Abstract
Lymphocytic choriomeningitis virus (LCMV) is a zoonotic pathogen of which mice are the natural reservoir. Different strains and clones of LCMV show different pathogenicity in mice. Here we determined the complete genomic sequences of 3 LCMV strains (OQ28 and BRC which were isolated from mice in Japan and WE(ngs) which was derived from strain WE). Strains OQ28 and BRC showed high sequence homology with other LCMV strains. Although phylogenetic analyses placed these 2 Japanese strains in different subclusters, they belonged to same cluster of LCMV isolates. WE(ngs) and WE had many sequence substitutions between them but fell into same subcluster. The pathogenicity of the 3 new LCMV isolates was examined by inoculating ICR mice with 102 and 104 TCID50 of virus. ICR mice infected with OQ28 or WE(ngs) exhibited severe clinical signs, and some of the infected mice died. In contrast, all ICR mice infected with BRC showed no clinical signs and survived infection. Virus was detected in the blood, organs, or both of most of the surviving ICR mice inoculated with either OQ28 or WE(ngs). However, virus was below the level of detection in all ICR mice surviving infection with strain BRC. Therefore, LCMV strains OQ28 and BRC were genetically classified in the same cluster of LCMV strains but exhibited very different pathogenicity.
Abbreviations: dpi, days postinfection; GP, viral glycoprotein; h, hydrophobic region; IFA, indirect fluorescent antibody assay; L, viral RNA-dependent RNA polymerase; LCMV, lymphocytic choriomeningitis virus; NP, nucleocapsid protein; UTR, untranslated region; Z, zinc-finger protein
Lymphocytic choriomeningitis virus (LCMV) is a member of the genus Arenavirus in the family Arenaviridae. The genus Arenavirus is divided into 2 groups (Old World and New World arenaviruses) according to genetic and antigenic characteristics.4 LCMV is a member of the Old World arenavirus group, which also includes Lassa, Mopeia, Mobala, and Ippy viruses.4,10 The LCMV genome contains 2 negative-sense single-stranded RNA segments, designated S RNA and L RNA, with approximate sizes of 3.4 kb and 7.2 kb, respectively.30,31 Each RNA segment has an ambisense coding strategy, encoding 2 different proteins in opposite orientations. S RNA encodes the nucleocapsid protein and glycoprotein, and L RNA encodes the viral RNA-dependent RNA polymerase and a small zinc finger protein.25,30
LCMV is a zoonotic agent that is transmitted to humans via urine or saliva of infected mice (Mus musculus), which are a natural reservoir of the virus.4 The prevalence of LCMV in mice is 7.0% to 25.9% in Japan and 4% to 9% in Europe.5,17,19,20,35 Mice are naturally infected by either vertical or horizontal transmission of the virus, and infected mice usually show no clinical signs. In contrast, experimentally infected mice inoculated intraperitoneally or intracerebrally can exhibit clinical signs such as ruffled fur, half-closed eyes, hunched posture, immobility, and neurologic deficits.4,12,19 Although human LCMV infections are generally either asymptomatic or mild, immunodeficient persons can develop spontaneous abortion, severe birth defects, aseptic meningitis, or fatal infections.1,2,13,22,27 Therefore, LCMV is an important agent that should be monitored in facilities housing and breeding mice.
LCMV strains Armstrong, Traub, and WE were isolated during the 1930s from laboratory mice and humans working in a mouse facility.4 Many other LCMV strains and clones used in research originated from these 3 isolates. Strains Aggressive and Docile are clones (variants) of strain UBC, which was derived from the parental strain WE, and strains E350, CA1371, 53b, and clone 13 were all derived from strain Armstrong.4 The lethality of strains Aggressive and Docile varies between mouse strains.38 Mice inoculated with 53b develop acute infections, whereas those inoculated with clone 13 mount chronic infections, even though both of the strains were derived from strain Armstrong.29 Furthermore, strain Armstrong produces more severe disease in C3H mice than do strains WE and Traub.4 Therefore, previous studies indicate that mice infected with different strains of LCMV exhibit differences in clinical signs and lethality.4,7 LCMV is a noncytolytic virus and causes immune-mediated viral disease.12 The clinical signs and lethal disease arise because virus-specific T cells attack infected cells on critical organs in infected mice.12
Here we report the characterization of 2 LCMV strains recently isolated in Japan (strains OQ28 and BRC) and a passaged isolate of strain WE. The complete genomic sequences of these 3 strains were determined, and their phylogenetic relationship to other LCMV strains was assessed. We also evaluated the pathogenicity in ICR mice of these isolates.
Materials and Methods
Viruses.
Strain OQ28 was isolated in 1990 from a wild mouse (M. musculus) captured in the port city of Osaka, Japan.19 This isolate was passaged once in Vero cells and once in ICR mouse brain. Strain BRC was isolated in our laboratory in 2005 from the kidneys of an MAI/Pas mouse (M. musculus), which had been imported in 2004 by the RIKEN BioResource Center (Japan).11 This isolate was passaged once in ICR mouse brain. Although strain WE (originally isolated in 1936 and kindly provided by Dr R Mori, Kyushu University) was used as an LCMV strain isolated outside of Japan, we designated this strain WE(ngs) because its exact passage history before provision in our laboratory is unknown. Strain WE(ngs) was passaged once in BALB/c mouse brain at our laboratory.
All LCMV isolates were passaged twice in newborn ICR mouse brains before use in pathogenicity experiments. Mouse brain homogenates (10%) were made by using a Multibeads Shocker (Yasuikikai, Osaka, Japan), and the supernatant was used for experimental inoculations. Virus titers of the supernatant were calculated as TCID50 by using an indirect fluorescent antibody assay (IFA) described previously.24 Ten-fold serial dilutions of the supernatant were used to inoculate Vero cells. Infected Vero cells were trypsinized at 5 d after infection (dpi), washed with PBS, and spotted on Teflon-coated glass slides. The slides were dried and fixed in cold acetone at 4 °C for 10 min and dried, and virus titers estimated by IFA using antiLCMV antiserum as previously described.28
Mice.
