Abstract
Forkhead box-containing protein o (Foxo) 1 is a key transcription factor in insulin and glucose metabolism. We identified a Foxo1-CoRepressor (FCoR) protein in mouse adipose tissue that inhibits Foxo1’s activity by enhancing acetylation via impairment of the interaction between Foxo1 and the deacetylase Sirt1 and via direct acetylation. FCoR is phosphorylated at Threonine 93 by catalytic subunit of protein kinase A and is translocated into nucleus, making it possible to bind to Foxo1 in both cytosol and nucleus. Knockdown of FCoR in 3T3-F442A cells enhanced expression of Foxo target and inhibited adipocyte differentiation. Overexpression of FCoR in white adipose tissue decreased expression of Foxo-target genes and adipocyte size and increased insulin sensitivity in Leprdb/db mice and in mice fed a high-fat diet. In contrast, Fcor knockout mice were lean, glucose intolerant, and had decreased insulin sensitivity that was accompanied by increased expression levels of Foxo-target genes and enlarged adipocytes. Taken together, these data suggest that FCoR is a novel repressor that regulates insulin sensitivity and energy metabolism in adipose tissue by acting to fine-tune Foxo1 activity.
Keywords: acetylation, Foxo1, FCoR, Sirt1
Introduction
Forkhead transcription factor (Foxo1) is a key transcription factor in insulin and glucose metabolism that is phosphorylated, subsequently exported to the cytoplasm, and inhibited by insulin/IGF1 in a PI3 kinase-dependent manner (Accili and Arden, 2004). Foxo1 plays an important role in mediating insulin action in several insulin-responsive tissues. Specifically, Foxo1 promotes glucose production in the liver, inhibits compensatory β-cell proliferation in insulin-resistant states, activates feeding by promoting orexigenic peptide expression in the hypothalamic arcuate nucleus, inhibits differentiation of preadipocytes and myoblasts into mature adipocytes or myotubes, and regulates energy storage and expenditure in adipose tissue (Nakae et al, 2008a, 2008b). Because of its involvement in so many physiological processes, it is important to elucidate the mechanism that regulates Foxo1 transcriptional activity.
Adipogenesis, during which preadipocytes differentiate into adipocytes, experiences several stages, including mesenchymal precursor, committed preadipocytes, growth-arrested preadipocyte, mitotic clonal expansion, terminal differentiation, and mature adipocyte (Lefterova and Lazar, 2009). Peroxisome proliferator-activated receptor-γ (PPARγ) regulates both the terminal differentiation and metabolism in mature adipocytes. Foxo1 is a PPARγ-interacting protein that antagonizes PPARγ activity (Dowell et al, 2003; Armoni et al, 2006; Fan et al, 2009). SirT2-mediated deacetylation of Foxo1 increases the association of Foxo1 with PPARγ, leading to inhibition of adipocyte differentiation (Jing et al, 2007). Furthermore, constitutively nuclear (CN) Foxo1 inhibits differentiation of the preadipocyte cell line 3T3-F442A cells by arresting the cell cycle that is required in the early stages of adipose conversion, whereas haploinsufficiency of Foxo1 restores the size of white adipocytes under high-fat diet (HFD) (Nakae et al, 2003; Kim et al, 2009). Overexpression of transactivation-defective Foxo1 in white adipose tissue (WAT) increases fat mass and number of small adipocytes. Overexpression of the same mutant Foxo1 in brown adipose tissue (BAT) increases oxygen consumption (Nakae et al, 2008a). Therefore, Foxo1 can be an attractive target to improve the energy homeostasis in adipose tissue. Targeting Foxo1 can increase energy store in WAT and increase energy expenditure in BAT.
Foxo1 is primarily regulated by post-translational modifications. In addition to Akt-induced phosphorylation, Foxo1 is acetylated by histone acetyltransferases (HATs) such as CREB-binding protein (CBP)/P300; Foxo1 deacetylation is mediated by class I–III deacetylases, including the nicotin amide adenine dinucleotide (NAD)-dependent histone deacetylase (HDAC), Sirt1 (Accili and Arden, 2004). Interaction with other proteins also regulates the transcriptional activity of Foxo family proteins (Foxos). For example, PPAR-γ co-activator 1α (PGC-1α) interacts with Foxo1 and stimulates gluconeogenesis in the liver (Puigserver et al, 2003). SMK-1 modulates the transcriptional response of DAF-16, the Foxo1 orthologue in C. elegans (Wolff et al, 2006), and the Drosophila Melted gene product interacts with both Tsc1 and FOXO to inhibit FOXO activity (Teleman et al, 2005).
In the present study, we identified a novel Foxo1-binding protein termed as Foxo1 CoRepressor (FCoR) in adipose tissue using a yeast two-hybrid screen of a mouse 3T3-L1 cDNA library. We demonstrated that FCoR inhibits Foxo1 transcriptional activity through increased Foxo1 acetylation, which is accompanied by preventing Foxo1 interaction with the deacetylase Sirt1 and by direct acetylation. Knockdown of FCoR in 3T3-F442A cells inhibited adipocyte differentiation, while knockout of Fcor led to a lean phenotype, glucose intolerance, and insulin resistance. In contrast, overexpression of FCoR in adipose tissue decreased adipocyte size, increased insulin sensitivity, and decreased PGC-1α expression in brown adipocytes, indicating that FCoR plays important roles in glucose and energy homeostasis.
Results
Identification of FCoR, a novel Foxo1-binding protein
To identify Foxo1-interacting proteins, we performed a yeast two-hybrid screen, using a GAL4-Foxo1 fragment (amino acids 1–154) as bait and a mouse 3T3-L1 cDNA library as prey. Screening of about 1.5 × 106 primary transformants yielded 224 clones. We selected 17 clones that met the following criteria: (1) they possessed a nuclear localization signal, (2) they encoded transcription factors, or (3) unknown proteins, and (4) they were restricted to or enriched in adipose tissue and/or differentiated 3T3-F442A cells. In all, 5 of 17 clones thus identified encoded partial transcripts of the RIKEN cDNA 2400009B08. The gene is predicted to encode a peptide with an Mr of 13.71 kDa LOC68234 (gb∣EDL21945.1∣mCG1048501). Because further characterization showed that it acted as a Foxo1 CoRepressor, it was termed as ‘FCoR’.
We performed 5′- or 3′-rapid amplification of cDNA ends (RACE) to determine the transcription start site (Supplementary Figure S1A). Sequencing of the 5′- and 3′-RACE products determined that FCoR is a 106-amino acid protein (Supplementary Figure S1B). cDNA cloning and in-vitro translation studies confirmed that the peptide has a molecular weight of 13 kDa (Figure 1A). Functional domain analysis using the Eukaryotic Linear Motif server ( http://elm.eu.org/; Teleman et al, 2005) revealed the presence of a forkhead-associated ligand domain (LIG_FHA_1) from amino acids 78 to 84 (Supplementary Figure S1B).
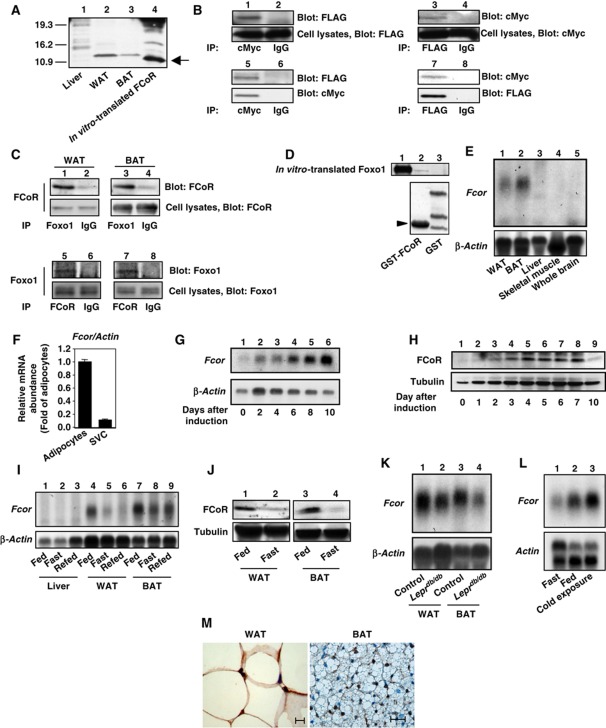
Figure 1.
