own translated proteins during the retrotransposition process (46, 81). The cis-preference of L1 makes it likely that RNA/protein interactions near the ribosome during translation are important in forming the appropriate RNP (Figure 3A) for retrotransposition of L1 to occur (14). In addition, the cis-preference may decrease the ability of other mRNAs to access the L1 ORF1p and ORF2p and explaining the lower retropseudogene copy numbers in the genome. Because multiple ORF1p molecules interact with the L1 RNA (80), and are present in higher numbers than ORF2p (50), it is reasonable to speculate that ORF1p outnumbers ORF2p in the L1 RNP. It is possible that these properties of ORF1p may be important to help “dislodge” the L1 transcript from the ribosomes (Figure 3A) and to protect the RNA from being diverted to RNA degradation pathways. Also, the ORF1p coating of the L1 RNA may prevent new reassembly of ribosomes and allow for the transcript to proceed to the nucleus. This may also be the reason why L1 and retropseudogenes that require ORF1p (82) for retrotransposition are kinetically slower in culture, while the non-translated RNAs, such as Alu, are both ORF1p independent (83, 84) and kinetically faster (53). After L1 RNP assembly, there are limited data on how L1, or any other transcripts such as SVA or processed transcripts (retropseudogenes), reach the nucleus for the insertion process to ensue.
Figure 3.
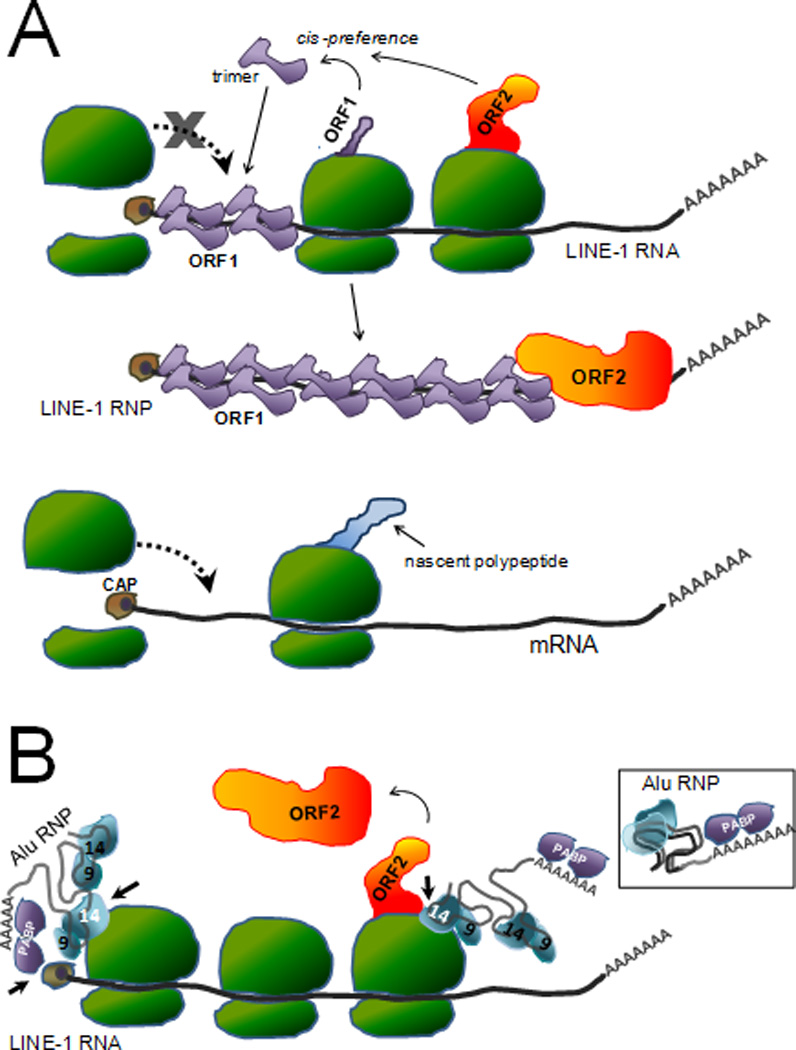
Proximity as a requirement for efficient retrotransposition. A. Proposed model explaining the reduced efficiency of retrotransposition of non-L1 mRNAs (retropseudogenes generation). Schematic representation of a full-length capped L1 transcript in a polyribosomal complex undergoing translation. As the L1 ORF1 protein is translated, the ORF1 monomers interact forming a trimer. Due to the cis-preference shown by L1 it is expected that the ORF1 trimer will bind the L1 RNA that generated it, likely driven by proximity. Although ORF2p is made in lower amounts, it also is thought to preferentially interact with the same L1 transcript. The increased number of ORF1p likely coats the L1 RNA, possibly preventing the reassembly of the ribosome and contributing to the “escape” from the translational machinery, allowing for the newly formed L1 RNP to progress through the retrotransposition cycle. In contrast, because mRNA from cellular genes will be undergoing translation in their own polyribosomes, these transcripts will be physically restricted from coming in contact with the L1 proteins. The lack of the ORF1p interaction with the mRNA will enable ribosomes to continue to assemble and continue translation, until the mRNA is targeted to the degradation pathway. [The CAP complex is represented as a brown circle at the 5’ end of the transcript. Ribosomes are shown in green, and the L1 ORF1 and ORF2 proteins are shown in purple and orange respectively.] B. Proposed model for Alu sequestration of ORF2 protein. Due to the lack of coding sequences, Alu transcripts are not engaged by the translation machinery. Thus the Alu transcript can exist as a “free” ribonucleoprotein (RNP) complex (see inset). The inset shows a liberal representation of the potential folding of the two 7SL derived monomers to form the propeller structure and the interaction with the SRP9 and SRP14 proteins (depicted in blue): only SRP9p is shown. In addition to SRP9/14p, the Poly-A binding protein (PABP, depicted in purple) is thought to interact with the A-tail of the Alu RNA. PABP is shown interacting with the 3’ A-tail of the Alu RNA. The structure is likely to fold to form a compact RNP. The bound SRP14p and PABP may play a role in targeting Alu RNPs to the ribosomes. SRP14p