SPF mice were purchased from Japan SLC (Japan). Excluded agents included mouse hepatitis virus, Sendai virus, pneumonia virus of mice, mouse adenovirus, LCMV, ectromelia virus, Mycoplasma pulmonis, Pseudomonas aeruginosa, Citrobacter rodentium, Salmonella spp., Pasteurella pneumotropica, Corynebacterium kutscheri, Clostridium piliforme, pinworms, intestinal protozoa, and ectoparasites. Newborn ICR outbred mice were used for passage of virus, and 3- to 4-wk old female ICR outbred and BALB/c inbred mice were used for experimental inoculations. All mice were maintained in a pathogen-free environment at 22 ± 2 °C and a humidity of 40% to 70%; food and water were available ad libitum. All experiments with infectious virus were performed under Animal Biosafety Level 3 biocontainment. Animal care and experimental procedures were performed in accordance with the Guidelines for Animal Experimentation of Nagasaki University with IACUC approval.
Sequence analysis of viral genomes.
Viral RNA was extracted by the single-step AGPC method as previously described, except that sodium acetate (pH 5.2) was used.23 cDNA was synthesized by using M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA) and 1 µM gene-specific primer. The cDNA products were purified by using MicroSpin S300 HR columns (GE Healthcare UK, Buckinghamshire, UK), and the DNA was eluted in water. PCR was performed by using Ex Taq polymerase (TaKaRa, Shiga, Japan) and 1 µM gene-specific primer, according to the manufacturer's instructions. End sequences of virus genomic RNA were determined by using a 5′/3′ RACE kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Sequences of primers used for RT–PCR were based on sequences of relatively conserved regions in S RNA and L RNA of the 3 strains (these sequences are not provided here due to the large number used but are available on request). PCR reactions were run on a TP600 PCR Thermal Cycler Dice Gradient (Takara) for 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 to 120 s at 72 °C; with a final elongation step of 5 min at 72 °C. PCR products were separated on a 1.0% agarose gel and purified by using a Gel Extraction Kit (LaboPass, Seoul, Korea). The products were sequenced with an automated DNA sequencer (models 3100 and 3130; ABI, Carlsbad, CA) using the PRISM Ready Reaction BigDye Terminator (version 3.1) and Cycle Sequencing Kit (version 1.1; ABI). Genomic sequences were assembled from sequences of overlapping PCR products. Complete sequences of S RNA and L RNA from strains OQ28, BRC, and WE(ngs) have been deposited into the DDBJ/EMBL/GenBank database (S RNA: AB627952, AB627953, and AB627951, respectively; L RNA: AB627955, AB627956, and AB627954, respectively).
Phylogenetic analyses.
Sequences for the S and L RNA of strains OQ28, BRC, and WE(ngs) were compared with published sequences available for other LCMV strains. The Vector NTI Advance 11 sequence analysis package (Invitrogen) was used to compare the sequence homology of the 4 coding regions (the nucleocapsid protein [NP], glycoprotein [GP], viral RNA-dependent RNA polymerase [L], and a small zinc finger protein [Z]). Phylogenetic and molecular evolutionary analyses of the NP coding region were conducted by using MEGA version 4.36 Distances were estimated by using the Tamura–Nei method with pairwise gap deletion, and phylogenetic trees were constructed by using the neighbor-joining method. Branch bootstrap values were determined by using 1000 iterations.
Experimental inoculation of mice with LCMV strains.
Groups of 5 or 6 3-wk-old female ICR mice were inoculated intraperitoneally with 102 or 104 TCID50 of virus. The inoculated mice were observed for clinical signs for 29 d, after which blood, lung, liver, spleen, and kidneys were removed from surviving mice and the viral genomic sequences in these tissues detected by RT–PCR. Serum IgG antibody titers in surviving mice were estimated by IFA as previously described.28
Three additional mouse experiments were performed with strains OQ28 and BRC. Six 3-wk-old female BALB/c mice were inoculated intraperitoneally with 104 TCID50 of strain OQ28 and were observed for 28 d for clinical signs. Female ICR mice (age, 4 wk) were inoculated intracerebrally with strain BRC and observed for clinical signs. In addition, 3-wk-old female ICR mice were inoculated intraperitoneally with 104 TCID50 of strain BRC, and virus distribution in tissues were examined in early phase of infection. At 7 dpi, blood, lung, liver, spleen, and kidneys were removed from surviving mice and viral genomic sequences in these tissues detected by RT–PCR.
Unless they died, mice on this protocol typically did not display clinical signs. To evaluate viral virulence and compare data appropriately with those in published studies, the IACUC approved death (or 29 dpi) as an endpoint for these studies, which used the minimal number of mice.
Virus distribution in tissues.
ICR mouse tissues were emulsified by using a Multibeads Shocker (Yasuikikai), and 200 µL of the 10% homogenate was used for RNA isolation. RNA was extracted by the single-step APGC method, and the isolated RNA was dissolved in 30 µL of diethylpyrocarbonate-treated water. Primers specific for the NP coding region of strains OQ28, BRC, and WE(ngs) were designed (Table 1) and used for viral genome detection by RT–PCR. Primers F01, F02, and R01 were used for the RT reaction; primers F02 and R01 were used in the first-round PCR reaction; and primers F03 and R02 were used in the second-round (nested) PCR reactions. The reaction solution for RT–PCR was as described earlier, with the primer used at 1 µM. The RT reaction used 10% total RNA of reaction solution as the template, and the cDNA products were purified by using MicroSpin S300 HR columns (GE Healthcare UK). First-round PCR reactions used 5% of the cDNA product as template, and second-round PCR reactions used 2.5% of the first-round PCR products as template. Cycle settings for PCR reactions were: 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 55 °C, 60 (first-round PCR) or 120 (second-round PCR) s at 72 °C; and a final elongation step of 5 min at 72 °C. PCR products were analyzed by electrophoresis on a 1.0% agarose gel.
Table 1.