Interaction between FCoR and Foxos and expression profiling of FCoR. (A) In-vitro translation of FCoR. Lysates from liver (lane 1), WAT (lane 2), and BAT (lane 3) from wild-type mice, along with in vitro-translated FCoR were analysed by western blotting using anti-mouse FCoR antiserum. (B) Interaction between exogenous FCoR and Foxo1. HEK293 cells were co-transfected with pFLAG-CMV2-WT Foxo1 or pFLAG-CMV2-Δ256 Foxo1 plus pCMV5-cMyc-WT FCoR and cultured in the presence of serum. At 48 h after transfection, cells were harvested and lysates were immunoprecipitated with anti-cMyc (lanes 1 and 5), anti-FLAG antibody (lanes 3 and 7), or normal mouse IgG (lanes 2, 4, 6, and 8) and blotted with anti-FLAG (lanes 1, 2, 5, and 6) or anti-cMyc antibody (lanes 3, 4, 7, and 8). (C) Interaction between endogenous Foxo1 and FCoR in WAT and BAT. Lysates from WAT (lanes 1, 2, 5, and 6) and BAT (lanes 3, 4, 7, and 8) were immunoprecipitated with anti-FOXO1 (lanes 1 and 3), anti-FCoR (lanes 5 and 6), or normal rabbit IgG (lanes 2, 4, 6, and 8) and blotted with anti-FCoR (lanes 1–4) or anti-FOXO1 (lanes 5–8), respectively. (D) Direct interaction of FCoR with Foxo1. GST-FCoR was subjected to a pull-down assay. Aliquots of in vitro-translated WT Foxo1 were incubated with glutathione-Sepharose beads coated with bacterially expressed GST-P13 (lane 2) or GST alone (lane 3) for 6 h at 4°C. The in vitro-translated Foxo1 proteins retained on the column were eluted and separated by SDS–PAGE followed by western blotting with anti-FOXO1 antibody. The bottom panel shows GST or GST-GST-P13 (10% of input, lane 3) blotted with anti-GST antibody. (E) Expression profiling of Fcor in various tissues. Total RNA isolated from WAT (lane 1), BAT (lane 2), liver (lane 3), skeletal muscle (lane 4), and whole brain (lane 5) of wild-type mice was subjected to northern blotting with Fcor (top panel) or β-actin (bottom panel). (F) Real-time PCR of Fcor using the adipocyte or stromal vascular fractions of fractionated WAT. (G) Northern blotting of 3T3-F442A cells during differentiation. Total RNA isolated from 3T3-F442A cells on the indicated day after induction of differentiation was subjected to Northern blotting with Fcor (top panel) or β-actin (bottom panel). (H) Western blotting of FCoR protein from 3T3-F442A cells during differentiation. Lysates from 3T3-F442A cells on the indicated day after induction of differentiation were subjected to western blotting using anti-FCoR polyclonal antiserum (top panel) or anti-tubulin monoclonal antibody (bottom panel). (I) Effects of the feeding state on Fcor gene expression. Total RNA from liver (lanes 1–3), WAT (lanes 4–6), and BAT (lanes 7–9) from C57Bl6J mice in the fed, fasting, or refed states was subjected to northern blotting with Fcor (top panel) or β-actin (bottom panel). (J) Western blotting of the FCoR protein from WAT (lanes 1 and 2) and BAT (lanes 3 and 4) from C57Bl6J mice in fed (lanes 1 and 3) or fasting state (lanes 2 and 4). Tissue lysates were subjected to western blotting using anti-FCoR polyclonal antiserum (top panel) or anti-tubulin monoclonal antibody (bottom panel). (K) Fcor gene expression in WAT and BAT from Leprdb/db mice. Total RNA isolated from WAT (lanes1 and 2) and BAT (lanes 3 and 4) of C57Bl6J (lanes 1 and 3) or Leprdb/db mice (lanes 2 and 4) was subjected to northern blotting with Fcor (top panel) or β-actin (bottom panel). (L) Effect of cold exposure on Fcor gene expression in BAT. Total RNA isolated from the BAT of C57Bl6J mice exposed to the cold (4°C for 6 h) was subjected to northern blotting with Fcor (top panel) or β-actin (bottom panel). (M) Immunohistochemistry of WAT (left panel) and BAT (right panel) from C57Bl6J mice using anti-FCoR anti-sera. Scale bars indicate 20 μm.
To confirm the interaction between Foxo1 and FCoR, we co-transfected HEK293 cells with FLAG-tagged Foxo1 and cMyc-tagged FCoR and performed reciprocal immunoprecipitation/immunoblotting experiments in the presence of serum using anti-FLAG and anti-cMyc antibodies. These experiments showed that FCoR interacted with Foxo1 (Figure 1B, lanes 1–4). To identify the FCoR-binding site of Foxo1, we co-transfected pFLAG-CMV2-Δ256 Foxo1, which encoded a Foxo1 mutant lacking the carboxyl terminal transactivation domain (Nakae et al, 2000), and pCMV5-cMyc-FCoR and performed immunoprecipitation studies. These experiments demonstrated that FLAG-Δ256 Foxo1 bound to cMyc-FCoR (Figure 1B, lanes 5–8), suggesting that FCoR binds the Foxo1 N-terminus.
Foxo family members, including Foxo1, Foxo3a, and Foxo4, are expressed in WAT and/or BAT. Foxo1 and Foxo3a can be expressed in both WAT and BAT but Foxo4 is mainly expressed in BAT (Supplementary Figure S2). To investigate whether endogenous Foxo1 associates with FCoR, mouse WAT and BAT extracts were immunoprecipitated with an anti-FOXO1 antibody or anti-FCoR antiserum, followed by immunoblotting with antibodies against FCoR or Foxo1. The results showed that endogenous Foxo1 associated with endogenous FCoR (Figure 1C). Taken together, these results suggest that FCoR interacts with Foxo1 in vivo. To determine whether the interaction between FCoR and Foxo1 was a direct interaction, we performed a GST-fusion protein pull-down assay. We detected an interaction between FCoR and Foxo1 (Figure 1D), indicating that the two proteins bind directly to one another.
FCoR is expressed in adipose tissue and differentiated 3T3-F442A cells
Fcor mRNA is expressed in mouse WAT and BAT, but not in liver, skeletal muscle, or brain (Figure 1E). Fractionation of WAT revealed that Fcor is expressed mainly in the adipocyte fraction (Figure 1F). Analysis of Fcor mRNA and protein levels indicated that FCoR is expressed in 3T3-F442A cells in a differentiation-dependent manner (Figure 1G and H).
To investigate whether FCoR expression was modulated in physiological conditions or insulin-resistant states, we examined Fcor mRNA regulation during fasting and feeding in C57bl6J mice. Fcor mRNA and FCoR protein levels in WAT and BAT decreased during fasting (Figure 1I and J), and Fcor mRNA expression levels were lower in the WAT and BAT of insulin-resistant Leprdb/db mice compared with control mice (Figure 1K). BAT is the main organ responsible for adaptive thermogenesis in rodents (Cannon et al, 1998). Interestingly, mouse BAT Fcor mRNA expression increased after 6 h of cold exposure (Figure 1L). Immunohistochemistry experiments demonstrated that the endogenous FCoR protein was expressed mainly in the nucleus in both WAT and BAT (Figure 1M). These data suggest that FCoR may have a functional role in adipose tissues in vivo.
FCoR inhibits the transcriptional activity of Foxo1 and Foxo3a, but not of FOXO4
We next addressed the question of whether FCoR affects Foxo1 transcriptional activity using reporter assays with the Foxo1-target genes Igfbp1 and G6pc (Nakae et al, 2006). A constitutively nuclear Foxo1 mutant (CN Foxo1) increased IGFBP1 or G6PC promoter activity by 3.5- and 22.5-fold, respectively, in the presence of 8-Br-cAMP/IBMX/dexamethasone (Figure 2A and B). FCoR had no effect on the activity of either promoter in basal conditions, but it inhibited the Foxo1-dependent promoter activity of both genes in a dose-dependent manner (Figure 2A and B). In the transactivation assay, FCoR repressed the forskolin-induced luciferase activity of a GAL4-Foxo1 fusion protein (Figure 2C). Furthermore, co-expression of FCoR inhibited the effect of CN Foxo1 on the expression of the endogenous Igfbp1 gene in SV40-transformed hepatocytes (Figure 2D). In these cells, FCoR localized to both the cytosol and nucleus (Supplementary Figure S3). These data indicate that FCoR and CN Foxo1 colocalize in the nucleus and that FCoR inhibits Foxo-dependent transcription.
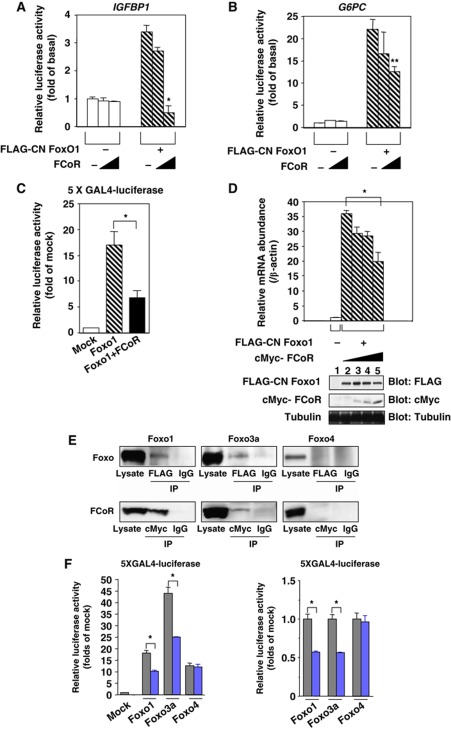
Figure 2.
FCoR inhibits Foxo1 transcriptional activity. FCoR inhibits CN Foxo1-induced IGFBP1 (A) and G6PC (B) promoter activity. After transient transfection with IGFBP1/luciferase (p925GL3) (A) or a G6Pase/luciferase reporter vector (PicaGene/human G6Pase promoter-luciferase) (B), SV40-transformed hepatocytes were infected with the indicated adenovirus. phRL-SV40 was used as an internal control for transfection efficiency. After overnight serum deprivation and induction with dexamethasone/8-Br-cAMP/IBMX, cells were harvested and luciferase activity was measured. Single and double asterisks indicate statistically significant difference between luciferase activity in the absence and the presence of FCoR (*P<0.005 and **P<0.05, respectively, by one-way ANOVA). Data represent the mean±s.e.m. from three independent experiments. (C) FCoR inhibits Foxo1-induced 5XGAL4-luciferase activity. After transient transfection, HEK293 cells were stimulated with forskolin (20 μM) for 6 h, harvested, and luciferase activity was measured. An asterisk indicates a statistically significant difference between luciferase activity in the absence and presence of FCoR (*P<0.02 by one-way ANOVA). Data represent the mean±s.e.m. from three independent experiments. (D) Effect of overexpression of FCoR on endogenous Igfbp1 gene expression. SV40-transformed hepatocytes were transduced with adenovirus encoding FLAG-CN Foxo1. After 2 h, cells were transduced again with adenovirus encoding cMyc-FCoR. After 36 h, cells were incubated with dexamethasone/cAMP/IBMX for 8 h and harvested. Total RNA was isolated from cells and subjected to real-time PCR to analyse the Igfbp1 and β-actin levels. Data were corrected using the β-actin expression level and then represented as relative mRNA abundance. Data represent the mean values±s.e.m. from three independent experiments. An asterisk indicates a statistically significant difference between endogenous Igfbp1 expression induced by FLAG-CN Foxo1 in the absence and presence of cMyc-FCoR (*P<0.001 by one-way ANOVA). (E) Interaction between Foxo1, Foxo3a or FOXO4 and FCoR. HEK293 cells were co-transfected with pCMV5-cMyc-Foxo1, pCMV5-cMyc-Foxo3a or pCMV5/cMyc-FOXO4 and pFLAG-CMV2-WT FCoR and immunoprecipitated with anti-FLAG, anti-cMyc or normal mouse IgG and blotted with anti-cMyc or anti-FLAG antibody. (F) FCoR inhibits Foxo1-induced and Foxo3a-induced, but not FOXO4-induced 5XGAL4-luciferase activity. After transient transfection with FCoR, HEK293 cells were stimulated with forskolin (20 μM) for 6 h, harvested, and luciferase activity was measured. The grey bar indicates mock-transfected cells and the blue bar indicates pCMV5-cMyc-WT FCoR-transfected cells. An asterisk indicates a statistically significant difference between luciferase activity in the absence and presence of FCoR (*P<0.01 by one-way ANOVA). Data represent the mean±s.e.m. from three independent experiments. Figure source data can be found with the Supplementary data.
