Abstract
Approximately 42% of the human genome is composed of endogenous retroelements, and the major retroelement component, long interspersed element-1 (L1), comprises ∼17% of the total genome. A single human cell has more than 5 × 105 copies of L1, 80∼100 copies of which are competent for retrotransposition (RTP). Notably, L1 can induce RTP of other retroelements, such as Alu and SVA, and is believed to function as a driving force of evolution. Although L1-RTP during early embryogenesis has been highlighted in the literature, recent observations revealed that L1-RTP also occurs in somatic cells. However, little is known about how environmental factors induce L1-RTP. Here, we summarize our current understanding of the mechanism of L1-RTP in somatic cells. We have focused on the mode of L1-RTP that is dependent on the basic helix–loop–helix/per–arnt–sim (bHLH/PAS) family of transcription factors. Along with the proposed function of bHLH/PAS proteins in environmental adaptation, we discuss the functional linking of L1-RTP and bHLH/PAS proteins for environmental adaptation and evolution.
Keywords: LINE-1, environmental factors, retrotransposition, bHLH/PAS family
Introduction
Approximately half of the human genome (∼45%) is composed of transposable elements, most of which are endogenous retroelements (∼42%; Lander et al., 2001; Bannert and Kurth, 2004). Notably, humans have more endogenous retroelements than do mice (∼37%) or fruit flies (∼3.6%), implying a possible role for endogenous retroelements in evolution (Kazazian, 2004). In humans, ∼8% of the genome comprises human endogenous retroviruses (HERV) that have gene structures similar to the long-terminal repeats (LTRs) of lentiviruses (Bannert and Kurth, 2004). By contrast, ∼34% of retroelements are of non-LTR types, half of which are long interspersed element-1 (L1). A single human cell contains more than 5 × 105 copies of L1, 80∼100 copies of which are competent for genome shuffling by retrotransposition (RTP; Brouha et al., 2003). Interestingly, ∼10% of such RTP-competent L1 sequences are “hot” and account for more than 80% of the total RTP activity (Brouha et al., 2003).
L1 is actively expressed in embryonic stem cells (Georgiou et al., 2009), and L1-RTP occurs in oocytes and during early embryonic development (van den Hurk et al., 2007; Kano et al., 2009). L1-RTP is also coupled with neuronal cell differentiation (Muotri et al., 2005), supporting the hypothetical role of L1-RTP in the plasticity of nerve cells. Moreover, L1-RTP was shown to be a critical event at an early stage of cell division in the fertilized egg, although its exact role remains elusive (Vitullo et al., 2012). While these lines of evidence strongly suggest that L1-RTP is pivotal for early embryogenesis, recent observations indicate that L1-RTP also occurs in somatic cells. Interestingly, some studies suggest that the L1 copy number is increased in the human brain (Muotri et al., 2005; Baillie et al., 2011), and greater L1 activity was detected in patients with defective ataxia telangiectasia mutated (ATM) genes (Coufal et al., 2011). Moreover, new somatic inserts were identified in the tumors, suggesting increased activity therein (Iskow et al., 2010; Ting et al., 2011). These observations support the idea that somatic cells possess the machinery that is involved in the induction of L1-RTP, but little is known about the cellular factors required for L1-RTP. Recently, we found that L1-RTP was induced by environmental compounds that included carcinogens (Okudaira et al., 2010, 2011; Ishizaka et al., 2012). L1-RTP induction by these compounds was dependent on members of the basic helix–loop–helix/per–arnt–sim (bHLH/PAS) protein family, which has been proposed to be associated with the environmental adaptation of living organisms (Beischlag et al., 2008; McIntosh et al., 2010). Here, we provide an overview of our current understanding of the mechanism of L1-RTP coupled with bHLH/PAS proteins, and discuss the role of L1-RTP in relation to environmental adaptation and revolution.
L1 and the Roles of L1-Encoded Proteins
L1 is ∼6 kb in length and is composed of a 5′-untranslated region (UTR), two non-overlapping open reading frames (ORFs), and a 3′ UTR with a poly(A) tail (Goodier and Kazazian, 2008). L1 has evolved along a single, unbroken lineage for the past 40 million years (myrs) in primates, and five subfamilies of L1 (L1PA1-5) have developed within the past 25 myrs in hominoid primates (Lee et al., 2007). Interestingly, species-specific L1 subfamilies emerged in Homo sapiens and Pan troglodytes after their divergence 6 myrs ago. In humans, active L1 (Ta-1) arose from the Ta-0 subfamily ∼4 myrs ago, and expanded as a dominant subfamily thereafter. Notably, it has also been shown that ∼69% of L1-inserted loci are polymorphic for the presence or absence of the Ta-1 insert, and that ∼90% of such loci possess Ta-1d inserts, the youngest subset of Ta-1, which arose ∼1.4 myrs ago (Boissinot et al., 2000). Of note, Ta-1d accounts for approximately two-third of the Ta-1 subfamily, indicating that it has been selectively expanded during the evolution of H. sapiens. Intriguingly, humans have higher numbers of RTP-competent L1 sequences (80∼100 copies) than do chimpanzees (∼5 copies), although there are similar numbers of species-specific inserts (1200–2000 copies) in humans and chimpanzees (Lee et al., 2007).