interacts with ribosome through direct contacts with amino acids located in its carboxy-terminus. The PABP bound to the Alu A-tail may also help direct the Alu RNP to the translating L1 transcript by interacting with the cap complex (brown circle) of the L1 RNA. These interactions (indicated by short black arrows) may favor the localization of Alu RNPs to translating ribosomes to increase their probability of coming in close proximity to newly synthesized ORF2p (in orange). For illustrative purposes, the L1 transcript bound to ribosome is shown as a linear molecule. However, the cap complex of the L1 RNA likely interacts with the PABP present in its own A-tail circularizing the RNA.
Although all retroelements use an RNA template in their process of amplification, there are fundamental differences in the insertion mechanism between LTR-retrotransposons and non-LTR retrotransposons (3). In general, LTR-retrotransposons reverse transcribe the RNA into a ds-cDNA molecule that is subsequently integrated into the genome. In contrast, L1 and its non-autonomous partners use a unique process to integrate new copies into the genome based on target primed reverse transcription (TPRT) of its RNA. The TPRT model is based on the in vitro studies of the amplification cycle of the R2 element in Bombyx mori (85) and has been adapted into a model for other elements that use the non-LTR insertion mechanism. Under this model, reverse transcription of the template RNA is primed by a nick at the chromosomal integration site. In the case of L1 and its parasites (such as Alu), the L1 ORF2 protein generates the initial nick (86). The ORF2p endonuclease preferentially cleaves the sequence 5’-TTTT/AA-3’, which resembles in vivo L1 integration sites (86, 87). The poly-A stretch at the 3’ end of LINEs, SINEs, and processed pseudogenes plays an important role for retrotransposition (88), likely by providing the annealing site for the genomic thymidine residues that prime reverse transcription (89) (Figure 4). Although very inefficiently, the retrotransposition of Alu transcripts without 3’ poly-A stretches can occur (83). In addition, the presence of a few “tailless” 3’ truncated Alu inserts in the human genome indicates the likely occurrence of internal priming during the TPRT step (90) (Figure 4). Also, a large number of retrotransposed copies of RNA pol III transcripts (tRNA, tRNA-related, and 4.5S RNA) that lack A-tails and referred to as tailless retropseudogenes have been described (29). Evaluation of the insertion sites demonstrated that the cleaved DNA strand which functions as a primer for reverse transcription by ORF2p presents perfect matched complementarity to the RNA template sequence (Figure 4). The cleavage consensus sequence for the tailless retropseudogenes varies from the one observed for Alu and L1 inserts, with TT/WX1-17 (29) as being more prevalent (Figure 4). On the other hand, in vitro studies show that structural parameters of the target DNA appear more important for recognition and determination of the nicking profiles of the L1 endonuclease (91). These data suggest that the consensus sequences derived from the computational studies of retrotransposed inserts are not exclusively determined by the cleavage preference of the ORF2p endonuclease, but also reflect the ability of the RNA to interact with the cleaved DNA to prime reverse transcription. ORF1p has been suggested to mediate nucleic acid strand transfer steps during L1 reverse transcription (78, 92). Because ORF1p is not strictly required for Alu retrotransposition, as the process can occur in chicken cells which are devoid of endogenous L1 ORF1p (84), it is difficult to define the role of ORF1p in Alu retrotransposition.