We also investigated whether other Foxo proteins (Foxo3a and Foxo4) co-immunoprecipitated with FCoR in the absence of serum and in the presence of forskolin. Transfection studies indicated that epitope-tagged Foxo3a also interacted with FCoR, but Foxo4 did not (Figure 2E). Furthermore, the transactivation assay demonstrated that FCoR repressed the forskolin-induced luciferase activity of a GAL4-Foxo1 fusion protein and a GAL4-Foxo3a fusion protein but did not repress the luciferase activity of a GAL4-FOXO4 fusion protein (Figure 2F). These data suggest that of the Foxo family members, FCoR interacts with Foxo1 and Foxo3a.
FCoR enhances the acetylation of Foxo1 through disruption of the interaction between Foxo1 and Sirt1 and through direct acetylation
Foxo is acetylated by CBP/P300 and deacetylated by Sirt1 or Sirt2 (Accili and Arden, 2004; Jing et al, 2007). We examined Foxo1 acetylation in HEK293 cells in the presence or absence of FCoR. Foxo1 was acetylated in the presence of H2O2 (Figure 3A, lane 1) and overexpression of FCoR enhanced Foxo1 acetylation (Figure 3A, lane 2). Sirt1 binds to acetylated Foxo1 (Brunet et al, 2004; Kitamura et al, 2005). Accordingly, Sirt1 bound to Foxo1 in the presence of H2O2 (Figure 3B, lane 1). However, co-transfection with FCoR decreased the amount of Sirt1 recovered in Foxo1 immunoprecipitates (Figure 3B, lane 2). These data suggest that FCoR disrupts the interaction between Foxo1 and Sirt1. Furthermore, we performed 5XGAL4-luciferase assays using ‘constitutively acetylated’ (6KQ) and ‘acetylation-defective’ (6KR) mutant Foxo1 (Kitamura et al, 2005). WT Foxo1 increased luciferase activity by ∼18-fold, while FCoR decreased WT Foxo1-induced luciferase activity by 40% (Figure 3C). However, FCoR failed to affect reporter activity induced by 6KQ or 6KR mutant Foxo1 (Figure 3C). These data suggest that acetylation is required for FCoR inhibition of Foxo1 transcriptional activity.
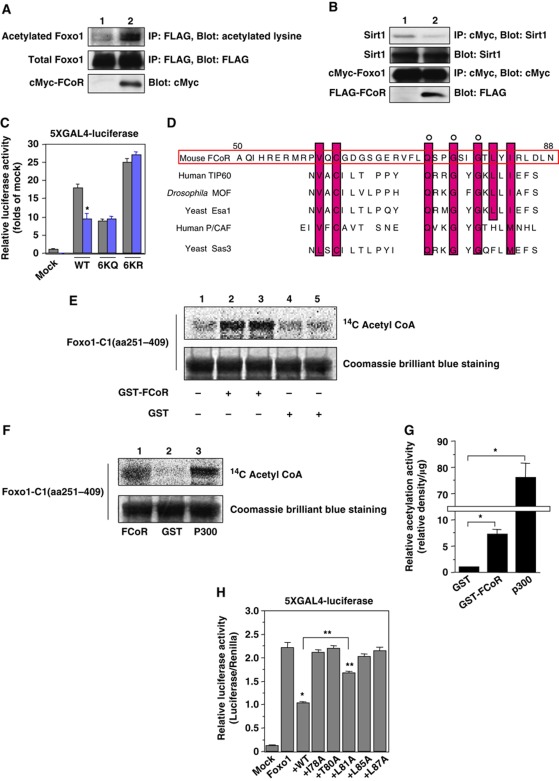
Figure 3.
FCoR enhances Foxo1 acetylation. (A) Effect of FCoR on Foxo1 acetylation. After transfection with pFLAG-CMV2-WT Foxo1 with or without pCMV5-cMyc-WT FCoR, HEK293 cells were incubated with H2O2 (500 μM), nicotinamide (NAM) (50 mM), and trichostatin A (TSA) (2 μM) for 3 h and harvested. Lysates were immunoprecipitated with anti-FLAG mouse monoclonal antibody (M2) and subjected to western blotting with the indicated antibodies. The bottom panel shows western blotting of the lysates with and anti-cMyc mouse monoclonal antibody. (B) FCoR disrupts the interaction between Foxo1 and Sirt1. After transfection with pCMV5-cMyc-WT Foxo1 and pTOPO-Sirt1 with or without pCMV5-cMyc-WT FCoR, HEK293 cells were incubated with H2O2 for 3 h and harvested. Cell lysates were immunoprecipitated with an anti-cMyc mouse monoclonal antibody and subjected to western blotting with the indicated antibodies. (C) 5XGAL4-luciferase assay of PM-WT, -6KQ, and -6KR Foxo1. At 36 h after transfection with pTAL-5XGAL4, phRL-SV40, and the indicated PM-Foxo1 with or without the FCoR expression vector, HEK293 cells were incubated with forskolin (20 μM) for 6 h and harvested. Luciferase activity was measured in the cell lysates. The grey bar indicates mock-transfected cells and the blue bar indicates pCMV5-cMyc-WT FCoR-transfected cells. Data represent the mean values±s.e.m. from three independent experiments. Asterisk indicates statistically significant difference (*P<0.005 by one-way ANOVA). (D) Sequence alignment of FCoR with various acetyltransferases. Residues that are identical or chemically similar to those in mouse FCoR are shown with a red background. FCoR has sequence similarity to the acetyl-CoA binding motifs of the MYST family of HATs (yeast Esa1, yeast Sas3, Drosophila MOF, human TIP60, and human P/CAF). (E) In-vitro acetylation assay of Foxo1. The GST-Foxo1-C1 (aa251–409) protein was subjected to in-vitro acetylation assays with GST (lanes 1, 4, and 5) or GST-FCoR (lanes 2 and 3) as described in ‘Materials and methods’. Reaction products were analysed by Coomassie brilliant blue staining and autoradiography (14C). (F) In-vitro acetylation assay of Foxo1. The GST-Foxo1-C1 (aa251–409) protein was subjected to in-vitro acetylation assays with GST-FCoR (lane 1), GST (lane 2), or recombinant p300 (lane 3) as described in ‘Materials and methods’. Reaction products were analysed by Coomassie brilliant blue staining and autoradiography (14C). (G) Quantification of acetylation activity of FCoR. The intensity of each acetylated band and Coomassie brilliant blue staining band was measured by NIH Image1.62. The relative intensity was corrected by the weight of GST-FCoR or recombinant p300. Data represent the mean values±s.e.m. from two independent experiments. An asterisk indicates statistically significant differences (*P<0.001 by one-way ANOVA). (H) 5XGAL4-luciferase assay of PM-Foxo1 with pCMV5/cMyc-WT, I78A, T80A, L81A, L85A, and L87A FCoR. At 36 h after transfection with pTAL-5XGAL4, phRL-SV40, and the indicated PM-Foxo1 with or without the FCoR expression vector, HEK293 cells were incubated with forskolin (20 μM) for 6 h and harvested. Luciferase activity was measured in the cell lysates. Data represent the mean values±s.e.m. from three independent experiments. Asterisks indicate statistically significant differences (*P<0.001 and **P<0.05 by one-way ANOVA). Figure source data can be found with the Supplementary data.
Interestingly, the amino-acid sequence of the carboxyl terminus of FCoR (amino acids 73–86) shows high sequence similarity to yeast Esa1 and other MYST members, including yeast Sas3, Drosophila MOF, human TIP60, and human P/CAF, which possess intrinsic HAT activity (Figure 3D). To investigate whether FCoR has intrinsic acetyltransferase activity, we performed an in-vitro acetylation assay using a GST-FCoR fusion protein and truncated Foxo1 (amino acids 251–409) as a substrate. This truncated Foxo1 includes four lysine residues that are acetylation sites (K259, K262, K271, and K291) (Daitoku et al, 2003; Kitamura et al, 2005; Qiang et al 2010). The in-vitro acetylation assay revealed that GST-FCoR acetylated truncated Foxo1 (Figure 3E). However, the relative activity of acetyltransferase of FCoR was ∼10% of recombinant p300 (Figure 3F and G). To confirm the importance and specificity of amino acid residues 78–87 for FCoR activity, we performed site-directed mutagenesis of FCoR (I78A, T80A, L81A, L85A, and L87A) and conducted 5XGAL4-luciferase assays. Although wild-type FCoR significantly suppressed Foxo1-induced luciferase activity, most mutations, except L81A, abolished the inhibition of Foxo1 (Figure 3H). Furthermore, this region, amino acids 78–87, corresponds to an LIG_FHA_1. These data suggest that FCoR can acetylate Foxo1 directly and that amino acids 78–87 are important for inhibition of Foxo1.
Phosphorylation of FCoR Threonine 93 regulates its subcellular localization
To investigate the mechanism by which FCoR inhibits the transcriptional activity of Foxo1, we next investigated the subcellular localization of FCoR. In HEK293 cells, FCoR localized mainly to the cytosol or mitochondria in the absence of forskolin (Figure 4A, and data not shown). However, forskolin induced FCoR nuclear localization (Figure 4A) in a time-dependent manner, with a T 1/2 of 15 min (Figure 4B). These data suggest that forskolin promotes FCoR nuclear localization in HEK293 cells. Furthermore, we investigated subcellular localization of endogenous FCoR in 3T3-F442A cells during differentiation. At 48 h after induction, in the presence of 3-isobutyl-1-methylxanthine (IBMX), which is a competitive non-selective phosphodiesterase inhibitor and raises intracellular cAMP and activates PKA, FCoR was localized in nucleus and, thereafter, moved to cytosol (Figure 4C). These data support the observation that PKA induces nuclear accumulation of FCoR.