L1 encodes two proteins: ORF1 and ORF2 (Goodier and Kazazian, 2008). ORF1 is a 40-kDa protein that acts on L1-mRNA in cis (Wei et al., 2001) and functions as its chaperone (Martin et al., 2005). ORF2 is a protein of about 150 kDa with both reverse transcription (RT; Mathias et al., 1991) and endonuclease (Feng et al., 1996) activities. ORF2 recognizes 5′-TTAAAA hexanucleotides in the genome and induces a nick between 3′-AA and TTTT in the complementary strand (Jurka, 1997; Gilbert et al., 2005). It has been proposed that first-strand DNA is synthesized by target site-primed RT, in which the poly(A) tail of L1 mRNA anneals to the poly-T stretch of nicked genomic DNA (Kazazian, 2004; Babushok and Kazazian, 2007; Goodier and Kazazian, 2008). ORF1 and ORF2 complete the entire process of L1-RTP and also transpose other elements, such as Alu and SVA (SINE–VNTR–Alus: short interspersed element, SINE; variable number of tandem repeats, VNTR; and Alu; Dewannieux et al., 2003; Wallace et al., 2008; Hancks and Kazazian, 2012; Raiz et al., 2012), indicating that L1 functions as a driving force of genome shuffling.
L1-RTP in Somatic Cells is Induced by Environmental Factors
Reported inducers
The environmental triggers, gamma irradiation (Farkash et al., 2006), and heavy metals (El-Sawy et al., 2005; Kale et al., 2005), such as mercury, cadmium, and nickel, have been shown to induce L1-RTP. Although it was proposed that L1-RTP induced by nickel occurs at the post-transcriptional level (El-Sawy et al., 2005), the precise mode of induction of L1-RTP remains to be clarified. Stribinskis and Ramos (2006) also reported that benzo[a]pyrene (B[a]P) induced L1-RTP. They found that B[a]P-induced L1-RTP depended on the aryl hydrocarbon receptor (AhR). Interestingly, however, it was shown that 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin), a well-known ligand of AhR (Beischlag et al., 2008; McIntosh et al., 2010), did not induce L1-RTP. In a proposed mechanism, ligand-bound AhR activates expression of metabolic enzymes encoded by cytochrome P450 (CYP) genes, which in turn convert B[a]P into a genotoxic compound that induces L1-RTP (Stribinskis and Ramos, 2006). By contrast, TCDD is not a genotoxic compound, and does not induce L1-RTP.
Newly identified environmental factors and novel modes of L1-RTP
We recently discovered that 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct and a putative physiological AhR ligand (Wincent et al., 2009), induces L1-RTP (Okudaira et al., 2010). Experiments using short-interfering RNA (siRNA) along with an AhR inhibitor revealed that the L1-RTP induced by FICZ was not dependent on AhR, but required AhR nuclear translocator-1 (ARNT1; Hoffman et al., 1991). Chromatin recruitment of ligand-bound AhR is dependent on the ARNT1 nuclear localization signal (Eguchi et al., 1997). Moreover, it was also shown that addition of FICZ initiated the molecular interaction between ARNT1 and ORF1, and promoted the recruitment of ORF1 to the chromatin-rich fraction (Okudaira et al., 2010). Given that there have been no reports indicating that ARNT1 by itself functions as a receptor for environmental compounds, it is likely that a novel cellular factor that functions as an FICZ receptor cooperates with ARNT1 in the induction of L1-RTP.
Further studies demonstrated that dimethylbenzoanthracene (DMBA), B[a]P, and 3-methylchoranthrene (3-MC) induced L1-RTP (Okudaira et al., 2011; Ishizaka et al., 2012). Interestingly, the results of siRNA-based experiments revealed that the induction of L1-RTP by these carcinogens depended on AhR. Moreover, the induction of L1-RTP by DMBA required ARNT1, whereas L1-RTP induced by B[a]P and 3-MC did not. Notably, ARNT1 siRNA blocked mRNA expression of the CYP gene CYP1A1. Additionally, it has been ascertained that the expression of CYP1A1 mRNA depends on a heterodimer of AhR and ARNT1 (AHRC, AhR complex). These observations indicate that ARNT1 siRNA efficiently attenuated the biological function of the AHRC, confirming that the induction of L1-RTP by B[a]P and 3-MC was independent of ARNT1. Moreover, the expression of CYP1A1 mRNA in response to environmental factors requires the chromatin recruitment of ligand-bound AhR, which is dependent on the ARNT1 nuclear localization signal (Eguchi et al., 1997), lending support to the notion that a cellular factor, the function of which was similar to that of ARNT1, could contribute to chromatin recruitment of ORF1. Since AhR forms a complex with estrogen receptor α (ERα; Ohtake et al., 2003), we examined whether ERα was involved in induction of L1-RTP. Notably, experiments using ERα siRNA indicated that L1-RTP induced by both B[a]P and 3-MC depended on ERα (Ishizaka et al., 2012).
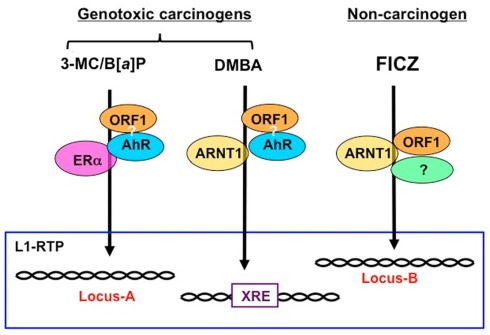
It has been shown that B[a]P increased the expression of L1 mRNA (Stribinskis and Ramos, 2006), whereas nickel chloride did not (El-Sawy et al., 2005). We also observed no apparent increase in L1 transcripts with FICZ (Okudaira et al., 2010). These observations suggest that the modes of L1-RTP triggered by environmental compounds are regulated differently at the transcriptional and post-transcriptional levels. It is conceivable that the cellular machinery that functions at the post-transcriptional level is regulated differently depending on the compound (Table 1). Although further study is required, the data support the idea that genotoxic carcinogens induce L1-RTP in an AhR-dependent manner. Moreover, the modes of L1-RTP induced by environmental compounds were different, suggesting that the integration sites of L1 differ depending on the trigger. We will discuss this possibility further later (Figure 1).