Figure 4.

Target primed reverse transcription (TPRT). A schematic representation of the TPRT step of the retrotransposition process is shown for L1, Alu, and tailless tRNA-derived transcripts. The genomic DNA is depicted as dark lines. The consensus 5’-TTAAAA-3’ is shown on the top strand of the insertion sites for the L1 and Alu, while a representative sequence of the 5’-TTWX1-17-3’ consensus is shown for the tailless retropseudogene. The bottom strand shows the nicked DNA with the exposed nucleotides (e.g. TTTT) thought to anneal with the RNA and provide the priming site for reverse transcription by ORF2p (depicted in orange). The Alu and tailless retropseudogene transcripts are shown with extended 3’ sequences (shown as Ns) which represent the unique region of Alu or part of the tRNA-related sequences followed by the RNA pol III terminator (shown as Us).
Finally, little is known about the retrotransposition process after ORF2p completes reverse transcription of the cDNA. How the second nick is created or how the second DNA strand is generated and ligation of the DNA occurs is still unknown. However, the hallmark tandem site duplications (TSDs) flanking the new retrotransposed copy are well-characterized.
4.2. Requirements for retrotransposition of non autonomous elements
Retrotransposition of the non autonomous elements requires two basic components: expression of the RNA template and accessibility to the L1 proteins. Although ORF1p and ORF2p are essential for L1 retrotransposition, this is not a universal requirement for the non-autonomous elements. As mentioned above, RNA pol II generated transcripts (L1 and retropseudogenes) require ORF1p (82), while RNA pol III transcripts (SINEs) only require ORF2p (53, 83, 84). Regardless, all non-autonomous retroelements require the interaction with the at least the L1 ORF2p to undergo retrotransposition. The physical interaction with ribosomes during translation probably limits other mRNA transcripts (retropseudogene source) from achieving the sufficient proximity to those ribosomes translating L1 to be able to sequester the required ORF1p and ORF2p for retrotransposition (Figure 3A). This limitation may explain the lower abundance of retropseudogenes, particularly when compared to the SINE or LINE copy numbers in the human genome. In contrast, Alu transcripts are not encumbered by the translation machinery. Still, Alu RNA must at least come in contact with ORF2p to facilitate trans-mobilization. However it is uncertain of how or where Alu transcripts interact with ORF2p.
Previously, the ability of the Alu transcript to come in close proximity to the translating L1, has been proposed as a model to explain how Alu efficiently competes for access to ORF2p (89). This proximity model is based on the ability of Alu RNA to target ribosomes through its interaction with two of the signal recognition particle (SRP) proteins. The Alu RNA retains the folding pattern of its ancestral 7SL transcript, which provides binding sites for SRP9p and SRP14p (93, 94) (Figure 3B). Because SRP14p normally targets the 7SL RNP to the ribosomes and causes the arrest of nascent chain elongation through direct contacts of the “Alu domain” of 7SL RNA and the ribosome (95), it is proposed that SRP9/14 proteins bound Alu RNA will have the same fate. Mutations altering the interaction of Alu transcripts with SRP9/14p diminish the retrotransposition capability, suggesting a potential role for these proteins in the process (96, 97). Further support for a critical role of SRP9/14p comes from a study using another 7SL-derived SINE, the rodent B1 element. A single nucleotide substitution in a key region for SRP9/14p-RNA interaction resulted in a significant increase in the retrotransposition rate of B1 (98). In addition, retrotransposition of Alu transcripts is about 10 fold more efficient than its monomeric form (83), possibly a reflection of having two sets of SRP9/14p proteins per Alu RNP (96) (Figure 3B). Another protein thought to help target the Alu RNP to the ribosomes is the cytoplasmic polyA binding protein (PABP). Because PABP binds SINEs that contain A-tails (99, 100), it is thought to form part of the Alu RNP complex. Due to the PABP interaction with eIF4G in the CAP complex (reviewed in 101), the PABP in the Alu complex may bring the RNP into proximity with newly synthesized ORF2p by binding the CAP complex of the translating L1 RNA(102) (Figure 3B). However, the role of PABP in SINE retrotransposition is unclear.