Primers used for RT-PCR detection of virus sequences.
| Primera | Positionb | |
| F01 | 5′ CGC ACC GGG GAT CCT AGG CT 3′ | 1–20 |
| F02 | 5′ AGG TGG AGA GTC AGG GAG GC 3′ | 1609–1628 |
| F03 | 5′ TGT TYT CCC ATG CYC TYC CCA CAA 3′ | 2328–2351 |
| R01 | 5′ CGC ACM GTG GAT CCT AGG C 3′ | 3357–3375 |
| R02 | 5′ GCT GAT CTK GAR AAG CTG AAR GC 3′ | 2984–3006 |
K, G or T; M, A or C; R, A or G; Y, C or T
Primers used for detection of virus genome sequences by RT-PCR were designed by using sequence data from strains OQ28, BRC, and WE(ngs).
Positions are relative to the S RNA sequence of strain WE (DDBJ/EMBL/GenBank accession no., M22138).
Results
Genetic comparison of LCMV strains.
S RNA and L RNA sequences of strains OQ28, BRC, and WE(ngs) were determined in the current study, and general characteristics of the S and L RNAs of these new isolates and the published strain WE (S RNA, M22138; L RNA, AF004519) are summarized in Table 2.9,26 The lengths of the 5′ and 3′ untranslated regions (UTR) and intergenic noncoding regions of both RNAs in strains OQ28, BRC, and WE(ngs) differed among the 4 strains, but the lengths of the coding regions (the NP, GP, L, and Z) were highly consistent among them. Strain WE(ngs), which had been passaged 3 times in our laboratory, differed from the parental strain WE only in the intergenic noncoding region of L RNA, where strain WE(ngs) had an insertion of 13 nucleotides relative to WE.
Table 2.
Comparison of S RNA and L RNA
| S RNA |
L RNA |
|||||||||||
| straina | size | 5′ UTR | GP | IGR | NP | 3′ UTR | size | 5′ UTR | Z | IGR | L | 3′ UTR |
| OQ28 | 3358 | 59 | 1497 | 64 | 1677 | 61 | 7248 | 101 | 273 | 212 | 6630 | 32 |
| BRC | 3380 | 80 | 1497 | 63 | 1677 | 63 | 7241 | 86 | 273 | 219 | 6630 | 33 |
| WE(ngs) | 3375 | 77 | 1497 | 64 | 1677 | 60 | 7232 | 88 | 273 | 209 | 6630 | 32 |
| WE | 3375 | 77 | 1497 | 64 | 1677 | 60 | 7219 | 88 | 273 | 196 | 6630 | 32 |
GP, glycoprotein; IGR, intergenic region; L, virus RNA-dependant RNA polymerase; NP, nucleoprotein; UTR, untranslated region; Z, zinc-finger protein.
We compared the genomic sequences of strains OQ28, BRC, and WE(ngs) with those of published LCMV strains and Old World arenaviruses (Table 3). The amino acid sequence of the NP coding region was more highly conserved than those of the other 3 coding regions. The NP, GP, L, and Z coding regions of strain WE(ngs) were found to have substitutions of 3 amino acids (1%), 13 amino acids (3%), 51 amino acids (2%), and 3 amino acids (2%), respectively, relative to the original WE strain.
Table 3.
Amino-acid sequence homology in LCMV coding regions
| NP |
GP |
L |
Z |
|||||||||
| OQ28 | BRC | WE(ngs) | OQ28 | BRC | WE(ngs) | OQ28 | BRC | WE(ngs) | OQ28 | BRC | WE(ngs) | |
| LCMV | ||||||||||||
| OQ28 | — | 94 | 97 | — | 90 | 95 | — | 79 | 90 | — | 77 | 89 |
| BRC | 94 | — | 94 | 90 | — | 90 | 79 | — | 79 | 77 | — | 78 |
| WE(ngs) | 97 | 94 | — | 95 | 90 | — | 90 | 79 | — | 89 | 78 | — |
| WE | 96 | 93 | 99 | 94 | 88 | 97 | 88 | 78 | 98 | 87 | 78 | 98 |
| Traub | 98 | 94 | 96 | 97 | 91 | 96 | 92 | 79 | 90 | 91 | 78 | 89 |
| Y | 97 | 92 | 96 | 93 | 88 | 93 | — | — | — | — | — | — |
| California | 98 | 93 | 97 | 96 | 90 | 94 | 84 | 78 | 83 | 82 | 79 | 89 |
| Michigan | 97 | 94 | 98 | 96 | 90 | 94 | 90 | 79 | 90 | 87 | 82 | 84 |
| Ohio | 97 | 92 | 97 | 93 | 89 | 93 | 90 | 80 | 91 | 83 | 81 | 90 |
| Rhode Island | 97 | 93 | 97 | 93 | 89 | 93 | 90 | 79 | 91 | 83 | 81 | 90 |
| Wisconsin | 96 | 93 | 96 | 95 | 91 | 94 | 89 | 79 | 88 | 90 | 79 | 87 |
| WHI | 97 | 94 | 97 | 94 | 90 | 93 | — | — | — | — | — | — |
| Armstrong 53b | 96 | 93 | 96 | 96 | 90 | 94 | 90 | 79 | 88 | 90 | 77 | 89 |
| Armstrong cl13 | 96 | 93 | 96 | 96 | 90 | 95 | 90 | 79 | 88 | 90 | 77 | 89 |
| Docile | 96 | 93 | 96 | 94 | 90 | 95 | 90 | 79 | 89 | 90 | 79 | 87 |
| Aggressive | 96 | 94 | 96 | 94 | 90 | 95 | 90 | 79 | 89 | 90 | 79 | 87 |
| UBC | 96 | 94 | 96 | 94 | 90 | 95 | 90 | 79 | 89 | 89 | 79 | 86 |
| Douglas | 96 | 94 | 97 | 95 | 90 | 94 | 91 | 79 | 90 | 84 | 81 | 90 |