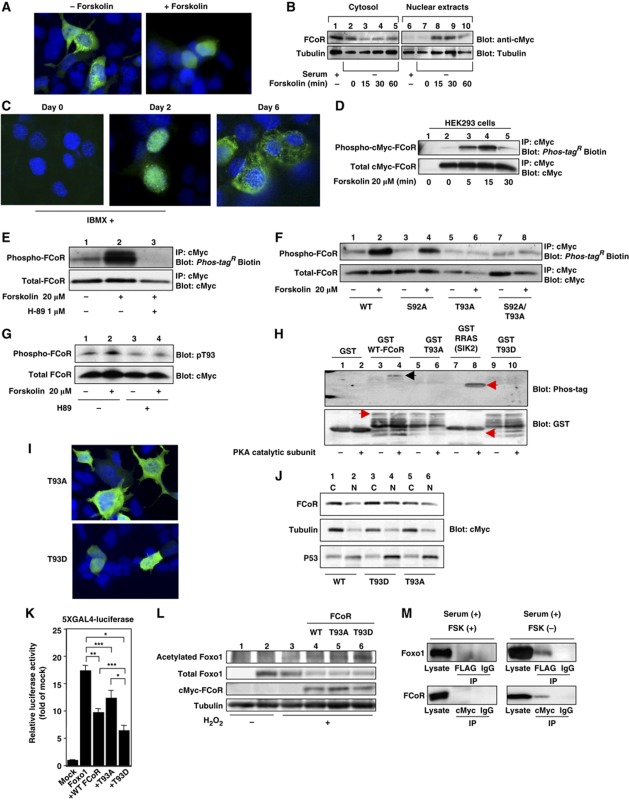
Figure 4.
FCoR is phosphorylated and translocated into the nucleus in a PKA-dependent manner. (A) Subcellular localization of exogenous FCoR in HEK293 cells. At 36 h after transfection with pCMV5-cMyc-WT FCoR, HEK293 cells were stimulated with forskolin (20 μM) for 30 min, fixed, and stained with anti-cMyc mouse monoclonal antibody. (B) Western blotting of cytosolic and nuclear extracts from HEK293 cells transfected with cMyc-FCoR. At 36 h after transfection with pCMV5-cMyc-WT FCoR, HEK293 cells were stimulated with forskolin (20 μM) at the indicated time and harvested. Lysates were fractionated into cytosolic and nuclear extracts and subjected to western blotting with the indicated antibodies. (C) Immunofluorescence of endogenous FCoR in 3T3-F442A cells. 3T3-F442A cells were cultured and induced differentiation into mature adipocytes as described in Materials and Methods except supplementation with 500 nM of IBMX for 48 h. Thereafter, cells were cultured in DMEM containing 10% fetal calf serum and 1.7 μM of insulin. Cells were fixed at the indicated days and stained with anti-FCoR rabbit polyclonal antibody. (D) Phosphorylation of FCoR by forskolin. After transfection with pCMV5-cMyc-WT FCoR, HEK293 cells were stimulated with forskolin (20 μM) at the indicated time and harvested. Cell lysates were immunoprecipitated with anti-cMyc mouse monoclonal antibody, subjected to western blotting, and phosphorylated FCoR was detected. (E) Inhibition of phosphorylation of FCoR by H89. At 48 h after transient transfection with pCMV5-cMyc-WT FCoR, HEK293 cells were incubated with or without H89 (1 μM) for 3 h and stimulated with forskolin (20 μM) for 15 min and harvested. Phosphorylated FCoR was detected as described above. (F) Phosphorylation of mutant FCoR. After transfection with pCMV5-cMyc-WT (lanes 1 and 2), S92A (lanes 3 and 4), T93A (lanes 5 and 6), or S92A/T93A (lanes 7 and 8) FCoR, HEK293 cells were stimulated with forskolin (20 μM) for 15 min. Phosphorylated FCoR was detected as described above. (G) Detection of phosphorylated Threonine 93 by anti-phospho-T93-specific (anti-pT93) antibody. At 48 h after transfection with pCMV5-cMyc-WT FCoR, HEK293 cells were incubated with or without H89 (1 μM) for 3 h and stimulated with or without forskolin (20 μM) for 15 min and harvested. Lysates were subjected to western blotting with anti-pT93. (H) In-vitro phosphorylation assay of FCoR. GST, GST-WT, GST-T93A, GST-T93D FCoR, and GST-RRAS, which is a short sequence of salt-inducible kinase (SIK) 2, was phosphorylated in vitro by incubating with a catalytic subunit from cAMP-dependent protein kinase as described in ‘Materials and methods’. Reaction products were subjected to western blotting, phosphorylated products were detected using Phos-tagR BTL-104 (top panel) and then blotted with anti-GST antibody (bottom panel). (I) Subcellular localization of exogenous T93A (the top panel) or T93D FCoR (the bottom panel) in HEK293 cells. At 36 h after transfection with pCMV5-cMyc-T93A or T93D FCoR, HEK293 cells were fixed as described in Materials and methods and stained with anti-cMyc mouse monoclonal antibody. (J) Western blotting of cytosolic and nuclear extracts from HEK293 cells transfected with cMyc-WT (lanes 1 and 2), T93D (lanes 3 and 4), or T93A (lanes 5 and 6) FCoR. At 36 h after transfection, HEK293 cells were harvested. Lysates were fractionated into cytosolic and nuclear extracts and subjected to western blotting with the indicated antibodies. (K) 5XGAL4-luciferase assay of PM-Foxo1 with pCMV5/cMyc-WT, T93A, and T93D FCoR. At 36 h after transfection with pTAL-5XGAL4, phRL-SV40, and the indicated PM-Foxo1 with or without the FCoR expression vector, HEK293 cells were incubated with forskolin (20 μM) for 6 h and harvested. Luciferase activity was measured in the lysates. Data represent the mean values±s.e.m. from three independent experiments. Asterisks indicate statistically significant difference (*P<0.001, **P<0.005, and ***P<0.05 by one-way ANOVA). (L) The T93D FCoR mutant acetylates Foxo1 the most. After transfection with pFLAG-CMV2-WT Foxo1 with or without pCMV5-cMyc-WT (lane 4), T93A (lane 5), or T93D (lane 6) FCoR, HEK293 cells were incubated with (lanes 3–6) or without H2O2 (lanes 1 and 2) (500 μM), nicotinamide (NAM) (50 mM), and trichostatin A (TSA) (2 μM) for 3 h and harvested. Lysates were immunoprecipitated with anti-FLAG mouse monoclonal antibody (M2) and subjected to western blotting with the indicated antibodies. (M) Interaction between Foxo1 and FCoR. HEK293 cells were co-transfected with pCMV5-cMyc-Foxo1 and pFLAG-CMV2-WTFCoR and culture at the indicated condition. Cell lysates were immunoprecipitated with anti-FLAG, anti-cMyc or normal mouse IgG and blotted with anti-cMyc or anti-FLAG antibody. Figure source data can be found with the Supplementary data.
We next examined whether forskolin induced FCoR phosphorylation. Sequence analysis revealed a consensus protein kinase A (PKA) phosphorylation site (R/K-X1–2-S/T-X) at Threonine 93 (Supplementary Figure S1B). Forskolin rapidly induced FCoR phosphorylation (Figure 4D) in a H-89-inhibitable manner (H89 inhibits PKA) (Figure 4E; Lochner and Moolman, 2006).
Site-directed mutagenesis of Serine 92 or/and Threonine 93 to Alanine (S92A, T93A, or S92A/T93A) indicated that Threonine 93, but not Serine 92, is phosphorylated in response to forskolin (Figure 4F). To confirm that Threonine 93 is phosphorylated by forskolin, we generated a site-specific phospho-T93 antibody (pT93). pT93 immunoreactivity increased in response to forskolin (Figure 4G), suggesting that the main site of FCoR that is phosphorylated in response to forskolin is Threonine 93. Furthermore, the PKA catalytic subunit phosphorylated the GST-WT FCoR fusion protein directly, but did not phosphorylate GST-T93A or -T93D FCoR fusion protein (Figure 4H). These data suggest that PKA directly phosphorylates Threonine 93.
FCoR has a predicted nuclear export signal (NES; amino acids 78–87) (Supplementary Figure S4A). Indeed, substitution of Leucine 81 with Alanine enhances FCoR nuclear translocation and inhibits Foxo1 activity in the absence of forskolin (Supplementary Figure S4B and C). This indicates that Leucine 81 is important for NES. We hypothesized that Threonine 93 phosphorylation affects the subcellular localization of FCoR. Immunofluorescence of cells transfected with the T93A and T93D FCoR mutants revealed that T93A FCoR localized mainly to the cytosol, while T93D FCoR was mainly in the nucleus (Figure 4I). Furthermore, Western blotting of cytosolic and nuclear extracts of HEK293 cells transfected with WT-, T93A-, or T93D-FCoR in the absence of forskolin supported these findings (Figure 4J). We performed 5XGAL4 luciferase assays using WT-, T93A-, or T93D-FCoR. FoxO1 increased 5XGAL4 luciferase activity by 17-fold, while co-transfection with WT FCoR decreased transcriptional activity by 40%. The T93D-FCoR mutant decreased transcriptional activity more markedly than WT or T93A-FCoR (Figure 4K). Inhibition of Foxo1 activity by FCoR reflects the Foxo1 acetylation level. An in-vivo acetylation assay in HEK293T cells demonstrated that T93D FCoR acetylated Foxo1 the most (Figure 4L). Taken together, these data indicate that phosphorylation of Threonine 93 induces FCoR nuclear localization and leads to the acetylation and inhibition of Foxo1.
