Abstract
EMBO J 31 10, 2296–2308 (2012); published online April 20 2012
Meiosis is one of the most dramatic differentiation programmes a cell can undertake, since it leads to an irreversible reduction of the cell’s genetic content. It therefore comes as no surprise that meiosis should be tightly regulated. In this issue Hiriart et al (2012) identify a new layer of regulation in which the RNA interference (RNAi) pathway dampens the expression of meiotic genes during vegetative growth in fission yeast. Their study converges with a recent publication by Zofall et al (2012) to reveal how components of the RNAi pathway, the Mmi1 RNA elimination system and chromatin modifications contribute to the maintenance of the vegetative state until a decision is made to enter meiosis.
Cells express different parts of their genome depending on their function in an organism. Their expression programs can be altered according to intrinsic cues or in response to changes in the surroundings. When starved for nitrogen, fission yeast cells prepare for meiosis. Hundreds of genes are induced in successive waves (Mata et al, 2002), driving sexual differentiation and a cascade of events that culminate in the production of four haploid spores. The Ste11 and Mei4 transcription factors are master regulators in this programme. The wide changes in transcript abundance in ste11 and mei4 mutants underscore the importance of transcriptional regulation in meiosis. However, a remarkable discovery in the last few years has been that a number of meiotic genes, including mei4, are actually transcribed in vegetative yeast cells. Their transcripts though are unstable. How RNA-processing factors, the nuclear exosome and possibly other RNA degradation pathways control the expression of meiotic genes is currently the subject of intense study.
One major source of meiotic transcript instability in vegetative yeast comes from the Mmi1 RNA elimination pathway (Harigaya et al, 2006; Figure 1). In this pathway the RNA-binding protein Mmi1 recognises DSR (determinant of selective removal) regions in a class of meiotic transcripts and orchestrates the recruitment of multiple factors including Red1, Pab2 and Rrp6 to target these transcripts for degradation (St-Andre et al, 2010; Yamanaka et al, 2010; Sugiyama and Sugioka-Sugiyama, 2011). At the onset of meiosis Mmi1 is sequestered into the Mei2 dot, a meiosis-specific subnuclear structure comprising the Mei2 protein and the non-coding meiRNA (Watanabe and Yamamoto, 1994). The resulting stabilization of DSR-containing transcripts is essential for meiotic progression. A large-scale expression array in the Hiriart et al (2012) work extends the number of known Mmi1 protein-coding targets in vegetative cells to 212, underscoring the importance of the system.
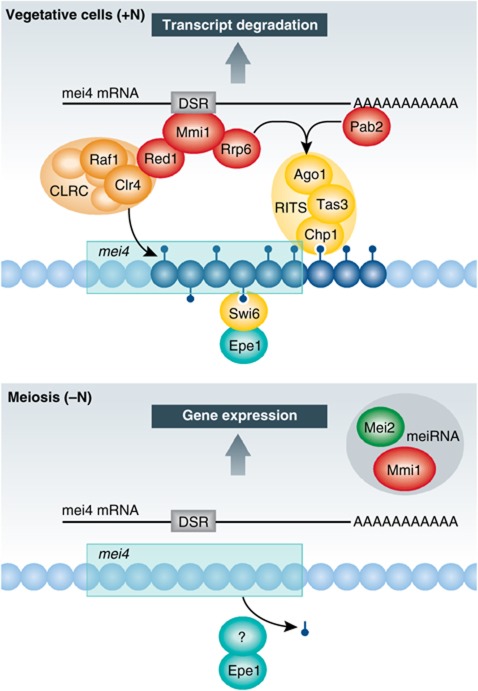
Figure 1.
Recruitment of RNAi and heterochromatin components to Mmi1 targets. During vegetative growth the Mmi1 protein targets DSR-containing transcripts for degradation. Mmi1 recruits the RNAi complex RITS and the Clr4-methyltransferase complex CLRC to participate in the repression of meiotic genes, as depicted in the case of mei4. During meiotic induction Mmi1 is sequestered in the Mei2 dot, causing dissociation of the complexes and expression of DSR-containing meiotic genes. The anti-silencing protein Epe1 helps remove H3K9me.