Alu sequence variations also affect activity of an individual element, as not all Alu transcripts are able to undergo retrotransposition. In addition to mutations that limit SRP9 and 14 binding, studies demonstrate that disruptions in the homopolymeric A-tail and increases in the unique 3’ sequence length of the Alu transcript (Figure 1) reduce retrotransposition rates (103). These sequence variations at the Alu 3’ end are likely to influence the annealing dynamics of the RNA and the cleaved DNA during the TPRT process (Figure 4), where mismatches and lengthy 3’ ends may destabilize the interaction.
Most SINE elements are not derived from 7SL, but instead share sequence homology to tRNAs. It is unlikely that SRP9p and SRP14p interact with tRNA derived SINEs or contribute to their retrotransposition. However, it is possible that alternate cellular proteins substitute for the role of SRP9/14p in the tRNA derived SINEs. Interestingly, the transcripts of these SINEs do not retain the folded cloverleaf structure of the tRNA, but instead form a structure that shares similar folding patterns between different SINEs (104). The conservation of this RNA structural similarity may reflect a binding motif for critical proteins that aid retrotransposition of these SINEs.
Currently, little is known about what other cellular factors may be required for either L1 or the non-autonomous elements. Some DNA repair proteins may play important roles in L1 retrotransposition. Components of the non-homologous end joining (NHEJ) pathway (105) and the damage sensor, ATM (106), appear necessary for promoting L1 retrotransposition. We expect that the role of these proteins may be elucidated when the remainder of the insertion process is better understood.
4.3. Cellular regulatory mechanisms of retrotransposition
Unregulated activity of mobile elements can cause significant genetic damage, thus leading cells to evolve mechanisms to minimize mobile element activity. Active elements cause disease predominantly through insertional mutagenesis and non-allelic homologous recombination (NAHR), resulting in loss of gene function (reviewed in 107). For example, L1 activity contributes to several types of genetic instability by causing inversions, insertions, deletions and chromosomal rearrangements. Recent studies also show that the expression of L1 or L1 ORF2p in human cells induces cell cycle arrest (106), apoptosis (108), and senescence (45, 109). Due to the highly damaging effects of mobile elements, cells have developed surveillance mechanisms to limit their impact.
The first line of defense against the damaging effects of retroelement activity is the prevention of their expression. Several observations demonstrate that methylation plays a major role in silencing retrotransposons. Deficiencies in the DNA methyltransferase 3-like gene (Dnmt3L) prevented de novo methylation of retrotransposons causing meiotic failure in the male germ line of mice (110). During malignant transformation, genome-wide hypomethylation of repetitive sequences occurs, altering the normal regulation of expression of these elements (111). Additionally, the piRNA pathway has been implicated in transposon control through methylation. Studies in mice showed that piRNAs and the Piwi proteins MILI and MIWI2 determine de novo methylation of retrotransposons in germ cells in mice (112, 113). siRNA activity appears to also regulate L1 expression (114).
Suppression of L1 retrotransposition is further controlled by mRNA processing through extensive splicing and premature polyadenylation (42–44). However, L1 RNA processing varies dramatically across tissues (45), with splicing being the potentially dominant mechanism of L1 RNA regulation in some tissues. However, processing of L1 mRNA does not fully mitigate L1-mediated damage because some L1 splice products are capable of generating active L1 ORF2p (45). Because SINEs only require ORF2p, tissues favoring L1 processing leading to the generation of ORF2p containing splice products could support Alu retrotransposition. Thus, Alu retroposition could be actively occurring even if no L1 activity is observed in those tissues.