Table 1.
Summary of cellular factors required for L1-RTP by environmental compounds.
| Cellular factors | Inducers | |||
|---|---|---|---|---|
| FICZ | B[a]P | 3-MC | DMBA | |
| AhR | – | ◯ | ◯ | ◯ |
| ARNT1 | ◯ | – | – | ◯ |
| ERα | N.T. | ◯ | ◯ | – |
| MAPKs | ||||
| SB202190 | ◯ | ◯ | ◯ | – |
| SP600125 | ◯ | ◯ | – | – |
◯, dependent; –, independent; N.T., not tested.
The induction of L1-RTP was examined by a PCR-based assay.
Figure 1.
Schematic modes of L1-RTP triggered by environmental compounds. Given that ORF1, which associates with retroelements, is recruited to the chromatin, regions of L1-RTP are likely determined by the binding partners of ORF1. As shown in Table 1, the induction of L1-RTP by environmental compounds depended on different sets of bHLH/PAS proteins. The induction of L1-RTP by DMBA depended on AhR and ARNT1, whereas that by 3-MC and B[a]P required AhR. Interestingly, L1-RTP by 3-MC and B[a]P did not require ARNT1, but depended on ERα. By contrast, FICZ-induced L1-RTP depending on ARNT1, whereas it did not require AhR. It is plausible that the regions where these compounds insert L1 are different: DMBA induces L1-RTP in the vicinity of XRE, whereas 3-MC and B[a]P induce L1-RTP in the region determined by AhR and ERα. The locus of L1-RTP by FICZ would differ from those determined by these genotoxic carcinogens. To prove this, it is necessary to identify the association of ORF1 and AhR and genome regions where L1 is inserted in response to each environmental compound.
Mitogen-activated protein kinases are required for RTP induced by environmental compounds
In mammalian cells, six groups of MAPKs – extracellular signal-regulated protein kinase1/2 (ERK1/2), ERK5, JNK, p38, ERK3/4, and ERK7/8 – have been identified and shown to be activated by intracellular and extracellular stimuli (Cargnello and Roux, 2011). Among these MAPKs, the cellular signal cascades activated by ERK1/2, p38, and JNK have been well characterized because of the availability of inhibitors. PD98059, SB202190, and SP600125 are inhibitors of ERK1/2, p38, and JNK, respectively. To examine the involvement of MAPKs in the induction of L1-RTP, the effects of these inhibitors on the induction of L1-RTP have been analyzed. SB202190 blocked FICZ-induced L1-RTP, and siRNA specific for cyclic-AMP responsive element binding protein (CREB) mRNA efficiently attenuated FICZ-induced L1-RTP (Okudaira et al., 2010). Additionally, FICZ-induced phosphorylation of CREB in an ARNT1-dependent manner, and SB202190 blocked the FICZ-induced phosphorylation of CREB and chromatin recruitment of ORF1. These observations verified that p38 is required for the induction of L1-RTP by FICZ at the step of chromatin recruitment of ORF1. Further analysis on the mode of L1-RTP induced by 3-MC and B[a]P revealed that the L1-RTP induced by 3-MC was coupled to CREB phosphorylation, and that both CREB siRNA and SB202190 abrogated L1-RTP induction by the compound. The effects of MAPK inhibitors are summarized in Table 1. The next challenge is to determine how MAPKs and the phosphorylation of MAPK substrates are involved in the induction of L1-RTP. One plausible role of these molecules is driving the chromatin recruitment of ORF1 or retroelements in response to environmental compounds (Chiu and Greene, 2008). Several studies suggest that retroelements, such as Alu, are present in a cytoplasmic high-molecular weight complex due to the function of APOBEC3 proteins, an innate restriction molecule (Chiu and Greene, 2008).
Functional Interaction of bHLH/PAS Proteins and L1-RTP
Biology of bHLH/PAS proteins
The bHLH/PAS family is composed of numerous transcriptional factors with PAS domains consisting of approximately 275 amino acids. Family members have sequence homology to the clock gene period (per) from Drosophila melanogaster, which is involved in the control of circadian rhythms; the arnt gene in mammals, which is required for signaling pathways activated in response to dioxin or polyaromatic hydrocarbons; and the single-minded (sim) gene, a neurodevelopmental regulator in flies (Beischlag et al., 2008; McIntosh et al., 2010). It is interesting to note that the bHLH/PAS family is involved in these three apparently different biological responses. The PAS domain has two sub-domains, PAS-A and PAS-B, each of which comprises approximately 70 amino acids. The PAS-A domain functions as a binding site for other PAS-A domain-containing proteins (McIntosh et al., 2010). In contrast, the PAS-B domain is involved in interactions with other classes of proteins, such as heat shock protein 90 and small molecules. One of the best-characterized functions of bHLH/PAS proteins is in the response to environmental pollutants, such as polyaromatic hydrocarbons (PAH; Beischlag et al., 2008). When PAH binds to the PAS-B domain of AhR, a heterodimeric complex of ligand-bound AhR and ARNT1 (AHRC) is formed. AHRC is then recruited to chromatin, where it recognizes a xenobiotic responsive element (XRE; Probst et al., 1993) and activates the expression of genes encoding xenobiotic metabolic enzymes (Beischlag et al., 2008). Interestingly, the AHRC also activates expression of AhR repressor (AhRR), which has both bHLH and PAS-A domains, but not a PAS-B domain. It has been proposed that the expression of AhRR down-regulates the activity of the AHRC by a feedback mechanism (Mimura et al., 1997).