Finally, to investigate regulation of the interaction between Foxo1 and FCoR, we performed co-immunoprecipitation studies with or without forskolin in the presence of serum. In the presence of forskolin, FCoR did not bind to Foxo1 (Figure 4M, left panel). In contrast, in the absence of forskolin, FCoR bound to Foxo1 (Figure 4M, right panel). Furthermore, in the absence of serum and the presence of forskolin, FCoR bound to Foxo1 (Figure 2E). These data indicate that the interaction between FCoR and Foxo1 depends on their subcellular localization.
FCoR is required for adipocyte differentiation
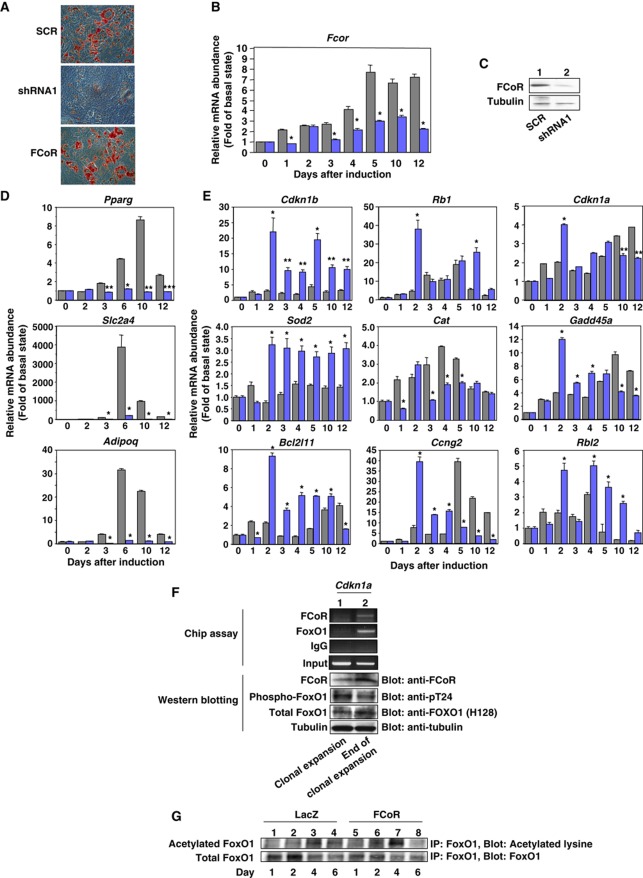
To investigate the physiological role of FCoR in adipocyte differentiation, we inhibited endogenous FCoR in 3T3-F442A cells using adenoviruses encoding two kinds of small hairpin (sh)RNA to decrease FCoR expression (Figure 5B and C; Supplementary Figure S5B and C). Oil-red O staining revealed that knockdown of endogenous FCoR inhibited the differentiation of 3T3-F442A cells (Figure 5A; Supplementary Figure S5A). Analysis of the expression of adipocyte-specific genes demonstrated reduced levels of Pparγ (Pparg), Glut4 (Slc2a4), and adiponectin (Adipoq) (Figure 5D; Supplementary Figure S5D). We showed previously that CN Foxo1 inhibits adipocyte differentiation (Nakae et al, 2003). If FCoR inhibits Foxo-dependent transcription, then knockdown of FCoR should enhance the expression of Foxo1-target genes. Indeed, real-time PCR analysis demonstrated that expression levels of several Foxo1-target genes increased in cells transduced with FCoR shRNA compared with scrambled shRNA-transduced cells (Figure 5E; Supplementary Figure S5E). To examine whether FCoR interacts with Foxo1 in vivo, we performed chromatin immunoprecipitation (Chip) assay using the p21 (Cdkn1a) promoter (Nakae et al, 2003). The Chip assay demonstrated that FCoR interacted with Cdkn1a in a differentiation time-dependent manner (Figure 5F). Furthermore, adenoviruses encoding FCoR significantly enhanced acetylation of endogenous Foxo1 at day 4 after induction of differentiation in 3T3-F442A cells (Figure 5G) and significantly increased Pparg expression at day 10 (Figure 5A; Supplementary Figure S6A).
Figure 5.
The effects of knockdown of FCoR on adipocyte differentiation. (A) Oil-red O staining of 3T3-F442A cells. At day 14 after induction of differentiation and transduction with adenoviruses encoding scrambled (the top panel), shRNA1 (the middle panel), and cMyc FCoR (the bottom panel), cells were stained with Oil-red O. (B) Knockdown of endogenous Fcor mRNA in 3T3-F442A cells during differentiation. 3T3-F442 cells were induced to differentiation into mature adipocytes as described in ‘Supplementary Experimental Procedures’ and transduced with adenovirus encoding shRNA1 (blue bar) or scrambled shRNA (grey bar). Total RNA was isolated from cells on the indicated day and subjected to real-time PCR of Fcor and β-actin. The data were corrected using the β-actin expression level and represent the mean values±s.e.m. from two independent experiments. Asterisks indicate statistically significant differences between Fcor expression levels in scrambles- and shRNA1-transduced cells (*P<0.001 by one-way ANOVA). (C) Western blotting of endogenous FCoR protein. On day 10 after transduction with adenovirus encoding scrambled or shRNA1, 3T3-F442A cells were harvested and the lysates were subjected to western blotting with the indicated antibodies. (D) Effects of knockdown of FCoR on the expression of adipocyte-specific genes during differentiation of 3T3-F442A cells. After induction of differentiation and transduction, total RNA was isolated on the indicated days and subjected to real-time PCR of Pparg (top panel), Slc2a4 (middle panel), and Adipoq (bottom panel). The grey bar indicates scrambled-transduced cells and the blue bar indicates shRNA1-transduced cells. The data were corrected using the β-actin expression level and represent the mean values±s.e.m. from two independent experiments. Asterisks indicate statistically significant difference between scrambled- and shRNA1-transduced cells (*P<0.001, **P<0.005, and ***P<0.01 by one-way ANOVA). (E) Effects of knockdown of FCoR on the expression levels of Foxo1-target genes during differentiation of 3T3-F442A cells. Total RNA from scrambled- (grey bar) or shRNA1-transduced cells (blue bar) on the indicated days was subjected to real-time PCR of the indicated Foxo1-target genes. The data were corrected using the β-actin expression level and represent the mean values±s.e.m. from two independent experiments. Asterisks indicate statistically significant difference between scrambled- and shRNA1-transduced cells (*P<0.001 and **P<0.005 by one-way ANOVA). (F) Chromatin immuunoprecipitation (Chip) assay of the Cdkn1a promoter. The Chip assay was performed using 3T3-F442A cells during (lane 1) or at the end of clonal expansion (lane 2) with the indicated antibodies. The lower panel shows western blotting of 3T3-F442A cells lysates at the indicated period with the indicated antibodies. (G) Acetylation of endogenous Foxo1 in 3T3-F442A cells transduced with adenoviruses encoding LacZ (lanes 1–4) or cMyc-FCoR (lanes 5–8) during differentiation. After induction of differentiation and transduction, cells were harvested on the indicated day, immunoprecipitated with anti-FOXO1 antibody, and blotted with the indicated antibodies.
Foxo1 inhibits the ligand-dependent transcriptional activation of PPARγ. Therefore, if FCoR inhibits Foxo1 activity, then FCoR should prevent Foxo1-mediated inhibition of PPARγ. A Luciferase assay using the J3-tk-Luc reporter vector, which contains three copies of the PPAR response element (PPRE) of the apolipoprotein A-II gene J site, demonstrated that FCoR did not prevent inhibition of PPARγ by Foxo1 (Supplementary Figure S6B). These data suggest that FCoR affects adipocyte differentiation, but not through the interaction of Foxo1 and PPARγ.
Overexpression of FCoR in WAT decreased adiposity and increased insulin sensitivity
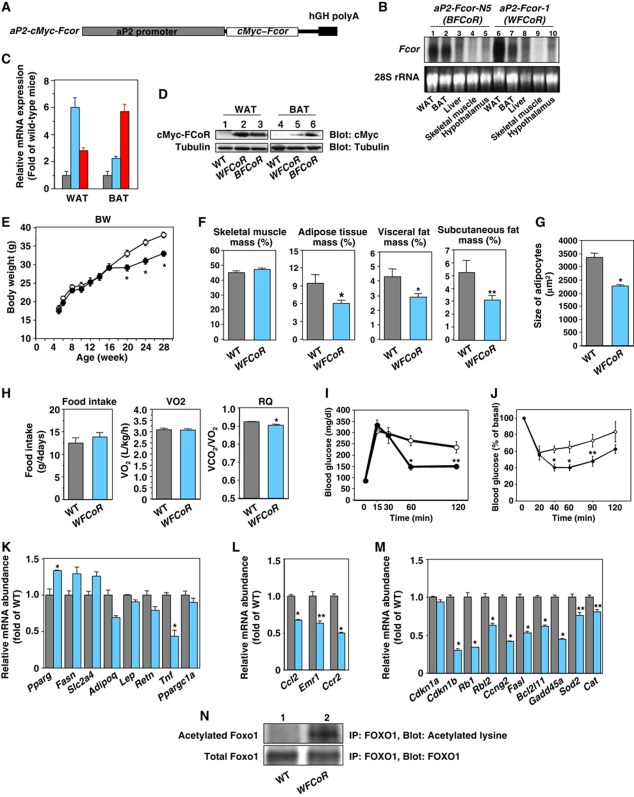
To study the function of FCoR in vivo, we generated transgenic mice that overexpressed FCoR in their adipose tissue (aP 2-cMyc-Fcor: RIKEN Acc. No. CDB0446T) using the 5.4-kb promoter-enhancer fragment of the mouse aP 2 (Figure 6A; Abel et al, 2001; Imai et al, 2001; He et al, 2003). We characterized two founder lines; line 1 (WFCoR) showed greater expression in WAT than in BAT, while the reverse was true for line N5 (BFCoR) (Figure 6B–D). We thus used WFCoR to study the role of FCoR in WAT and BFCoR to study the role of FCoR in BAT.
Figure 6.