Other cellular processes function as control mechanisms of retroelements. The members of the apolipoprotein B mRNA editing complex 3 (APOBEC3) have been implicated in the post-transcriptional regulation of L1 and Alu retrotransposition (115, 116). However, not all the proteins in the APOBEC3 gene family universally inhibit retrotransposons; instead, some show specificity to individual elements. For example, APOBEC3G has been shown to specifically inhibit Alu but not L1 activity (117). Interestingly, the ability of the different APOBEC3 proteins to inhibit retrotransposition is independent of their RNA editing function. Currently, the mechanism by which APOBEC3 inhibits retrotransposition remains unclear (118). However, recent advances on APOBEC 3A proteins indicate that they may function through the inhibition of the elongation of cDNA by reverse transcriptase (reviewed in 119). Another example, the Nucleotide Excision Repair endonuclease, ERCC1/XPF, has been demonstrated to limit L1 activity (120). The authors propose that L1 retrotransposition triggers a DNA damage signal which causes the recruitment of ERCC1/XPF and removal of the L1 insertion intermediate, preventing completion of the process. Although yet to be discovered, it is highly likely that other cellular factors participate in the regulation of retrotransposons.
5. ACTIVITY AND DISTRIBUTION OF NON-LTR RETROELEMENTS
5.1. Activity profiles during evolution
Throughout evolution, different mobile elements have dominated the genomic landscape. L1 elements have been active for over 100 million years (1, 121). Not surprisingly, the non-autonomous elements such as Alu which depend on L1, appeared much later (122). The peak Alu amplification rate occurred between 55 and 35 million years ago, generating approximately 80% of the Alu elements currently present in the human genome (18). During this same time period, peak activity was also observed for both the RNA pol II derived retropseudogenes (123) and the RNA pol III derived tailless retropseudogenes (29) possibly reflecting a time period favorable for non-autonomous element expansion. However, SVA does not follow the same pattern of expansion as it origins dates back only 18 to 25 million years ago (22).
5.2. Evidence of current activity
The presence of polymorphic elements between different individual attests to the current activity of non-LTR retroelements in humans. The extent of genomic diversity contributed by these elements was highlighted by research from several laboratories (4–6) and corroborated further with the analyses from the 1000 human genome project (7). However, the calculated retrotransposition rates for the different elements vary, with an estimated one insertion per every 21, 212, and 916 births for Alu, L1, and SVA, respectively (124). Even more compelling are the multiple de novo cases of human retroelement induced diseases where Alu contributes for the majority of documented cases (107).
Although the impact of the insertional mutagenesis by mobile elements is well documented, data on the activity at somatic level are scarce. Transgenic mouse models have shown that somatic activity occurs (40, 125–127). There are a few reports of cancers describing LINE-1 insertions disrupting tumors suppressors (128, 129) or present at a translocation breakpoint (130). However, only one of these examples appears to be an LINE-1 that inserted through retrotransposition. More recently, data from high throughput DNA sequence analyses of lung tumors detected nine somatic L1 insertions specific to the tumor DNA that were absent from the adjacent normal samples (4). As the field and methodology have advanced, the amount of evidence that these elements may have a significant role at the somatic level has increased. Further studies will provide a better understanding of the somatic role of these elements.
5.3. Insertional distribution
Retrotransposons are distributed in a non-random manner in the human genome. Although L1 and its non-autonomous parasites use ORF2p cleavage sites for insertion, they do not share the same distribution throughout the genome (1). Alu elements are enriched in GC-rich regions while L1s are found at higher densities in AT-rich regions. Data indicate that although the initial insertion distribution of these elements is comparable, selection through time contributes to differential fixation of Alu and L1 elements (1). Changes in the distribution of retrotransposons in the human genome through time indicate that recombination between chromosomes is likely an important mechanism for their redistribution (131). In addition, the distribution between the sex chromosomes and autosomal chromosomes also varies. Genomic analyses demonstrate that young Alu elements are more dense in chromosome Y relative to chromosome X (3 fold) and to autosomes (2 fold) (132, 133). However, the density of Alu elements increase over time in chromosome X relative to autosomal densities (133). Retropseudogenes are also significantly higher on the human X chromosome (134) while underrepresented on the Y chromosome. Human diseases caused by insertion of these elements, which represent recent events with a limited exposure to selection processes, also show a skewed distribution, with Alu and L1 insertions highly overrepresented on the X chromosome (107). However, it is difficult to determine if there is a biological component driving the enrichment of these elements on the X chromosome or if ascertainment bias of disease detection contributes to the observed numbers.