Chromatin recruitment of ORF1 is dependent on bHLH/PAS proteins
Experiments involving the forced expression of a plasmid DNA encoding ORF1 cDNA revealed that ORF1 is present in cytoplasmic stress granules in ribonucleoprotein complexes (Hohjoh and Singer, 1996; Goodier et al., 2007). Given that ORF1 acts in cis on L1-mRNA, it is conceivable that ORF1 is recruited from the cytoplasm to chromatin. Consistently, we observed that FICZ-induced enrichment of ORF1 in the chromatin-rich fraction and also triggered the physical association of ORF1 and ARNT1 (Okudaira et al., 2010). Additional experiments revealed that carcinogen-induced L1-RTP was dependent on AhR (Stribinskis and Ramos, 2006; Okudaira et al., 2011). These observations led us to hypothesize that ORF1 is functionally associated with bHLH/PAS proteins, and that both bHLH/PAS proteins and MAPK are involved in the chromatin recruitment of ORF1.
The importance of bHLH/PAS proteins in L1-RTP suggests that L1-RTP in the genome induced by environmental compounds may be directed. As summarized in Table 1, environmental carcinogens induce L1-RTP in an AhR-dependent manner. By contrast, FICZ-induced L1-RTP was not dependent on AhR. Instead, it requires ARNT1 and an additional cellular factor. Interestingly, the dependence of carcinogen-induced L1-RTP on ARNT1 differed among compounds: L1-RTP induced by DMBA required ARNT1, whereas that induced by 3-MC and B[a]P depended on AhR and ERα. These observations suggest that DMBA induces L1-RTP in the vicinity of XRE, which the AHRC targets for induction of mRNA expression of genes, such as CYP genes. In contrast, 3-MC and B[a]P would induce L1-RTP in the genome region determined by AhR and ERα. Moreover, the region in which FICZ-induced L1-RTP occurs differs from that in which carcinogen-induced L1-RTP occurs (Figure 1). It is important to analyze the regions of the newly integrated L1 to obtain novel information on the roles of L1-RTP in carcinogen-induced genetic alternations.
L1-RTP is Linked to the Development of Various Diseases
Genetic errors
L1-RTP accidentally disrupts structures of functional genes and gives rise to inborn errors (Goodier and Kazazian, 2008). Since the discovery of aberrant insertions of L1 in the gene encoding factor VIII in 2 of 240 sporadic hemophilia A patients (Kazazian et al., 1988), at least 20 genetic disorders have been identified as resulting from L1 insertional mutagenesis (Hancks and Kazazian, 2012). Including genetic alterations by Alu and SVA, 96 genetic disorders have been so far identified (Hancks and Kazazian, 2012). In addition, abnormal L1 insertion was detected in a patient with branchio-oto-renal syndrome, in which L1 disrupted the locus responsible for the disease (Morisada et al., 2010). Of note, a recent study of 18 unrelated patients with neurofibromatosis type 1 (NF1) identified insertions of 14 Alu, three L1, and one poly(T) stretch within the NF1 gene, indicating the retrotransposon insertions account for ∼0.4% of all NF1 mutations (Wimmer et al., 2011). Importantly, inserted L1, which was responsible for NF1 disruption, was Ta-1, which is the youngest subfamily of human L1 (Boissinot et al., 2000; Lee et al., 2007; Wimmer et al., 2011). Moreover, six different insertions were identified within the 1.5-kb region between exons 21 and 23. As the NF1 gene is 280 kb in length, it was proposed that the insertions of endogenous retroelements into the NF1 gene occurred non-randomly (Wimmer et al., 2011).
Tumorigenesis
During early studies of the alterations of cancer-related genes, aberrant L1 insertions in the c-myc and APC genes were found in breast and colon carcinoma, respectively (Morse et al., 1988; Miki et al., 1992). It was recently proposed that L1 functions as a natural mutagen for genetic alterations, and L1-RTP has been detected in lung carcinomas and pancreatic adenocarcinomas (Iskow et al., 2010; Ting et al., 2011). Interestingly, L1-RTP is associated with the expression of major satellite repeats and malignant tumor phenotypes (Ting et al., 2011). L1-RTP induces a variety of genetic alternations, including gene deletions, inversions, and inter/intra-chromosome translocations (Gilbert et al., 2002, 2005; Symer et al., 2002). Moreover, ORF2 induces DNA damage (Gasior et al., 2006). In addition to these direct effects of L1, the human genome is susceptible to genetic alterations by Alu–Alu non-allelic homologous recombination (NAHR; Belancio et al., 2010; Konkel and Batzer, 2010). For example, it was shown that Alu–Alu NAHR induced structural alterations in the tumor suppressor gene BRCA1, and the MLL-1 and Myb genes, in cancers cells (Strout et al., 1998; Mazoyer, 2005; O’Neil et al., 2007). Notably, the frequency of deletion via Alu–Alu NAHR under conditions of p53 deficiency was 40–300 times higher than that observed in the presence of wild-type p53 (Gebow et al., 2000), suggesting the importance of Alu–Alu NAHR during multistep carcinogenesis (Hanel and Moll, 2012). Environmental stimuli possibly function as risk factors for carcinogenesis by modulating genetic alternations via L1-RTP.