Generation of transgenic mice expressing wild-type FCoR driven by the aP2 promoter. (A) Diagram of the transgenic construct. In addition to 5.4 kb of the aP2 promoter, the construct contains the cMyc-WT Fcor cDNA and the human GH polyadenylation sequences. (B) Tissue survey of transgene expression in WAT (lanes 1 and 6), BAT (lanes 2 and 7), liver (lanes 3 and 8), skeletal muscle (lanes 4 and 9), and hypothalamus (lanes 5 and 10) in line N5 (lanes 1–5) and line 1 (lanes 6–10). Total RNA was isolated from the tissues indicated at the bottom of the autoradiogram and analysed by northern blotting using Fcor or β-actin cDNA probe. (C) Real-time PCR of transgene expression in WAT and BAT from transgenic lines. Data represent the mean values±s.e.m. of RNA samples from each of five transgenic mice for each tissue. The grey, sky blue, and red bars indicate wild-type, aP2-FCoR-1 (WFCoR), and aP2-FCoR-N5 (BFCoR), respectively. (D) Western blotting of transgenic cMyc-FCoR expression in WAT (lanes 1–3) and BAT (lanes 4–6). Lysates from the tissue of the indicated types of mice were subjected to western blotting with the indicated antibodies. (E) Body weight of wild-type and transgenic mice (WFCoR) fed a normal chow diet. Data represent the mean values±s.e.m. of 15–20 male mice for each genotype. The open and closed circles indicate wild-type and WFCoR mice, respectively. An asterisk indicates a statistically significant difference between wild-type and WFCoR (*P<0.01 by one-way ANOVA). (F) Adiposity of 5-month-old WFCoR transgenic mice fed a normal chow diet. Skeletal muscle mass, adipose tissue mass (sum of visceral and subcutaneous fat mass), visceral fat mass, and subcutaneous fat mass were calculated. The data are reported as percentage of body weight and represent the mean±s.e.m. of five mice of each genotype. Asterisks indicate statistically significant differences between wild-type and WFCoR mice (*P<0.02 and **P<0.05 by one-way ANOVA). (G) Average of size of adipocytes from epididymal fat of 5-month-old wild-type (grey bar) and WFCoR (sky blue bar) mice fed a normal chow diet. An asterisk indicates a statistically significant difference between wild-type and WFCoR mice (*P<0.001 by one-way ANOVA). (H) Food intake, oxygen consumption, and respiratory quotient of 5-month-old wild-type (grey bar) and WFCoR (sky blue bar) mice fed a normal chow diet. An asterisk indicates a statistically significant difference between wild-type and WFCoR mice (*P<0.001 by one-way ANOVA). (I) Intraperitoneal glucose tolerance test in wild-type (n=10) and WFCoR transgenic mice (n=10) fed a normal chow diet. The open and closed circles indicate wild-type and WFCoR mice, respectively. Data represent the mean values±s.e.m. Asterisks indicate statistically significant differences between wild-type and WFCoR mice (*P<0.005 and **P<0.02 by one-way ANOVA). (J) Intraperitoneal insulin tolerance test in wild-type (n=8) and WFCoR (n=10) mice fed a normal chow diet. Data represent the mean values±s.e.m. Asterisks indicate statistically significant differences between wild-type and WFCoR mice (*P<0.005 and **P<0.05 by one-way ANOVA). (K) Real-time PCR of WAT-specific genes using WAT from wild-type (grey bar) and WFCoR transgenic mice (sky blue bar) fed a normal chow diet. Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. An asterisk indicates a statistically significant difference between wild-type and WFCoR transgenic mice (*P<0.05 by one-way ANOVA). (L) Real-time PCR of Ccl2, Emr1, and Ccr2 genes using WAT from wild-type (grey bar) and WFCoR transgenic mice (sky blue bar) fed a normal chow diet. Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. Asterisks indicate statistically significant differences between wild-type and WFCoR transgenic mice (*P<0.001 and **P<0.005 by one-way ANOVA). (M) Real-time PCR of Foxo1-target genes using WAT from wild-type (grey bar) and WFCoR transgenic mice (sky blue bar). Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. Asterisks indicate statistically significant differences between wild-type and WFCoR transgenic mice (*P<0.001 and **P<0.005 by one-way ANOVA). (N) Acetylation of endogenous Foxo1 in WAT from 5-month-old wild-type (lane 1) and WFCoR transgenic mice (lane 2). Tissue lysates containing nicotinamide (NAM) (50 mM) and trichostatin A (TSA) (2 μM) were immunoprecipitated with anti-FOXO1 antibody and blotted with the indicated antibodies.
WFCoR mice showed an adult-onset decrease in weight loss (Figure 6E). CT scan revealed that WFCoR mice had decreased adipose mass, including the visceral and subcutaneous depots (Figure 6F). Histological analysis of WAT from these animals demonstrated that the adipocytes in the WAT of WFCoR mice were similar to WT mice at 12 weeks but the adipocytes in the WAT of WFCoR mice were smaller than those from WT mice at 24 weeks (Figure 6G; Supplementary Figure S7A and B). Although food intake, locomotor activity, and oxygen consumption were similar to wild-type mice, the respiratory quotient of WFCoR mice was significantly reduced (Figure 6H, and data not shown). These data indicate increased usage of lipid as an energy source, resulting in lower body weight.
To investigate the effects of FCoR overexpression in WAT on glucose metabolism, we performed intraperitoneal glucose (IPGTT) and insulin tolerance test (ITT). Two-month-old WFCoR mice showed the same glucose tolerance and insulin sensitivity as wild-type mice (data not shown). However, 4-month-old mice showed evidence of increased glucose (Figure 6I) and insulin tolerance (Figure 6J). Insulin secretion in WFCoR mice was similar to that of wild-type mice during IPGTT (Supplementary Figure S8). These data suggest that overexpression of FCoR in WAT decreases adiposity and increases insulin sensitivity in an age-dependent manner.
To investigate the mechanism by which FCoR increases insulin sensitivity, we examined the expression levels of adipose tissue-specific and Foxo1-target genes in WAT. FCoR transgenic mice showed increased Pparg and decreased Tnf-α(Tnf), Mcp-1 (Ccl2), and Ccr2 gene expression (Figure 6J and K). Expression of several Foxo1-target genes (Greer and Brunet, 2005) was decreased, including the expression of p27 (Cdkn1b), Rb1, p130 (Rbl2), Cyclin G2 (Ccng2), fas ligand (fasl), Bcl2l11, Gadd45 (Gadd45a), Sod2, and Cat was decreased (Figure 6L). Acetylation of endogenous Foxo1 in WAT was significantly increased compared with wild-type mice (Figure 6M). These data suggest that overexpression of FCoR in WAT inhibits the expression of Foxo1-target genes, decreases the size of adipocytes in WAT, and increases insulin sensitivity.
We crossed Leprdb/+-WFCoR with Leprdb/+ mice and generated Leprdb/db-WFCoR mice in order to investigate whether overexpression of FCoR in WAT improves glucose metabolism in Leprdb/db mice. Leprdb/db-WFCoR mice showed a significant decrease in body weight compared with Leprdb/db mice (Supplementary Figure S9A and B). IPGTT and ITT demonstrated that Leprdb/db-WFCoR mice have improved glucose tolerance and insulin sensitivity (Supplementary Figure S9C and D). CT scans revealed decreased visceral fat mass in Leprdb/db-WFCoR compared with Leprdb/db mice, despite similar total adipose tissue mass (Supplementary Figure S9E). Measurement of adipocyte size revealed that white adipocytes from Leprdb/db-WFCoR mice were significantly smaller than those from Leprdb/db (Supplementary Figure S9F–H). Although the food intake and respiratory quotient of Leprdb/db-WFCoR mice were similar to Leprdb/db mice, oxygen consumption was significantly increased (data not shown; Supplementary Figure S9I). Gene expression analysis demonstrated that the expression levels of Ccl2 and several Foxo1-target genes were decreased significantly compared with levels in Leprdb/db mice (Supplementary Figure S9J–L). These data suggest that the FCoR in WAT can improve glucose metabolism in Leprdb/db mice.
We next examined the effects of overexpression of FCoR in mice with diet-induced obesity. The WFCoR mice had a lean phenotype compared with wild-type mice fed a HFD and had smaller adipocytes in WAT (Supplementary Figure S10A and B). IPGTT and ITT revealed that HFD WFCoR mice had improved glucose tolerance and insulin sensitivity compared with wild-type mice (Supplementary Figure S10C and D). These findings are in accordance with the idea that FCoR in WAT improves insulin sensitivity.
FCoR in BAT affects thermoregulation
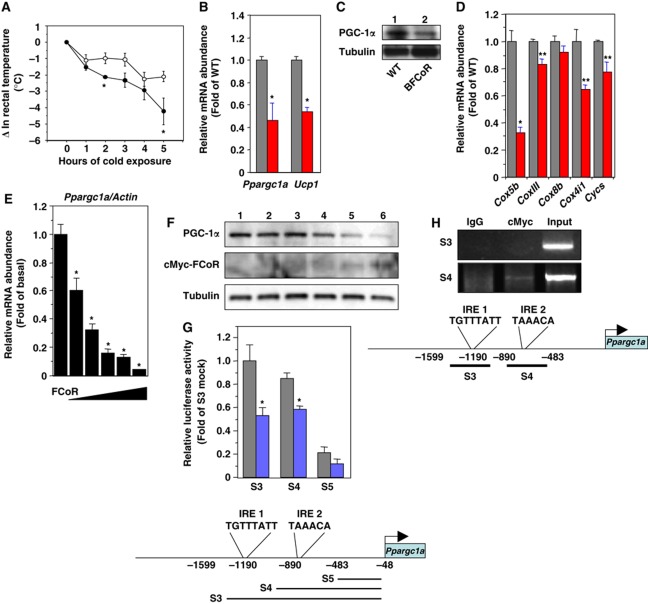
We also analysed BAT function in BFCoR mice (Figure 6B–D). BFCoR mice have normal body weight (Supplementary Figure S11A) and IPGTT (Supplementary Figure S11B). Insulin sensitivity was similar to wild-type mice (Supplementary Figure S11C). We also measured oxygen consumption to investigate the effects of overexpression of FCoR on the physiological function of BAT. Oxygen consumption and the respiratory quotient of BFCoR mice were similar to wild-type mice (Supplementary Figure S11D and E). The BFCoR mice exhibited significantly reduced cold tolerance compared with wild-type mice (Figure 7A). We detected ∼60% and ∼50% decreases of Ppargc1a and Ucp1, respectively (Figure 7B), in BFCoR mice, along with a commensurate decrease in PGC-1α protein (Figure 7C). Furthermore, BFCoR mice exhibited reduced expression levels of mitochondrial components (Figure 7D). These data indicate that overexpression of FCoR in BAT suppresses thermoregulation by suppressing PGC-1α expression.