6. SUMMARY AND PERSPECTIVE
The impact of mobile element activity on the human genome became clearly evident after determining that these elements have contributed to almost half of its mass. As the field progresses in understanding the biology of the currently active elements, further data demonstrate their significant role in human disease. Among the currently active human transposable elements, LINE-1 takes a central role by not only providing factors for its own retrotransposition, but also enabling the trans-mobilization of non-autonomous elements such as Alu and SVA. Because non-autonomous elements depend on LINE-1 for the active components/protein(s) for their mechanism, it is reasonable to presume that the parasitic element would benefit from a constant and close proximity to an active LINE-1 element i.e. a requirement for “flocking together”. However, the amplification behavior and impact of these parasitic non-autonomous elements do not parallel that of LINE-1. For example, diseases caused by de novo Alu inserts outnumber those caused by LINE-1 inserts by about 2 to 1. Although it is still unclear why a non-autonomous element would outnumber autonomous elements, ongoing studies demonstrate that these elements exhibit different expression patterns, cellular localization, retrotransposition requirements and possibly insertional preferences that may help explain differences in amplification success rates. The recent discovery that some tissues can express transcripts that generate ORF2p only further demonstrates the unique genetic impact that each family of non-LTR retrotransposons may have and underscores the importance of evaluating these elements individually. To be able to comprehend the collective impact of the active mobile elements on the human genome, the particularities regarding each type of element must first be elucidated.
ACKNOWLEDGEMENTS
This publication was made possible by Grants Number R01GM079709A and P20RR020152 to AMR-E from the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Boeke J, Garfinkel DJ, Styles CA, Fink CR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 3.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 Retrotransposition Activity in Human Genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, Wheelan SJ, Ji H, Boeke JD, Burns KH. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The 1000 Genomes Project Consortium: A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. Embo J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc.Natl.Acad.Sci.U.S.A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA.Biol. 2010;7:67–72. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 13.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. Embo J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Human Molecular Genetics. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 15.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol.Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol.Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nigumann P, Redik K, Matlik K, Speek M. Many Human Genes Are Transcribed from the Antisense Promoter of L1 Retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 18.Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol.Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- 19.Weiner A, Deininger P, Efstradiatis A. The Reverse Flow of Genetic Information: pseudogenes and transposable elements derived from nonviral cellular RNA. Annual Reviews of Biochemistry. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 20.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 21.Fuhrman S, Deininger PL, LaPorte P, Friedmann T, Geiduschek P. Analysis of Transcription of the Human Alu Family of Ubiquitous Repeating Element by Eukaryotic RNA Polymerase III. Nucleic Acid.Res. 1981;9:6439–6455. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol.Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goncalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khachane AN, Harrison PM. Strong association between pseudogenization mechanisms and gene sequence length. Biol Direct. 2009;4:38. doi: 10.1186/1745-6150-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullu E, Weiner AM. Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 1984;3:3303–3310. doi: 10.1002/j.1460-2075.1984.tb02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buzdin A, Gogvadze E, Kovalskaya E, Volchkov P, Ustyugova S, Illarionova A, Fushan A, Vinogradova T, Sverdlov E. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perreault J, Noel JF, Briere F, Cousineau B, Lucier JF, Perreault JP, Boire G. Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res. 2005;33:2032–2041. doi: 10.1093/nar/gki504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz J, Churakov G, Zischler H, Brosius J. A novel class of Mammalian-specific tailless retropseudogenes. Genome Res. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlicek A, Paces J, Elleder D, Hejnar J. Processed Pseudogenes of Human Endogenous Retroviruses Generated by LINEs: Their Integration, Stability, and Distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc.Natl.Acad.Sci.U.S.A. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, Cordon-Cardo C, Levine AJ, Tang LH. Association of Nuclear Localization of a Long Interspersed Nuclear Element-1 Protein in Breast Tumors with Poor Prognostic Outcomes. Genes Cancer. 2010;1:115–124. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bratthauer GL, Cardiff RD, Fanning TG. Expression of LINE-1 retrotransposons in human breast cancer. Cancer. 1994;73:2333–2336. doi: 10.1002/1097-0142(19940501)73:9<2333::aid-cncr2820730915>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol.Res. 1996;8:239–247. [PubMed] [Google Scholar]
- 35.Bratthauer GL, Fanning TG. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7:507–510. [PubMed] [Google Scholar]