Although there is no model system to evaluate the association of L1-RTP with tumorigenesis, we recently showed that L1-RTP was common in skin tumors induced by DMBA and 12-O-tetradecanoylphorbol-13-acetate (TPA; Balmain et al., 1984; Okudaira et al., 2011). Of 15 skin tumors, 13 were positive for L1-RTP. It has been proposed that DMBA induces H-ras activation (Nelson et al., 1992), whereas TPA promotes the growth of transformed cells in a manner depending on TPA susceptibility locus (Psl) that is located on chromosome 9 (Angel et al., 1997). Recently, glutathione S-transferase α4 was identified as a candidate Psl gene (Abel et al., 2010). Interestingly, it was recently reported that activation of the H-ras gene in normal cells activates an ATM-dependent growth-arrest signal (Bartkova et al., 2006; Di Micco et al., 2006). Since ATM-dependent signaling causes senescence, an additional genetic alteration is required for tumor development; otherwise, tumor growth would be prevented. It remained elusive to clarify how TPA-induced L1-RTP is linked with the functional modification of the Psl gene product and attenuation of the ATM-dependent growth-arrest signal. Although it remains unclear whether L1-RTP is really involved in tumorigenesis, DMBA/TPA-induced skin carcinogenesis would be a good animal model for clarification of the role of L1-RTP in tumor development.
Autoimmune diseases
Intriguingly, a positive link between L1-RTP and the development of autoimmune diseases was proposed recently (Crow, 2010). Originally, heterozygous mutations of Trex1, a gene that encodes the 3′ repair exonuclease 1, were detected in familial chilblain lupus and Aicardi–Goutières syndrome (Rice et al., 2007). Interestingly, Trex1-deficient mice provided evidence that Trex1 functions as a negative regulator of the interferon-regulatory DNA response (Stetson et al., 2008). Without Trex1 function, mice die due to severe autoimmunity. However, such lethal effects of Trex1 deficiency were attenuated in mice with IRF3, IFNαR1, and RAG2 gene deficiencies (Stetson et al., 2008). Moreover, single-stranded DNA derived from endogenous retroelements accumulated in Trex1-deficient cells, and Trex1 overexpression blocked RTP of L1 (Stetson et al., 2008). Consistent with these experimental data, Trex1 activity was reduced in the synovial fibroblasts of rheumatoid arthritis patients (Neidhart et al., 2010). These observations suggest that RTP-induced production of DNA derived from endogenous retroelements is a potential molecular mechanism for the development of autoimmune disorders.
Future Perspectives
L1-RTP depends on three steps: transcription, RT, and integration. It is known that the expression of L1 is regulated by the methylation of CpG islands in the 5′ UTR region (Hata and Sakaki, 1997; Woodcock et al., 1997; Muotri et al., 2010). Moreover, the 5′ UTR of L1 contains a ubiquitously active antisense promoter that encodes siRNAs that effectively suppress L1-RTP (Yang and Kazazian, 2006). Although these reports indicate that L1 activity is regulated at the transcriptional level, our observations and those of others indicate that L1-RTP is also regulated post-transcriptionally. Environmental compounds induce the chromatin recruitment of ORF1 via bHLH/PAS proteins, suggesting that the functional coupling of L1-RTP and bHLH/PAS proteins is another pivotal step in the regulation of L1-RTP.
Since bHLH/PAS proteins are transcription factors that strictly recognize cis elements, it is plausible that the induction of L1-RTP in the genome depends on the selection of bHLH/PAS proteins by the individual compounds. DMBA-induced L1-RTP depends on both AhR and ARNT1, and it is likely that L1-RTP is induced in the vicinity of the XRE (Figure 1). Further study is required to identify a target locus for the induction of L1-RTP, which would provide novel information regarding the biological relevance of L1-RTP in somatic cells.
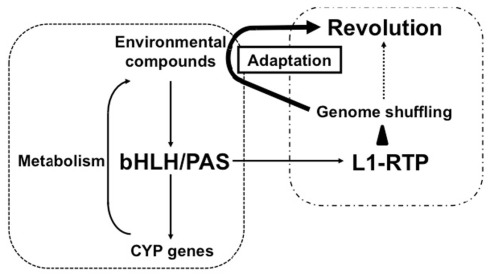
It is interesting to note that Candida albicans possesses a non-LTR-type retroelement with a structure similar to that of human L1 (Dong et al., 2009). This was shown to be functional in Saccharomyces cerevisiae, indicating that an endogenous retroelement similar to human L1 is functional in lower eukaryotes. Conversely, AhR homologs are present in the genomes of lower eukaryotes such as D. melanogaster and Caenorhabditis elegans (Hahn, 2002). Combined with our observation that L1-RTP and bHLH/PAS are linked functionally, these results suggest that genome shuffling by bHLH/PAS-dependent L1-RTP may facilitate adaptation to environmental changes. It is tempting to speculate that bHLH/PAS molecules recognize environmental pollutants and promote genome shuffling by RTP to generate novel cellular properties that can overcome changes in the environment (Figure 2). Such property of L1 might contribute to revolution.