An essential contribution of Hiriart et al (2012) is to establish a functional connection between Mmi1-mediated RNA elimination and another repressive pathway, RNAi. RNAi in S. pombe has been dissected in great detail in connection with its role in the formation of constitutive heterochromatin at centromeres and telomeres, and in the mating-type region. Central to the pathway at these locations are the ribonuclease Dicer and the RITS complex consisting of Ago1, Tas3, and Chp1 (reviewed by Creamer and Partridge, 2011). Dicer processes double-stranded RNA molecules transcribed from heterochromatic regions into 21–23-nucleotide siRNAs. siRNAs loaded onto Ago1 are believed to guide RITS to heterochromatic transcripts through base-pairing. One of the RITS-interacting complexes, CLRC, contains the Clr4 methyltransferase. By methylating histone H3 at lysine 9 (H3K9me), Clr4 creates a binding site for the chromodomain protein Chp1, thereby providing an alternate path of association for RITS to chromatin. The combined action of RNAi and histone modifications silences gene expression in constitutive heterochromatin both transcriptionally and post-transcriptionally.
Here, Hiriart et al (2012) find that in addition to being present in constitutive heterochromatin, RITS is also associated with Mmi1 targets in vegetative cells. The association of RITS with meiotic genes is documented for the three subunits of RITS at mei4 and for Chp1 at numerous other targets. The physical association of RITS with meiotic genes suggests that RITS participates in the repression of these genes. Consistently, the mei4 and ssm4 steady-state transcript levels double in mutants lacking a RITS subunit and in dcr1Δ mutants. Furthermore, the association of RITS with Mmi1 targets is lost when cells are induced to undergo sexual differentiation by nitrogen starvation, coinciding with the sequestration of Mmi1 in the Mei2 dot.
These findings extend the work recently published by Zofall et al (2012) on facultative heterochromatic islands. H3K9me was clearly detected at approximately 30 chromosomal locations in the study by Zofall et al (2012) and possibly in a latent state at a far greater number of locations. According to their diverse dependency on trans-acting factors, H3K9me islands do not represent one homogenous group corresponding to a single biological function. One subset corresponds to the Mmi1-regulated loci characterized here by Hiriart et al (2012). Nitrogen starvation reduces H3K9me at these islands, which include mei4 and ssm4, as it reduces their association with RITS. A picture emerges where the combined action of RNAi and heterochromatin regulates Mmi1 targets.
The mechanism of association of RITS with Mmi1 targets differs quite strikingly from its association with constitutive heterochromatin, permitting and perhaps facilitating the dynamic changes that occur during differentiation. At centromeres the association of RITS depends very strongly on Dcr1 and Clr4, but at meiotic genes it rather relies on the Mmi1 protein and its co-factors, Rrp6, Red1 and Pab2 (Figure 1). Red1 facilitates the association of RITS with chromatin—perhaps through direct binding of Red1 and Clr4 and resulting H3K9me as suggested by Zofall et al (2012)—while Pab2 appears to facilitate association through RNA. A pending question is the extent to which siRNAs from meiotic transcripts contribute to the association of RITS with meiotic genes and the biogenesis of these siRNAs.
How does RITS help silence its meiotic targets? H3K9me, the histone deacetylase Clr3 and Mit1 are detected at these targets (Hiriart et al, 2012; Zofall et al, 2012), suggesting that RITS might bring the same mechanisms of silencing to meiotic genes as to constitutive heterochromatin. Consistent with this scenario, a previous study found increased expression of meiotic genes in histone deacetylase and clr4 mutants (Hansen et al, 2005). The amplitude of the repression by RITS and heterochromatin is notably small compared to the changes occurring in a meiotic time course (Mata et al, 2002; Hansen et al, 2005) and unlike Hiriart et al (2012), Zofall et al (2012) did not detect increased mei4 or ssm4 transcript in ago1Δ cells. However, Hiriart et al (2012) provide genetic evidence that the contribution of RNAi to the repression of meiosis is physiologically important, by showing that deletion of chp1 or dcr1 partially suppresses the meiotic block in sme2Δ cells defective in producing meiRNA. Hiriart et al (2012) also identify meiRNA as one of the most sensitive Mmi1 targets, which reveals that the transition between vegetative state and meiosis is regulated through a double-negative loop where Mmi1 and meiRNA inhibit each other. RNAi and heterochromatin stabilize the Mmi1-dependent state in this bistable system, their repressive effects adding up with several other mechanisms (i.e. Cremona et al, 2011) for a tight and robust repression of meiotic genes in vegetative cells.
Footnotes
The authors declare that they have no conflict of interest.
References
- Creamer KM, Partridge JF (2011) RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip Rev RNA 2: 632–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona N, Potter K, Wise JA (2011) A meiotic gene regulatory cascade driven by alternative fates for newly synthesized transcripts. Mol Biol Cell 22: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bähler J, Thon G (2005) Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 25: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat-Todescini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, Lachuer J, Perazza D, Yamamoto M, Verdel A (2012) Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J 31: 2296–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147 [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F (2010) Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem 285: 27859–27868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R (2011) Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30: 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M (1994) S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78: 487–498 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M (2010) Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29: 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]