| Pasteur | 96 | 93 | 96 | 94 | 91 | 95 | 90 | 79 | 89 | 89 | 79 | 91 |
| MX | 95 | 92 | 95 | 93 | 90 | 94 | 88 | 78 | 88 | 87 | 78 | 89 |
| CH-5871 | 96 | 92 | 96 | 94 | 90 | 92 | — | — | — | — | — | — |
| CH-5692 | 96 | 93 | 96 | 94 | 91 | 93 | 88 | 78 | 87 | 88 | 78 | 84 |
| Massachusetts | 97 | 94 | 96 | 95 | 91 | 94 | 92 | 79 | 90 | 88 | 83 | 89 |
| Marseille no.12 | 95 | 93 | 95 | 95 | 90 | 95 | 89 | 79 | 88 | 80 | 80 | 78 |
| M1 | 94 | 99 | 94 | 90 | 99 | 89 | 79 | 100 | 80 | 77 | 100 | 78 |
| Dandenong | 94 | 97 | 95 | 91 | 95 | 90 | 80 | 88 | 80 | 81 | 94 | 82 |
| LE | 93 | 95 | 93 | 87 | 95 | 88 | — | — | — | — | — | — |
| Georgia | 92 | 94 | 92 | 88 | 90 | 89 | 80 | 79 | 80 | 80 | 79 | 82 |
| Bulgaria | 94 | 94 | 94 | 90 | 92 | 90 | 80 | 81 | 80 | — | — | — |
| GR01 | 90 | 90 | 90 | 83 | 83 | 83 | — | — | — | — | — | — |
| CABN | 90 | 89 | 91 | 80 | 82 | 80 | — | — | — | — | — | — |
| SN05 | 90 | 89 | 90 | 80 | 82 | 80 | — | — | — | — | — | — |
| Merino Walk virus | 67 | 66 | 67 | 56 | 57 | 57 | 46 | 46 | 46 | 55 | 57 | 57 |
| Ippy virus | 65 | 64 | 65 | 60 | 61 | 60 | 47 | 46 | 47 | 57 | 56 | 54 |
| Lassa virus | 63 | 63 | 63 | 61 | 61 | 60 | 47 | 47 | 47 | 53 | 54 | 55 |
| Mobala virus | 64 | 64 | 63 | 60 | 58 | 58 | 48 | 46 | 47 | 52 | 53 | 53 |
| Mopeia virus | 66 | 65 | 65 | 58 | 59 | 58 | 47 | 47 | 46 | 48 | 54 | 49 |
| Morogoro virus | 66 | 66 | 66 | 58 | 59 | 58 | — | — | — | 51 | 58 | 50 |
Values represent the percentage of amino acid sequence identity in the 4 coding regions of strains BRC, OQ28, and WE(ngs) with those of other LCMV isolates and Old World arenaviruses. Sequences of the NP, GP, L, and Z coding region analyzed here cover all 558, 498, 2209, and 90 amino acids (excluding the stop codon), respectively. However, the NP coding regions of strains Y and LE lack the first 64 and 195 amino acids, respectively, and the GP coding region of strain LE lacks the final 4 amino acids.
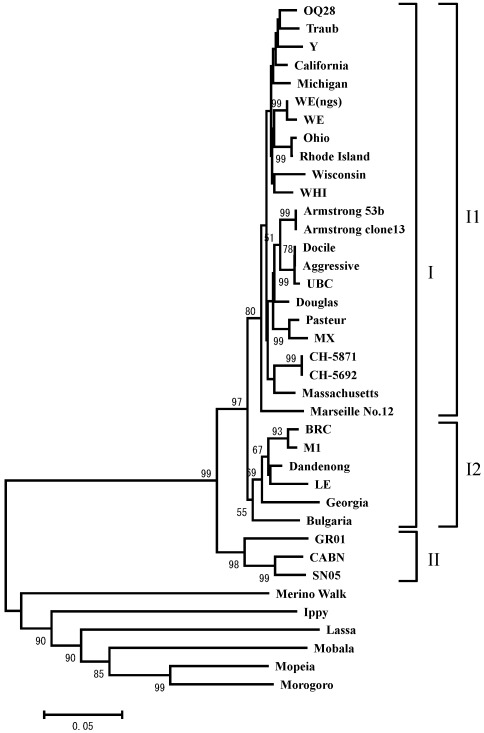
A phylogenetic tree based on the amino acid sequences of the NP coding region was constructed by using the neighbor-joining method and the MEGA program.36 Accordingly, LCMV strains were grouped into 2 clades (I and II; Figure 1), and 2 subclusters (I1 and I2) were evident within cluster I. Strains OQ28 and WE(ngs) were grouped with the original WE strain into subcluster I1, whereas strain BRC was assigned to subcluster I2, with strain M1 which was isolated from the same mouse strain (MAI/Pas).11 These results are consistent with the known origins of LCMV strains and demonstrate that serial passage of LCMV can result in considerable amino acid substitution (1% to 3%) in coding regions.
Figure 1.
Phylogenetic comparison of LCMV strains OQ28, BRC, and WE(ngs) to other Old World arenaviruses. The phylogenetic tree was based on amino acid sequences of NP coding region available in the GenBank database and was constructed with the neighbor-joining method by using MEGA version 4. The phylogenetic tree is based on 559-residue amino acid sequences of the NP coding region, although strain Y lacks the first 64 residues and strain LE lacks the first 195 amino acids. The numbers at the nodes are bootstrap values based on 1000 replications. A value greater than 50 is considered significant for a branch point. The scale bar represents 0.05 substitutions per amino acid position.
Virus titer used experimental inoculations.
The virus titer of 3 strains passaged in newborn ICR mouse brains was measured by IFA before performing mouse inoculation studies. Whereas WE(ngs) formed large plaques in Vero cells, strains OQ28 and BRC formed small plaques or foci of infection that were not visible without IFA staining. Therefore, viral titers were determined as TCID50 values by using IFA instead of on the basis of numbers of plaque-forming units. The viral titers of OQ28 and WE(ngs) grown in Vero cells were comparable (2 × 106.5 and 2 × 107.5 TCID50/mL, respectively), but BRC grew less well (2 × 104.3 TCID50/mL).
Clinical signs and mortality in mice.