Figure 2.
Possible functional link between bHLH/PAS proteins and L1-RTP in environmental adaptation and evolution. As a well-characterized biological function, bHLH/PAS proteins of AhR and ARNT1 induce gene expression of CYP genes in response to various compounds. The induced CYP proteins metabolize the environmental compounds and detoxify them. By contrast, L1-RTP induces genome shuffling, and random mutagenesis increases the chance of the emergence of living organisms that possess novel characteristics. As shown here, bHLH/PAS proteins are involved in the induction of L1-RTP, indicating the functional link between these two well-conserved cellular activities. The novel properties can overcome the disadvantageous effects of environmental compounds, enabling living organisms to survive under the selective pressure in the altered environment and contributing to evolution.
After the divergence of humans and chimpanzees about 6 myrs ago, both L1s continued to propagate, and Ta-1d emerged as the major subfamily in H. sapiens (Boissinot et al., 2000). Together with our observation that L1-RTP is induced in somatic cells by environmental factors, this affords the opportunity to speculate about novel aspects of L1 biology. Since human- and chimpanzee-specific L1 copy numbers are similar (Lee et al., 2007), it is likely that L1-RTP in the germ cells of both species is strictly regulated. However, the observation that H. sapiens has more copies of active L1 than do chimpanzees suggests that “hot” L1 offers an unidentified advantage to human activity. Interestingly, L1-RTP was induced in the dentate gyrus of the hippocampus when mice performed voluntary exercise (Muotri et al., 2009), and some data suggest that L1 copy numbers are increased in the human hippocampus (Coufal et al., 2011). One plausible direction for future research on L1 would be to focus on its role in the central nervous system (Hancks and Kazazian, 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in parts by a Grant-in-Aid for Research from the Ministry of Health, Labour, and Welfare of Japan (09156296), a research grant for the Log-range Research Initiative (LRI) from Japan Chemical Industry Association (JCIA) and a Grant-in-Aid from the Tokyo Biochemical Research Foundation. Mr. Noriyuki Okudaira is an applicant supported by the Grant-in-Aid from the Tokyo Biochemical Research Foundation.
References
- Abel E. L., Angel J. M., Riggs P. K., Langfield L., Lo H. H., Person M. D., Awasthi Y. C., Wang L. E., Strom S. S., Wei Q., DiGiovanni J. (2010). Evidence that Gsta4 modifies susceptibility to skin tumor development in mice and humans. J. Natl. Cancer Inst. 102, 1663–1675 10.1093/jnci/djq392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel J. M., Beltrán L., Minda K., Rupp T., DiGiovanni J. (1997). Association of a murine chromosome 9 locus (Psl1) with susceptibility to mouse skin tumor promotion by 12-O-tetradecanoylphorbol-13-acetate. Mol. Carcinog. 20, 162–167 [DOI] [PubMed] [Google Scholar]
- Babushok D. V., Kazazian H. H., Jr. (2007). Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28, 527–539 10.1002/humu.20486 [DOI] [PubMed] [Google Scholar]
- Baillie J. K., Barnett M. W., Upton K. R., Gerhardt D. J., Richmond T. A., De Sapio F., Brennan P. M., Rizzu P., Smith S., Fell M., Talbot R. T., Gustincich S., Freeman T. C., Mattick J. S., Hume D. A., Heutink P., Carninci P., Jeddeloh J. A., Faulkner G. J. (2011). Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 10.1038/nature10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A., Ramsden M., Bowden G. T., Smith J. (1984). Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature 307, 658–660 10.1038/307658a0 [DOI] [PubMed] [Google Scholar]
- Bannert N., Kurth R. (2004). Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U.S.A. 101, 14572–14579 10.1073/pnas.0404838101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V. P., Roy-Engel A. M., Deininger P. L. (2010). All y’all need to know ‘bout retroelements in cancer. Semin. Cancer Biol. 20, 200–210 10.1016/j.semcancer.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S., Chevret P., Furano A. V. (2000). L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17, 915–928 10.1093/oxfordjournals.molbev.a026372 [DOI] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H., Jr. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M., Roux P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y. L., Greene W. C. (2008). The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26, 317–353 10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- Coufal N. G., Garcia-Perez J. L., Peng G. E., Marchetto M. C., Muotri A. R., Mu Y., Carson C. T., Macia A., Moran J. V., Gage F. H. (2011). Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 20382–20387 10.1073/pnas.1100273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M. K. (2010). Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity 43, 7–16 10.3109/08916930903374865 [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. (2003). LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P. G., Bensimon A., Maestro R., Pelicci P. G., d’Adda di Fagagna F. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Dong C., Poulter R. T., Han J. S. (2009). LINE-like retrotransposition in Saccharomyces cerevisiae. Genetics 181, 301–311 10.1534/genetics.108.096636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi H., Ikuta T., Tachibana T., Yoneda Y., Kawajiri K. (1997). A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1beta is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J. Biol. Chem. 272, 17640–17647 10.1074/jbc.272.28.17640 [DOI] [PubMed] [Google Scholar]