- 36.Bratthauer GL, Fanning TG. LINE-1 retrotransposon expression in pediatric germ cell tumors. Cancer. 1993;71:2383–2386. doi: 10.1002/1097-0142(19930401)71:7<2383::aid-cncr2820710733>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 37.Su Y, Davies S, Davis M, Lu H, Giller R, Krailo M, Cai Q, Robison L, Shu XO. Expression of LINE-1 p40 protein in pediatric malignant germ cell tumors and its association with clinicopathological parameters: A report from the Children's Oncology Group. Cancer Lett. 2006 doi: 10.1016/j.canlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 38.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci U S A. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 41.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J.Biol.Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 42.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Research. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perepelitsa-Belancio V, Deininger PL. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 45.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol.Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodier JL, Mandal PK, Zhang L, Kazazian HH., Jr Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum.Mol Genet. 2010 doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, Gilbert N. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 Protein Localizes in Stress Granules with Other RNA-Binding Proteins, Including Components of RNA Interference RNA-Induced Silencing Complex. Mol.Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Human Molecular Genetics. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 51.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 52.Versteeg R, van Schaik BDC, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AHC. The Human Transcriptome Map Reveals Extremes in Gene Density, Intron Length, GC Content, and Repeat Pattern for Domains of Highly and Weakly Expressed Genes. Genome Research. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA Polymerase Dictates ORF1 Requirement and Timing of LINE and SINE Retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maraia RJ, Driscoll CT, Bilyeu T, Hsu K, Darlington GJ. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol.Cell Biol. 1993;13:4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li TH, Schmid CW. Alu's dimeric consensus sequence destabilizes its transcripts. Gene. 2004;324:191–200. doi: 10.1016/j.gene.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 56.Paulson KE, Schmid CW. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic.Acids.Res. 1986;14:6145–6158. doi: 10.1093/nar/14.15.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinnett D, Richer C, Deragon JM, Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J.Biol.Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- 58.Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic.Acids.Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jasinska A, Krzyzosiak WJ. Repetitive sequences that shape the human transcriptome. FEBS Letters. 2004;567:136–141. doi: 10.1016/j.febslet.2004.03.109. [DOI] [PubMed] [Google Scholar]
- 60.Li TH, Kim C, Rubin CM, Schmid CW. K562 cells implicate increased chromatin accessibility in Alu transcriptional activation. Nucleic Acids Res. 2000;28:3031–3039. doi: 10.1093/nar/28.16.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol.Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 62.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chesnokov I, Schmid CW. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J.Mol.Evol. 1996;42:30–36. doi: 10.1007/BF00163208. [DOI] [PubMed] [Google Scholar]
- 66.Roy AM, West NC, Rao A, Adhikari P, Alemán C, Barnes AP, Deininger PL. Upstream flanking sequences and transcription of SINEs. J Mol.Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- 67.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic.Acids.Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubin CM, VandeVoort CA, Teplitz RL, Schmid CW. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic.Acids.Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/s0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- 70.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic.Acids.Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panning B, Smiley JR. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol.Cell Biol. 1993;13:3231–3244. doi: 10.1128/mcb.13.6.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panning B, Smiley JR. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J.Mol.Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 73.Russanova VR, Driscoll CT, Howard BH. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol.Cell Biol. 1995;15:4282–4290. doi: 10.1128/mcb.15.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 75.Bantysh OB, Buzdin AA. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochemistry (Mosc.) 2009;74:1393–1399. doi: 10.1134/s0006297909120153. [DOI] [PubMed] [Google Scholar]