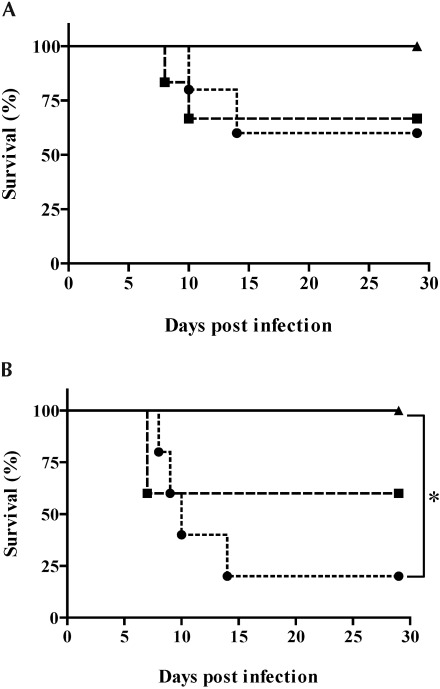
To investigate and compare the pathogenicity of the 3 LCMV strains, ICR mice were inoculated intraperitoneally with different virus doses of strains OQ28, BRC, and WE(ngs) (Figure 2). Among mice inoculated with 102 TCID50 of virus (Figure 2 A), those infected with strains OQ28 or WE(ngs) showed no clinical signs until 6 dpi, at which time they developed ruffled fur, half-closed eyes, a hunched posture, and lethargy. In addition, 33% to 40% of infected mice showing clinical signs died between 8 and 14 dpi. The remaining mice recovered gradually after 14 dpi and had no clinical signs at 29 dpi. In contrast, no mice inoculated with 102 TCID50 of strain BRC showed any clinical signs, and all survived through 29 dpi. Responses of mice infected with 104 TCID50 of virus (Figure 2 B) were similar to those of mice infected with 102 TCID50; those infected with strain BRC did not develop any clinical signs of infection, whereas 40% to 80% of mice infected with 104 TCID50 OQ28 and WE(ngs) died between 7 and 14 dpi. Survival analyses showed a statistically significant difference (P = 0.0133) between strains BRC and WE(ngs) in mice infected with 104 TCID50.
Figure 2.
Survival analysis of ICR mice inoculated with (A) 102 or (B) 104 TCID50 of LCMV strain OQ28 (squares), BRC (triangles), or WE(ngs) (circles). Survival was significantly (*, P = 0.0133, log-rank test) different between strains BRC and WE(ngs) in the 104 TCID50 inoculation group.
In additional experiments, BALB/c mice that were inoculated intraperitoneally with 104 TCID50 OQ28 developed clinical signs similar to those noted in ICR mice. Only one BALB/c mouse that developed clinical signs died (17%; at 12 dpi). All ICR mice inoculated intracerebrally with 104 TCID50 BRC developed clinical neurologic signs and died by 7 dpi.
Virus distribution in surviving mice.
All surviving ICR mice in the 102 and 104 TCID50 groups developed IgG antibody titers of 3200 to 25,600 after inoculation, suggesting that the animals experienced infection despite their survival. Differences in antibody titers did not correlate with either the dose or strain of virus. To investigate the tissue distribution of virus in ICR mice that survived infection, we used RT–PCR to detect viral sequences in blood, lung, liver, spleen, and kidneys (Table 4). Viral genomic sequences were detected in the organs or blood of 75% to 100% of the mice surviving infection with 102 or 104 TCID50 OQ28. In contrast, viral sequences were not detected in any of the organs or blood of any mice infected with BRC. In surviving mice that were infected with strain WE(ngs), viral sequences were detectable only in the kidneys of mice inoculated with 102 TCID50 and in the lung and kidneys of the sole mouse that survived infection with 104 TCID50. These results suggest that OQ28 causes a viremia in mice that persists for at least 29 dpi whereas BRC appears to be completely eliminated from the organs of inoculated mice by 29 dpi.
Table 4.
Antibody titers and virus distribution in survived mice
| No. that survived/ total no. |
RT-PCRb |
|||||||
| Strain | TCID50 | Antibody titera | Bloodc | Lung | Liver | Spleen | Kidneys | |
| OQ28 | 102 | 4/6 | 12,800–25,600 | 2/4 | 3/4 | 2/4 | 3/4 | 3/4 |
| 104 | 3/5 | 3200–12,800 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | |
| BRC | 102 | 6/6 | 3200–12,800 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| 104 | 5/5 | 12,800 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |
| WE(ngs) | 102 | 3/5 | 6400–12,800 | 0/3 | 0/3 | 0/3 | 0/3 | 2/3 |
| 104 | 1/5 | 12,800 | 0/1 | 1/1 | 0/1 | 0/1 | 1/1 | |
Groups (n = 5 or 6) of 3-wk-old female ICR mice were inoculated intraperitoneally with 102 or 10 TCID50 of strains OQ28, BRC, or WE(ngs).
Serum of surviving mice was collected at 29 dpi, and IgG antibody titers in serum were measured by indirect fluorescent antibody assay.
Virus distribution in mice surviving to 29 dpi was investigated by RT followed by nested PCR by using primers listed in Table 1.
Data are given as number of surviving mice that tested positive / total number of survivors tested.
In an additional experiment, blood, lung, liver, spleen, and kidneys were removed at 7 dpi from ICR mice that had been inoculated intraperitoneally with 104 TCID50 BRC. RT–PCR detected viral sequences in the liver, spleen, and kidneys.
Discussion
We have shown here that the LCMV strains OQ28 and BRC lie in different phylogenetic subclusters, according to their NP coding regions. In addition, inoculation of ICR mice with these 2 strains yielded contrasting results. ICR mice infected with strain OQ28 showed clinical signs and lethality, and virus could still be detected at 29 dpi in the blood, lung, liver, spleen, and kidneys of most surviving mice. In contrast, all ICR mice infected with strain BRC showed no clinical signs, and no virus was detected at 29 dpi in any organ. Furthermore, the clinical signs and lethality of ICR mice infected with strain WE(ngs), the passaged isolate of strain WE, were similar to those observed for strain OQ28. Selection of death as an endpoint allowed us to accurately compare lethality among mice infected with the various isolates and to investigate virus distribution among tissues at a very late stage of infection.