Figure 7.
FCoR in BAT affects thermoregulation through suppression of Ppargc1a expression. (A) Changes in the rectal temperature of 6-month-old wild-type (open circle) and BFCoR (closed circle) mice after cold exposure. Data represent the mean values±s.e.m. (n=6 for each genotype). Asterisks indicate statistically significant differences between wild-type and BFCoR transgenic mice (*P<0.05 by one-way ANOVA). (B) Real-time PCR of Ppargc1a and Ucp1 in BAT from wild-type (grey bar) and BFCoR transgenic mice (red bar). The data were corrected using the β-actin expression level and represent the mean values±s.e.m. of three independent experiments (n=6 for each genotype). Asterisks indicate statistically significant differences between wild type and BFCoR transgenic mice (*P<0.05 by one-way ANOVA). (C) Western blotting of PGC-1α and UCP-1 protein in BAT from wild-type (lane 1) and BFCoR transgenic mice (lane 2). Lysates (200 μg) of BAT from wild-type and BFCoR transgenic mice were subjected to western blotting with the indicated antibodies. (D) Real-time PCR of mitochondrial component genes using BAT from wild-type (grey bar) and BFCoR transgenic mice (red bar). Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. Asterisks indicate statistically significant differences between wild-type and BFCoR transgenic mice (*P<0.001 and **P<0.005 by one-way ANOVA). (E) The effects of overexpression of cMyc-FCoR on endogenous Ppargc1a gene expression in T37i cells. T37i cells were transduced with adenovirus encoding LacZ or cMyc-FCoR. Cells were harvested 48 h after transduction. Total RNA was isolated and subjected to real-time PCR of Ppargc1a. Data were corrected using the β-actin expression level, shown as ‘fold change′ compared with LacZ-transduced cells, and represent the mean values±s.e.m. from three independent experiments. An asterisk indicates a statistically significant difference between LacZ- and cMyc-FCoR-transduced cells (*P<0.001 by one-way ANOVA). (F) PGC-1α protein expression was decreased in a dose-dependent manner by overexpression of FCoR in T37i cells. (G) Effect of FCoR on Ppargc1a promoter activity. Data were obtained in five experiments and are represented as the mean values±s.e.m. of fold change compared with mock vector-transfected activity in T37i cells transfected with truncated Ppargc1a (S3). *P<0.001 (one-factor ANOVA of cells transfected with pCMV5/cMyc and with pCVM5/cMyc-FCoR vector). (H) Chip assays of T37i cells transduced with an adenovirus encoding cMyc-FCoR and harvested 36 h after transduction. The PCR primers used to amplify the mouse Ppargc1a promoter sequence as indicated as S3 and S4. PCRss with total input chromatin are shown as controls. Figure source data can be found with the Supplementary data.
FCoR regulates PGC-1α gene expression
To examine whether FCoR can suppress Ppargc1a expression at the cellular level, we transduced the brown adipocyte cell line T37i cells with adenovirus encoding LacZ or cMyc-FCoR and examined endogenous Ppargc1a expression. Real-time PCR showed that overexpression of FCoR suppressed endogenous Ppargc1a expression in T37i cells in an FCoR dose-dependent manner (Figure 7E). The PGC-1α protein level was also decreased by FCoR expression in T37i cells (Figure 7F). To investigate whether FCoR suppresses Ppargc1a promoter activity, we performed a luciferase reporter assay using truncated versions of Ppargc1a promoters. The Ppargc1a promoter has two insulin-responsive elements, IRE1 and IRE2 (Figure 7G). To characterize the FCoR-target site in the Ppargc1apromoter, we constructed different versions of the mouse Ppargc1a promoter by progressively deleting portions of its upstream region. The transcriptional activity of each mutant promoter in response to FCoR was examined in T37i cells (Figure 7G). Mutant promoters with deletions up to –890 nt in the Ppargc1a promoter, which includes IRE1 and/or IRE2, still responded to FCoR. However, further deletion up to –483 nt, which has no IRE, completely abolished the responsiveness of the mutant promoter to FCoR suppression (Figure 7G). Thus, the FCoR-target site was confined to a proximal in the mouse Ppargc1a promoter and FCoR suppressed Ppargc1a promoter activity. These data indicate that FCoR inhibits Ppargc1a expression. Using Chip assays, we investigated two IREs in the Ppargc1a promoter. Chip assays using both promoter regions revealed that FCoR bound the proximal IRE2 but not the distal IRE1 (Figure 7H). These data suggest that FCoR binds to the Ppargc1a promoter via IRE2.
Fcor knockout mice exhibited insulin resistance
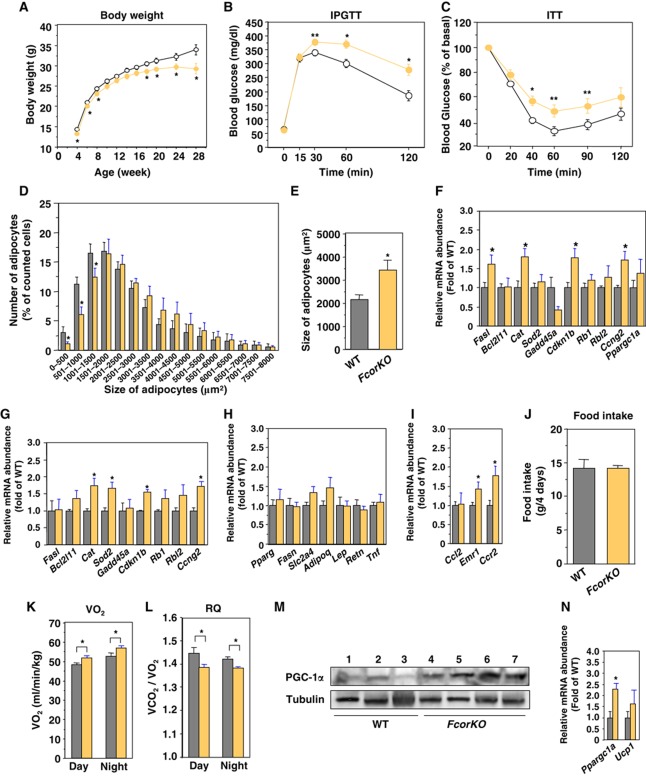
To investigate the physiological roles of FCoR, we generated Fcor knockout (FcorKO) mice (Supplementary Figure S12A–E). FcorKO mice had a lean phenotype, glucose intolerance, and insulin resistance on a normal chow diet (Figure 8A–C). CT scan revealed that FcorKO mice had adipose mass that was similar to that of WT mice, including the visceral and subcutaneous depots (data not shown). In contrast to our findings that FCoR is overexpressed in WAT, the size of epididymal fat adipocytes was larger in FcorKO mice than in wild-type mice (Figure 8D and E). These data suggest that there may be fewer adipocytes in FcorKO mice than in WT mice. The expression of several Foxo-target genes was significantly increased in both WAT and BAT in FcorKO mice (Figure 8F and G). The expression levels of WAT-specific genes were similar in FcorKO mice as in wild-type mice, but the expression levels of inflammatory genes, including Emr1 and Ccr2, were significantly increased (Figure 8H and I), leading to insulin resistance (Olefsky and Glass, 2010).
Figure 8.
Deletion of Fcor induces insulin resistance. (A) Body weight of wild-type and FcorKO mice fed a normal chow diet. Data represent the mean values±s.e.m. of 20–30 male mice for each genotype. The open and yellow closed circles indicate wild-type and FcorKO mice, respectively. An asterisk indicates statistically significant difference between wild-type and FcorKO (*P<0.01 by one-way ANOVA). (B) Intraperitoneal glucose tolerance test in wild-type (n=10) and FcorKO mice (n=10) under normal chow diet. The open and yellow closed circles indicate wild-type and FcorKO mice, respectively. Data represent mean values±s.e.m. Asterisks indicate statistically significant differences between wild-type and FcorKO mice (*P<0.001 and **P<0.02 by one-way ANOVA). (C) Intraperitoneal insulin tolerance test in wild-type (n=10) and FcorKO (n=10) fed a normal chow diet. Data represent the mean values±s.e.m. Asterisks indicate statistically significant differences between wild-type and FcorKO mice (*P<0.005 and **P<0.05 by one-way ANOVA). (D) Distribution of adipocyte size. The sizes of single adipocytes from 5-month-old wild-type (grey bar) or FcorKO (yellow bar) mice fed a normal chow diet were measured as described in ‘Supplementary Experimental Procedures’. The data represent the mean values±s.e.m. of the percentage of all adipocytes with the indicated size. Asterisks indicate statistically significant differences between wild-type and FcorKO mice (*P<0.05 by one-way ANOVA). (E) The average of size of adipocytes from (D). An asterisk indicates a statistically significant difference between wild-type and FcorKO mice (*P<0.005 by one-way ANOVA). (F, G) Real-time PCR of Foxo1-target genes using WAT (F) and BAT (G) from wild-type (grey bar) and FcorKO (yellow bar) mice. Data represent the mean±s.e.m. of RNA samples from 10 mice of each genotype. Asterisks indicate statistically significant differences between wild-type and WFCoR transgenic mice (*P<0.05 by one-way ANOVA). (H) Real-time PCR of WAT-specific genes using WAT from wild-type (grey bar) and FcorKO (yellow bar) mice fed a normal chow diet. Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. (I) Real-time PCR of Ccl2, Emr1, and Ccr2 genes using WAT from wild-type (grey bar) and FcorKO (yellow bar) mice. Data represent the mean values±s.e.m. of RNA samples from five mice of each genotype. Asterisks indicate statistically significant differences between wild-type and FcorKO mice (*P<0.05 by one-way ANOVA). (J) Food intake of 5-month-old wild-type (grey bar) and FcorKO (yellow bar) mice. Data represent the mean values±s.e.m. of food intake for 4 days. (K, L) The effects of deletion of Fcor on energy expenditure. Average oxygen consumption (K) and respiratory quotient (L) of 5-month-old wild-type (grey bar) and FcorKO (yellow bar) mice fed a normal diet. Data represent the mean values±s.e.m. of six male mice for each genotype. Measurements of oxygen consumption were performed for 72 h after allowing the mice to acclimate to the cage environment. An asterisk indicates a statistically significant difference between wild-type and FcorKO mice (*P<0.05 by one-way ANOVA). (M) Western blotting of PGC-1α (top panel) and tubulin (bottom panel) in BAT from wild-type (lanes 1–3) and FcorKO (lanes 4–7) mice. (N) Real-time PCR of Ppargc1a and Ucp1 using BAT from wild-type (grey bar) and FcorKO (yellow bar) mice. Data represent the mean values±s.e.m. of RNA samples from 10 mice of each genotype. Asterisks indicate statistically significant differences between wild-type and WFCoR transgenic mice (*P<0.05 by one-way ANOVA). Figure source data can be found with the Supplementary data.