- El-Sawy M., Kale S. P., Dugan C., Nguyen T. Q., Belancio V., Bruch H., Roy-Engel A. M., Deininger P. L. (2005). Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 354, 246–257 10.1016/j.jmb.2005.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash E. A., Kao G. D., Horman S. R., Prak E. T. (2006). Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 34, 1196–1204 10.1093/nar/gkj522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J. V., Kazazian H. H., Jr., Boeke J. D. (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 10.1016/S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- Gasior S. L., Wakeman T. P., Xu B., Deininger P. L. (2006). The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebow D., Miselis N., Liber H. L. (2000). Homologous and nonhomologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol. Cell Biol. 20, 4028–4035 10.1128/MCB.20.11.4028-4035.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou I., Noutsopoulos D., Dimitriadou E., Markopoulos G., Apergi A., Lazaros L., Vaxevanoglou T., Pantos K., Syrrou M., Tzavaras T. (2009). Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum. Mol. Genet. 18, 1221–1228 10.1093/hmg/ddp022 [DOI] [PubMed] [Google Scholar]
- Gilbert N., Luts S., Morrisj T. A., Moran J. V. (2005). Multiple fates of L1 retrotransposition intermideaites in cultured human cells. Mol. Cell Biol. 25, 7780–7795 10.1128/MCB.25.17.7780-7795.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., Moran J. V. (2002). Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 10.1016/S0092-8674(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Goodier J. L., Kazazian H. H., Jr. (2008). Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- Goodier J. L., Zhang L., Vetter M. R., Kazazian H. H., Jr. (2007). LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell Biol. 27, 6469–6483 10.1128/MCB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., Jr. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 1–13 10.1016/j.gde.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel W., Moll U. M. (2012). Links between mutant p53 and genomic instability. J. Cell Biochem. 113, 433–439 10.1002/jcb.23400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. E. (2002). Aryl hydrocarbon receptors: diversity and evolution. Chem. Biol. Interact. 141, 131–160 10.1016/S0009-2797(02)00070-4 [DOI] [PubMed] [Google Scholar]
- Hata K., Sakaki Y. (1997). Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189, 227–234 10.1016/S0378-1119(96)00856-6 [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. (1991). Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252, 954–958 10.1126/science.1712121 [DOI] [PubMed] [Google Scholar]
- Hohjoh H., Singer M. F. (1996). Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639 [PMC free article] [PubMed] [Google Scholar]
- Ishizaka Y., Okudaira N., Okamura T. (2012). Regulation of retrotransposition of long interspersed element-1 by mitogen-activated protein kinases. In Protein kinase (in press). [Google Scholar]
- Iskow R. C., McCabe M. T., Mills R. E., Torene S., Pittard W. S., Neuwald A. F., Van Meir E. G., Vertino P. M., Devine S. E. (2010). Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 10.1016/j.cell.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. (1997). Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. U.S.A. 94, 1872–1877 10.1073/pnas.94.5.1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S. P., Moore L., Deininger P. L., Roy-Engel A. M. (2005). Heavy metals stimulate human LINE-1 retrotransposition. Int. J. Environ. Res. Public Health 2, 14–23 10.3390/ijerph2005010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M. R., Gerton G. L., Ostertag E. M., Kazazian H. H., Jr. (2009). L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H., Jr. (2004). Mobile elements: drivers of genome evolution. Science 303, 1626–1632 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, Wong C., Youssoufian H., Scott A. F., Phillips D. G., Antonarakis S. E. (1988). Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Konkel M. K., Batzer M. A. (2010). A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 20, 211–221 10.1016/j.semcancer.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., McPherson J. D., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W. R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., McLysaght A., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J. International Human Genome Sequencing Consortium. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee J., Cordaux R., Han K., Wang J., Hedges D. J., Liang P., Batzer M. A. (2007). Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene 390, 18–27 10.1016/j.gene.2006.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S. C., Hodges R. S., Williams M. C. (2005). LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348,549–561 10.1016/j.jmb.2005.03.015 [DOI] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Jr., Boeke J. D., Gabriel A. (1991). Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810 10.1126/science.1722352 [DOI] [PubMed] [Google Scholar]
- Mazoyer S. (2005). Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum. Mutat. 25, 415–422 10.1002/humu.20169 [DOI] [PubMed] [Google Scholar]
- McIntosh B. E., Hogenesch J. B., Bradfield C. A. (2010). Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 10.1146/annurev-physiol-021909-135922 [DOI] [PubMed] [Google Scholar]
- Miki Y., Nishisho I., Horii A., Miyoshi Y., Utsunomiya J., Kinzler K. W., Vogelstein B., Nakamura Y. (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645 [PubMed] [Google Scholar]
- Mimura J., Yamashita K., Nakamura K., Morita M., Takagi T. N., Nakao K., Ema M., Sogawa K., Yasuda M., Katsuki M., Fujii-Kuriyama Y. (1997). Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2, 645–654 10.1046/j.1365-2443.1997.1490345.x [DOI] [PubMed] [Google Scholar]
- Morisada N., Rendtorff N. D., Nozu K., Morishita T., Miyakawa T., Matsumoto T., Hisano S., Iijima K., Tranebjaerg L., Shirahata A., Matsuo M., Kusuhara K. (2010). Branchio-oto-renal syndrome caused by partial EYA1 deletion due to LINE-1 insertion. Pediatr. Nephrol. 25, 1343–1348 10.1007/s00467-010-1445-x [DOI] [PubMed] [Google Scholar]