Other investigators have shown that LCMV strains can be divided into 3 large groups in light of phylogenetic analyses of the S RNA.1 Previous work has shown that the NP coding region is the most conserved region of arenaviruses.8 Here we have shown that the new LCMV strains OQ28, BRC, and WE(ngs) are closely related to strains Traub, M1, and WE, respectively. In addition, UBC is a high-passage variant of strain WE, and strains Aggressive and Docile are clones (variants) of UBC.4 Although strains UBC, Aggressive, and Docile are grouped with WE in subcluster I1 according to phylogenetic analyses of the NP coding region, they fell into a separate sister clade from strain WE; we obtained a similar result from analyses of the Z coding region (data not shown). However, UBC, Aggressive, Docile, and WE all fell into same clade when analyses were based on the GP or L coding regions (data not shown). Therefore, the increased number of amino acid substitutions in the NP and Z coding regions of strains WE and UBC erroneously cause them to be assigned to different clades.
Whereas subcluster I1 contains most of the LCMV strains isolated in the United States,1 subcluster I2 consists of European LCMV isolates, and cluster II comprises viruses isolated in Spain from wood mice (Apodemus sylvaticus).14 The classification of strains WE(ngs) and BRC into subclusters I1 and I2, respectively, is consistent with the geographic origins of the isolates. Strain OQ28 was isolated from a mouse of mixed breed (M. musculus molossinus, M. m. domesticus, and M. m. castaneus) in the port city of Osaka (Japan).19 Because M. m. domesticus and M. m. castaneus are not indigenous to Japan, strain OQ28 may have invaded from outside of Japan.19 Therefore, the classification of OQ28 into subcluster I1 is consistent with the possibility that this LCMV strain came from mice that originated in North America. Although LCMV is typically considered to be an Old World arenavirus, many strains are found in the New World. Furthermore, M. m. domesticus is found in Western Europe, Africa, and the United States. In addition, many LCMV strains that were isolated in Europe form a different subcluster (I2) from those isolated in North America. Whether the invasion of LCMV was simultaneous with the invasion of mice is unknown. However, our data raise the possibility that LCMV invaded North America in antiquity. In phylogenetic analyses of Hantavirus isolates, distinct clusters are formed by isolates obtained from different host species.33 Similarly, many LCMV strains of clusters I and II evaluated in the current study were isolated from the generea Mus and Apodemus, respectively.1,14 This finding suggests that the 2 major clusters of LCMV isolates may reflect the host species, as appears to be the case for Hantavirus.
The LCMV genome can rapidly accumulate mutations, as seen for clones 13 and 53b of strain Armstrong, which differ by 2 amino acid substitutions.18 These 2 closely related clones also exhibit differences in their pathogenicity.18 Therefore, that strains BRC and WE(ngs) exhibit different characteristics from their parental strains (M1 and WE, respectively) is unsurprising. Sequencing of the coding regions confirmed amino acid substitutions of 1% to 3% in the NP and GP coding regions between the strains originally isolated from mice and those that had been passaged (Table 3). In Theiler murine encephalomyelitis virus, the noncoding region is an important determinant of virulence.16 The noncoding regions of OQ28 had a large deletion (5′UTR of S RNA) and insertion (5′UTR of L RNA) relative to those of strains WE(ngs) and BRC. Similar additions and omissions of sequence occur in similar positions in other LCMV strains, including California, Traub, OH, Rhode Island, GR01, CABN, and SN05.1,14 However, whether these deletions or insertions in noncoding regions are related to the pathogenesis of LCMV in mice is unknown.
The LCMV strains Aggressive and Docile are clones (variants) of UBC and exhibit the same pathogenicity (lethality) in ICR mice.38 We therefore selected ICR mice for the infection experiments in the current study. However, BRC was detected at 7 dpi in the liver, spleen, and kidneys of ICR mice inoculated intraperitoneally but not at 29 dpi in ICR mice inoculated intraperitoneally. Despite similar passage, strain BRC propagated less well (that is, to lower titer) in newborn ICR mouse brains than did the other 2 LCMV strains. This finding together with the complete clearance of the virus in infected mice, suggests that strain BRC may have decreased affinity for organs of mice. Amino acid substitutions in the GP signal peptide coding region can lead to this effect.34 The GP signal peptide coding region comprises amino acids 1 through 58 at the N-terminus of the GP coding sequence, including an N-terminal region, hydrophobic region (h1 and h2 are separated by a lysine residue position at 33), and C-terminal region.34 Several single amino acid substitutions in the GP signal peptide coding region influence virus infectivity and cleavage of viral glycoproteins (GP1 and GP2).32,34 However, because BRC, OQ28, and WE(ngs) did not have any substitutions at previously identified as being influential (amino acids 2, 4, 12, 16, 17, 20, 22, 29, 33, 37, 46, 49, and 54), 32,34 we searched for commonality among the GP signal peptide sequences of virus strains of subcluster I2; we found common amino acid substitutions at amino acid 5 in the h1 region and residue 50 in the h2 region. Whereas amino acids 5 and 50 of virus strains in subcluster I1 and cluster II were valine and leucine, those of subcluster I2 were isoleucine and cysteine, respectively. Because the h1 and h2 regions are associated with cell infectivity and cleavage of GP1 and GP2, respectively, amino acids 5 and 50 in the GP signal peptide may be responsible for the low infectivity and propagation of the strain BRC.34 Although the mortality for mice of LCMV strains in subcluster I2 is unknown, strains Dandenong and LE led to mortality in immunodeficient individuals and a fetus, respectively.13,22 Therefore, the hypotheses that amino acids 5 and 50 in the GP signal peptide coding region are determinants of pathogenicity and propagation should be considered.