- 76.Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Research. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, Ruan Y, Wei CL, Gingeras TR, Guigo R, Harrow J, Gerstein MB. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17:839–851. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol.Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin SL, Cruceanu M, Branciforte D, Wai-Lun LP, Kwok SC, Hodges RS, Williams MC. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol.Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 80.Basame S, Wai-Lun LP, Howard G, Branciforte D, Keller D, Martin SL. Spatial Assembly and RNA Binding Stoichiometry of a LINE-1 Protein Essential for Retrotransposition. J Mol.Biol. 2006;357:351–357. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 81.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat.Struct.Mol.Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 82.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat.Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 83.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat.Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 84.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 86.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 87.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc.Natl.Acad.Sci.U.S.A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 89.Boeke JD. LINEs and Alus--the polyA connection. Nat.Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 90.Srikanta D, Sen SK, Conlin EM, Batzer MA. Internal priming: an opportunistic pathway for L1 and Alu retrotransposition in hominins. Gene. 2009;448:233–241. doi: 10.1016/j.gene.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Repanas K, Zingler N, Layer LE, Schumann GG, Perrakis A, Weichenrieder O. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007;35:4914–4926. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin SL. The ORF1 Protein Encoded by LINE-1: Structure and Function During L1 Retrotransposition. J Biomed.Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hsu K, Chang DY, Maraia RJ. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J Biol Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- 94.Chang DY, Hsu K, Maraia RJ. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas Y, Bui N, Strub K. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res. 1997;25:1920–1929. doi: 10.1093/nar/25.10.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sarrowa J, Chang DY, Maraia RJ. The decline in human Alu retroposition was accompanied by an asymmetric decrease in SRP9/14 binding to dimeric Alu RNA and increased expression of small cytoplasmic Alu RNA. Mol.Cell Biol. 1997;17:1144–1151. doi: 10.1128/mcb.17.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol.Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 99.West N, Roy-Engel A, Imataka H, Sonenberg N, Deininger P. Shared Protein Components of SINE RNPs. J.Mol.Biol. 2002;321:423–432. doi: 10.1016/s0022-2836(02)00542-9. [DOI] [PubMed] [Google Scholar]
- 100.Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Huttenhofer A, Tiedge H, Brosius J. Poly(A)-binding Protein is Associated with Neuronal BC1 and BC200 Ribonucleoprotein Particles. J.Mol.Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 101.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu.Rev.Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 102.Roy-Engel AM, Salem AH, Oyeniran OO, Deininger L, Hedges DJ, Kilroy GE, Batzer MA, Deininger PL. Active alu element "A-Tails": size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun FJ, Fleurdepine S, Bousquet-Antonelli C, Caetano-Anolles G, Deragon JM. Common evolutionary trends for SINE RNA structures. Trends in Genetics. 2007;23:26–33. doi: 10.1016/j.tig.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 105.Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, Takeda S, Okada N. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J.Mol.Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 108.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 111.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat.Struct.Mol.Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 115.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc.Natl.Acad.Sci.U.S.A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PTN, Yu XF. Differential Inhibition of Long Interspersed Element 1 by APOBEC3 Does Not Correlate with High-Molecular-Mass-Complex Formation or P-Body Association. The Journal of Virology. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aguiar RS, Peterlin BM. APOBEC3 proteins and reverse transcription. Virus Research. 2008;134:74–85. doi: 10.1016/j.virusres.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 120.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic.Acids.Res. 1992;20:3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH., Jr L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.An W, Han JS, Schrum CM, Maitra A, Koentgen F, Boeke JD. Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis. 2008;46:373–383. doi: 10.1002/dvg.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc.Natl.Acad.Sci.U.S.A. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333:87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 129.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 130.Liu J, Nau MM, Zucman-Rossi J, Powell JI, Allegra CJ, Wright JJ. LINE-I element insertion at the t(11, 22) translocation breakpoint of a desmoplastic small round cell tumor. Genes Chromosomes.Cancer. 1997;18:232–239. doi: 10.1002/(sici)1098-2264(199703)18:3<232::aid-gcc10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 131.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jurka J, Krnjajic M, Kapitonov VV, Stenger JE, Kokhanyy O. Active Alu elements are passed primarily through paternal germlines. Theor.Popul.Biol. 2002;61:519–530. doi: 10.1006/tpbi.2002.1602. [DOI] [PubMed] [Google Scholar]
- 133.Jurka J, Kohany O, Pavlicek A, Kapitonov VV, Jurka MV. Duplication, coclustering, and selection of human Alu retrotransposons. Proc.Natl.Acad.Sci.U.S.A. 2004;101:1268–1272. doi: 10.1073/pnas.0308084100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Drouin G. Processed pseudogenes are more abundant in human and mouse X chromosomes than in autosomes. Mol Biol Evol. 2006;23:1652–1655. doi: 10.1093/molbev/msl048. [DOI] [PubMed] [Google Scholar]
- 135.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5' end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shumyatsky G, Shimba S, Reddy R. Capping signals correspond to the 5' end in four eukaryotic small RNAs containing gamma-monomethylphosphate cap structure. Gene Expr. 1994;4:29–41. [PMC free article] [PubMed] [Google Scholar]