- Morse B., Rotherg P. G., South V. J., Spandorfer J. M., Astrin S. M. (1988). Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 333, 87–90 10.1038/333087a0 [DOI] [PubMed] [Google Scholar]
- Muotri A. R., Chu V. T., Marchetto M. C., Deng W., Moran J. V., Gage F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Muotri A. R., Marchetto M. C., Coufal N. G., Oefner R., Yeo G., Nakashima K., Gage F. H. (2010). L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446 10.1038/nature09544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A. R., Zhao C., Marchetto M. C., Gage F. H. (2009). Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19, 1002–1007 10.1002/hipo.20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhart M., Karouzakis E., Schumann G. G., Gay R. E., Gay S. (2010). Trex-1 deficiency in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 62, 2673–2679 10.1002/art.27567 [DOI] [PubMed] [Google Scholar]
- Nelson M. A., Futscher B. W., Kinsella T., Wymer J., Bowden G. T. (1992). Detection of mutant Ha-ras genes in chemically initiated mouse skin epidermis before the development of benign tumors. Proc. Natl. Acad. Sci. U.S.A. 89, 6398–6402 10.1073/pnas.89.8.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F., Takeyama K., Matsumoto T., Kitagawa H., Yamamoto Y., Nohara K., Tohyama C., Krust A., Mimura J., Chambon P., Yanagisawa J., Fujii-Kuriyama Y., Kato S. (2003). Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423, 545–550 10.1038/nature01606 [DOI] [PubMed] [Google Scholar]
- Okudaira N., Iijima K., Koyama T., Minemoto Y., Kano S., Mimori A., Ishizaka Y. (2010). Induction of long interspersed nucleotide element-1 (L1) retrotransposition by 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct. Proc. Natl. Acad. Sci. U.S.A. 107, 18487–18492 10.1073/pnas.1001252107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudaira N., Goto M., Yanobu-Takanashi R., Tamura M., An A., Abe Y., Kano S., Hagiwara S., Ishizaka Y., Okamura T. (2011). Involvement of retrotransposition of long interspersed nucleotide element-1 in skin tumorigenesis induced by 7,12-dimethylbenz[a]anthracene and 12-O-tetradecanoylphorbol-13-acetate. Cancer Sci. 102, 2000–2006 10.1111/j.1349-7006.2011.02060.x [DOI] [PubMed] [Google Scholar]
- O’Neil J., Tchinda J., Gutierrez A., Moreau L., Maser R. S., Wong K. K., Li W., McKenna K., Liu X. S., Feng B., Neuberg D., Silverman L., DeAngelo D. J., Kutok J. L., Rothstein R., DePinho R. A., Chin L., Lee C., Look A. T. (2007). Alu elements mediate MYB gene tandem duplication in human T-ALL. J. Exp. Med. 204, 3059–3066 10.1084/jem.20070876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst M. R., Reisz-Porszasz S., Agbunag R. V., Ong M. S., Hankinson O. (1993). Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol. Pharmacol. 44, 511–518 [PubMed] [Google Scholar]
- Raiz J., Damert A., Chira S., Held U., Klawitter S., Hamdorf M., Löwer J., Strätling W. H., Löwer R., Schumann G. G. (2012). The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 10.1093/nar/gkr863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G., Newman W. G., Dean J., Patrick T., Parmar R., Flintoff K., Robins P., Harvey S., Hollis T., O’Hara A., Herrick A. L., Bowden A. P., Perrino F. W., Lindahl T., Barnes D. E., Crow Y. J. (2007). Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 80, 811–815 10.1086/513443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V., Ramos K. S. (2006). Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 66, 2616–2620 10.1158/0008-5472.CAN-05-3478 [DOI] [PubMed] [Google Scholar]
- Strout M. P., Marcucci G., Bloomfield C. D., Caligiuri M. A. (1998). The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 95, 2390–2395 10.1073/pnas.95.5.2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., Parmigiani G., Boeke J. D. (2002). Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- Ting D. T., Lipson D., Paul S., Brannigan B. W., Akhavanfard S., Coffman E. J., Contino G., Deshpande V., Iafrate A. J., Letovsky S., Rivera M. N., Bardeesy N., Maheswaran S., Haber D. A. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 10.1126/science.1200801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk J. A., Meij I. C., Seleme M. C., Kano H., Nikopoulos K., Hoefsloot L. H., Sistermans E. A., de Wijs I. J., Mukhopadhyay A., Plomp A. S., de Jong P. T., Kazazian H. H., Cremers F. P. (2007). L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592 10.1093/hmg/ddm108 [DOI] [PubMed] [Google Scholar]
- Vitullo P., Sciamanna I., Baiocchi M., Sinibaldi-Vallebona P., Spadafora C. (2012). LINE-1 retrotransposon copies are amplified during murine early embryo development. Mol. Reprod. Dev. 79, 118–127 10.1002/mrd.22003 [DOI] [PubMed] [Google Scholar]
- Wallace N., Wagstaff B. J., Deininger P. L., Roy-Engel A. M. (2008). LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene 419, 1–6 10.1016/j.gene.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V. (2001). Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 21, 1429–1439 10.1128/MCB.21.22.7714-7720.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K., Callens T., Wernstedt A., Messiaen L. (2011). The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 7, e1002371. 10.1371/journal.pgen.1002371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wincent E., Amini N., Luecke S., Glatt H., Bergman J., Crescenzi C., Rannug A., Rannug U. (2009). The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J. Biol. Chem. 284, 2690–2696 10.1074/jbc.M808321200 [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Lawler C. B., Linsenmeyer M. E., Doherty J. P., Warren W. D. (1997). Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem. 272, 7810–7816 10.1074/jbc.272.12.7810 [DOI] [PubMed] [Google Scholar]
- Yang N., Kazazian H. H., Jr. (2006). L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771 10.1038/nsmb1041 [DOI] [PubMed] [Google Scholar]