LCMV is a noncytolytic virus, and clinical signs and lethal disease due to LCMV infection generally are associated with immunopathology rather than direct tissue damage by the virus.12 This outcome results from the attack by virus-specific T cells of infected cells in critical organs in mice.12 Although both ICR and BALB/c mice infected with strain OQ28 showed clinical signs and mortality, intracerebral inoculation of BALB/c mice with same virus strain has been described as nonlethal.19 In addition, although ICR mice infected intraperitoneally with strain BRC in the current study showed no clinical signs or mortality, 4-wk-old ICR mice infected intracerebrally showed neurologic deficits (clonic convulsions) and died. These results and those of other investigators15 indicate that LCMV strains can display markedly different pathogenicity depending on the route of inoculation, virus dose, and mouse strain.
Because all mice that died of infection with strains OQ28 and WE(ngs) had detectable levels of viral RNA in their blood at the time of death (not shown), we felt it likely that all organs would also be positive for virus and therefore did not test them directly. Mice surviving infection with strains OQ28 and WE(ngs) had virus in organs or blood or both, indicating that despite surviving the acute infection, these mice were not able to eliminate the virus. Strain Armstrong 53b and clone 13 (which is derived from Armstrong) cause acute compared with chronic infection, respectively, in mice.29 Genetic analysis of these 2 clones revealed amino acid substitutions at position 260 of the GP coding region and position 1079 of the L coding region, suggesting that these residues are associated with acute or chronic infection and the cytotoxic T cell response.18,29 Furthermore, mice infected with strain Armstrong clone 13 continue to express programmed death 1 (PD1), which is involved in the negative regulation of immune responses, in CD8+ cells, and dysfunction or immune exhaustion of antigen-specific T cells is thought to be involved in chronic infection.3,6,21,37 Like strains OQ28 and WE(ngs), Armstrong clone 13 persists in the blood and organs until 30 dpi.6 The 2 amino acid positions associated with chronic infection in strain Armstrong (GP260 and L1079) were maintained in strains OQ28, BRC, and WE(ngs), indicating that more than just these 2 amino acids must be responsible for the differences in pathogenicity among these 3 LCMV strains. In addition, persistence of the virus was detected only in mice infected with strains OQ28 and WE(ngs). These results suggest that chronic infection may not be specific to these 2 amino acids but may also involve other factors, as suggested by other investigators.18
The current study determined the phylogenetic relationship between OQ28 and BRC, strains newly isolated in Japan, and other LCMV strains and characterized their pathogenicities in ICR mice. However, because mouse strain, virus strain, and route of inoculation all influence the clinical signs, lethality, and virus distribution in mice infected with various strains of LCMV, infection of mouse strains other than ICR and BALB/c with OQ28 and BRC may show different characteristics.4 The pathogenic characteristics of strains OQ28 and BRC should be further investigated through experimental infection of other mouse strains.
Acknowledgments
We thank R Eberle (Oklahoma State University Center for Veterinary Health Sciences) and H Yamanaka (Nagasaki University) for their critical comments and assistance with this manuscript. We also acknowledge the invaluable support of all our laboratory members. This work was supported by a Grant-in-Aid for Scientific Research (B) (grant 15300143, 19300148) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (HS and KO).
References
- 1.Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, Hui J, Briese T, St George K, Ksiazek TG, Lipkin WI, Nichol ST. 2010. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerg Infect Dis 16:1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amman BR, Pavlin BI, Albariño CG, Comer JA, Erickson BR, Oliver JB, Sealy TK, Vincent MJ, Nichol ST, Paddock CD, Tumpey AJ, Wagoner KD, Glauer RD, Smith KA, Winpisinger KA, Parsely MS, Wyrick P, Hannafin CH, Bandy U, Zaki S, Rollin PE, Ksiazek TG. 2007. Pet rodents and fatal lymphocytic choriomeningitis in transplant patients. Emerg Infect Dis 13:719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439:682–687 [DOI] [PubMed] [Google Scholar]
- 4.Barthold SW, Smith AL. 2007. Lymphocytic choriomeningitis virus, 179–213. In: Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, Smith AL. The mouse in biomedical research, 2nd ed, vol 2. Waltham (MA): Academic Press. [Google Scholar]
- 5.Becker SD, Bennett M, Stewart JP, Hurst JL. 2007. Serological survey of virus infection among wild house mice (Mus domesticus) in the UK. Lab Anim 41:229–238 [DOI] [PubMed] [Google Scholar]
- 6.Blackburn SD, Crawford A, Shin H, Polley A, Freeman GJ, Wherry EJ. 2010. Tissue-specific differences in PD1 and PD-L1 expression during chronic viral infection: implications for CD8 T-cell exhaustion. J Virol 84:2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buesa-Gomez J, Teng MN, Oldstone CE, Oldstone MB, de la Torre JC. 1996. Variants able to cause growth hormone deficiency syndrome are present within the disease-nil WE strain of lymphocytic choriomeningitis virus. J Virol 70:8988–8992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui HH, Botten J, Fusseder N, Pasquetto V, Mothe B, Buchmeier MJ, Sette A. 2007. Protein sequence database for pathogenic arenaviruses. Immunome Res 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djavani M, Lukashevich IS, Salvato MS. 1998. Sequence comparison of the large genomic RNA segments of 2 strains of lymphocytic choriomeningitis virus differing in pathogenic potential for guinea pigs. Virus Genes 17:151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emonet S, Lemasson JJ, Gonzalez JP, de Lamballerie X, Charrel RN. 2006. Phylogeny and evolution of Old World arenaviruses. Virology 350:251–257 [DOI] [PubMed] [Google Scholar]
- 11.Ike F, Bourgade F, Ohsawa K, Sato H, Morikawa S, Saijo M, Kurane I, Takimoto K, Yamada YK, Jaubert J, Berard M, Nakata H, Hiraiwa N, Mekada K, Takakura A, Itoh T, Obata Y, Yoshiki A, Montagutelli X. 2007. Lymphocytic choriomeningitis infection undetected by dirty-bedding sentinel monitoring and revealed after embryo transfer of an inbred strain derived from wild mice. Comp Med 57:272–281 [PubMed] [Google Scholar]
- 12.Jacoby RO, Fox JG, Davisson M. 2002. Biology and diseases of mice, lymphocytic choriomeningitis virus (LCMV) infection, 66–69. In: Fox JG, Andersen LC, Loew FM, Quimby FW. Laboratory animal medicine, 2nd ed. Waltham (MA): Academic Press. [Google Scholar]
- 13.Meritet JF, Krivine A, Lewin F, Poissonnier MH, Poizat R, Loget P, Rozenberg F, Lebon P. 2009. A case of congenital lymphocytic choriomeningitis virus (LCMV) infection revealed by hydrops fetalis. Prenat Diagn 29:626–627 [DOI] [PubMed] [Google Scholar]
- 14.Ledesma J, Fedele CG, Carro F, Lledó L, Sánchez-Seco MP, Tenorio A, Soriguer RC, Saz JV, Domínguez G, Rosas MF, Barandika JF, Gegúndez MI. 2009. Independent lineage of lymphocytic choriomeningitis virus in wood mice (Apodemus sylvaticus), Spain. Emerg Infect Dis 15:1677–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leist T, Althage A, Haenseler E, Hengartner H, Zinkernagel RM. 1989. Major histocompatibility complex-linked susceptibility or resistance to disease caused by a noncytopathic virus varies with the disease parameter evaluated. J Exp Med 170:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipton HL, Calenoff M, Bandyopadhyay P, Miller SD, Dal Canto MC, Gerety S, Jensen K. 1991. The 5′ noncoding sequences from a less-virulent Theiler's virus dramatically attenuate GDVII neurovirulence. J Virol 65:4370–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lledó L, Gegúndez MI, Saz JV, Bahamontes N, Beltrán M. 2003. Lymphocytic choriomeningitis virus infection in a province of Spain: analysis of sera from the general population and wild rodents. J Med Virol 70:273–275 [DOI] [PubMed] [Google Scholar]
- 18.Matloubian M, Kolhekar SR, Somasundaram T, Ahmed R. 1993. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol 67:7340–7349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita C, Matsuura Y, Fujii H, Joh K, Baba K, Kato M, Hisada M. 1991. Isolation of lymphocytic choriomeningitis virus from wild house mice (Mus musculus) in Osaka Port, Japan. J Vet Med Sci 53:889–892 [DOI] [PubMed] [Google Scholar]
- 20.Morita C, Matsuura Y, Kawashima E, Takahashi S, Kawaguchi J, Iida S, Yamanaka T, Jitsukawa W. 1991. Seroepidemiological survey of lymphocytic choriomeningitis virus in wild house mouse (Mus musculus) in Yokohama Port, Japan. J Vet Med Sci 53:219–222 [DOI] [PubMed] [Google Scholar]
- 21.Ou R, Zhang M, Huang L, Moskophidis D. 2008. Control of virus-specific CD8+ T-cell exhaustion and immune-mediated pathology by E3 ubiquitin ligase Cbl-b during chronic viral infection. J Virol 82:3353–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358:991–998 [DOI] [PubMed] [Google Scholar]
- 23.Puissant C, Houdebine LM. 1990. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate– phenol–chloroform extraction. Biotechniques 8:148–149 [PubMed] [Google Scholar]
- 24.Reed LJ, Muench H. 1938. A simple method of estimating 50% endpoints. Am J Hyg 27:493–497 [Google Scholar]
- 25.Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Dutko FJ, Oldstone MB. 1985. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J Virol 53:966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanowski V, Matsuura Y, Bishop DH. 1985. Complete sequence of the S RNA of lymphocytic choriomeningitis virus (WE strain) compared to that of Pichinde arenavirus. Virus Res 3:101–114 [DOI] [PubMed] [Google Scholar]
- 27.Rousseau MC, Saron MF, Brouqui P, Bourgeade A. 1997. Lymphocytic choriomeningitis virus in southern France: 4 case reports and a review of the literature. Eur J Epidemiol 13:817–823 [DOI] [PubMed] [Google Scholar]
- 28.Sato H, Miyata H. 1986. Detection of lymphocytic choriomeningitis virus antibody in colonies of laboratory animals in Japan. Jikken Dobutsu 35:189–192 [DOI] [PubMed] [Google Scholar]
- 29.Salvato M, Borrow P, Shimomaye E, Oldstone MB. 1991. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J Virol 65:1863–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salvato MS, Shimomaye EM. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1–10 [DOI] [PubMed] [Google Scholar]
- 31.Salvato MS, Shimomaye E, Southern P, Oldstone MB. 1988. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, clone 13 (CTL–). Virology 164:517–522 [DOI] [PubMed] [Google Scholar]
- 32.Saunders AA, Ting JP, Meisner J, Neuman BW, Perez M, de la Torre JC, Buchmeier MJ. 2007. Mapping the landscape of the lymphocytic choriomeningitis virus stable signal peptide reveals novel functional domains. J Virol 81:5649–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmaljohn C, Hjelle B. 1997. Hantaviruses: a global disease problem. Emerg Infect Dis 3:95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrempf S, Froeschke M, Giroglou T, von Laer D, Dobberstein B. 2007. Signal peptide requirements for lymphocytic choriomeningitis virus glycoprotein C maturation and virus infectivity. J Virol 81:12515–12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagliapietra V, Rosà R, Hauffe HC, Laakkonen J, Voutilainen L, Vapalahti O, Vaheri A, Henttonen H, Rizzoli A. 2009. Spatial and temporal dynamics of lymphocytic choriomeningitis virus in wild rodents, northern Italy. Emerg Infect Dis 15:1019–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 37.Williams MA, Bevan MJ. 2006. Immunology: exhausted T cells perk up. Nature 439:669–670 [DOI] [PubMed] [Google Scholar]
- 38.Zinkernagel RM, Leist T, Hengartner H, Althage A. 1985. Susceptibility to lymphocytic choriomeningitis virus isolates correlates directly with early and high cytotoxic T cell activity, as well as with footpad swelling reaction, and all 3 are regulated by H2D. J Exp Med 162:2125–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]