Although the food intake of FcorKO mice was similar to that of wild-type mice, oxygen consumption was significantly increased and respiratory quotient was significantly decreased (Figure 8J–L). These data indicate that FcorKO mice had increased energy expenditure and increased usage of fat that led to a lean phenotype.
Overexpression of FCoR suppressed PGC-1α expression in BFCoR mice and T37i cells. Therefore, deletion of Fcor would be expected to increase PGC-1α expression. Indeed, the expression levels of Ppargc1a and the PGC-1α protein in the BAT of FcorKO mice were significantly increased compared with wild-type mice (Figure 8M and N). These data confirm that FCoR suppresses the transcriptional activity of Foxo1 and PGC-1α expression.
Discussion
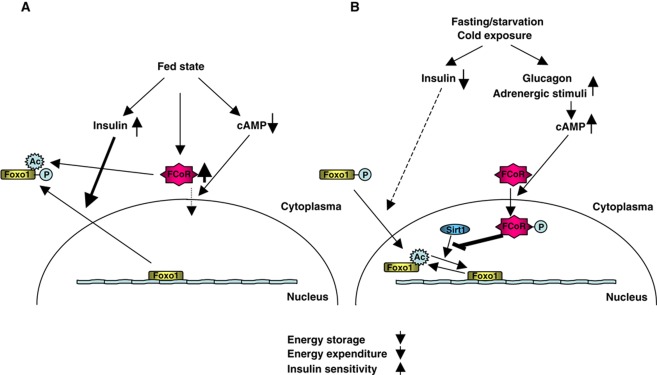
Using a two-hybrid screen, we identified a Foxo1-CoRepressor termed as FCoR that is expressed in adipose tissue, inhibits Foxo1 transcriptional activity, and is phosphorylated by PKA and then translocated into the nucleus. FCoR is activated in a fasting or starved state, or during cold exposure, all conditions in which PKA is activated by glucagon or adrenergic stimuli. Thus, FCoR may be an important metabolic regulator that coordinates the insulin/cAMP response by interacting with Foxo. However, at fasting state, expression level of activated FCoR protein is decreased. We speculate that the active, presumed phosphorylated, FCoR protein is unstable and turned over rapidly.
Experiments showed that FCoR prevents Sirt1 binding to Foxo1, thus enhancing Foxo1 acetylation and inhibiting its transcriptional activity. Several proteins have been reported to affect Foxo1 and Sirt1. In C. elegans, two 14-3-3 proteins bind to SIR-2.1 and DAF-16 and are required for SIR-2.1-induced transcriptional activation of DAF-16 (Berdichevsky et al, 2006). Furthermore, four-and-a-half LIM 2 (FHL2) also interacts with FOXO1; FHL2 enhances the interaction between FOXO1 and SIRT1 and therefore enhances the deacetylation of FOXO1 (Yang et al, 2005). To date, FCoR is the only protein that is known to inhibit the interaction of Sirt1 and Foxo1.
Recently, it has been reported that class IIa HDACs 4/5/7 and class I HDAC (HDAC3) are critical components of the transcriptional response to fasting in liver, shuttling into the nucleus in response to forskolin or glucagon. HDAC4 and 5 induces the acute transcription of gluconeogenic enzymes such as G6Pase via deacetylation and activation of Foxo family transcription factors (Mihaylova et al, 2011; Wang et al 2011). In the present study, we used trichostatin A (TSA) for the prevention of deacetylation of Foxo1. Therefore, we cannot exclude the possibility that FCoR works on the regulatory complex with the class IIa and I HDACs. It is established that in hepatocytes, Foxo is actively promoting transcription of its target genes after forskolin, glucagon, or cAMP treatment. However, Fcor is expressed little in liver of wild-type mice under normal chow diet. If FCoR is expressed in liver, then the predicted acetylation and inhibition of Foxo may run counter to the known activation of Foxo by forskolin/cAMP agonists.
Interestingly, FCoR itself has intrinsic acetyltransferase activity. Acetylation represents a fail-safe mechanism for preventing excessive Foxo1 activity (Banks et al, 2011). Therefore, FCoR may maintain Foxo1 acetylation by direct acetylation in the cytosol in the fed state and by interruption of the association of Foxo1 with Sirt1 in the nucleus in a fasting or starved state or during cold exposure. This is consistent with the idea that FCoR acts to fine-tune Foxo1 activity (Figure 9). FCoR can also acetylate other transcription factors and cofactors (Nakae et al, unpublished observation). Therefore, the sudden decline of body weight in WFCoR mice may be due to acetylation of other binding partners of FCoR. Further investigation of FCoR-binding proteins is needed to identify such partners.
Figure 9.
A model for the roles of FCoR in fine-tuning of Foxo1 activity. (A) When mice are in the fed state, Foxo1 is phosphorylated and FCoR expression is increased. Both proteins remain in the cytosol. FCoR acetylates Foxo1 directly and keeps it in the acetylated state. (B) When mice are in the fasting state, insulin level decreases and Foxo1 remains in the nucleus and is activated. At the same time, glucagon from pancreatic α-cells is increased and activates cAMP-dependent protein kinase (PKA). Furthermore, upon exposure to the cold, adrenergic stimuli also activate PKA. These events lead to the nuclear localization of FCoR, which increases acetylation of Foxo1 by interrupting the association between Foxo1 and Sirt1. FCoR thus regulates energy storage, energy expenditure, and insulin sensitivity by fine-tuning Foxo1 function.
The finding that knockdown of endogenous FCoR inhibited the differentiation of 3T3-F442A cells suggests that FCoR is indispensable for adipocyte differentiation. This is in keeping with our previous findings that overexpression of CN Foxo1 (ADA) inhibits the differentiation of 3T3-F442A cells (Nakae et al, 2003). Jing et al reported that the KR mutant of Foxo1, which is activated and thus mimics the deacetylated protein (Banks et al, 2011), inhibits adipocyte differentiation (Jing et al, 2007). Therefore, knockdown of FCoR may accelerate Foxo1 deacetylation and so inhibit adipogenesis.
The present study demonstrated that while FCoR overexpression decreased the adipocyte size in transgenic mice, FcorKO mice exhibited increased adipocyte size, consistent with findings for Foxo1 haploinsufficiency or transgenic mice overexpressing a dominant-negative Foxo1 mutant in fat cells (aP2-FLAG-Δ256) (Nakae et al, 2003, 2008a; Kim et al, 2009). Namely, inhibition of Foxo1 leads to smaller adipocytes. Therefore, inhibition of Foxo1 by overexpression of FCoR reduces adipocyte size. On the other hand, activation of Foxo1 by deletion of FCoR increases adipocyte size. Foxo1 is involved in the early stages of adipose conversion (Nakae et al, 2003). Therefore, it is speculated that generation of adipocytes in FcorKO mice is decreased due to activation of Foxo1. Previous reports showed that Foxo1 inhibits PPARγ activity (Dowell et al, 2003; Armoni et al, 2006; Fan et al, 2009). Knockdown of FCoR should enhance Foxo1 activity, leading to inhibition of PPARγ and adipogenesis. However, in the present study, FCoR did not prevent Foxo1-induced inhibition of PPARγ activity. More than that, knockdown of Fcor suppressed Pparg gene expression in 3T3-F442A cells and overexpression of FCoR in WAT significantly increased Pparg expression. Foxo1 represses transcription from either Pparg1 or Pparg2 promoter (Armoni et al, 2006). Therefore, inhibition of Foxo1 by FCoR may work on Pparg promoter and increase Pparg expression, leading to enhanced adipogenesis and smaller adipocytes.
We found that FCoR expression in BAT of transgenic mice led to cold intolerance and to decreased expression levels of Ppargc1a and Ucp1, suggesting that FCoR regulates brown adipocyte function. FCoR expression is induced by cold exposure. Therefore, FCoR in BAT may restrict mitochondrial biogenesis during cold exposure by inhibition of Ppargc1a expression. However, the findings are inconsistent with our previous report that aP2-FLAG-Δ256 mice, in which a transactivation-defective Foxo1 (Δ256) is expressed in both WAT and BAT, showed increased oxygen consumption accompanied by increased expression of PGC-1α, uncoupling protein (UCP)-1, UCP-2, and β3-adrenergic receptor (β3-AR) (Nakae et al, 2008a). If FCoR works by inhibiting Foxo1 activity in BAT, then overexpression of FCoR should enhance the thermogenic function of BAT, leading to increased expression of Ppargc1a and increased energy expenditure. Therefore, FCoR may inhibit Ppargc1a expression via other partners but not via inhibition of Foxo1 in BAT. However, in liver, Ppargc1a is a transcriptional target that is induced by the dephosphorylation mutant of Foxo1 (Daitoku et al, 2003; Matsumoto et al, 2006). Further investigation is needed to clarify the molecular target of FCoR in the Ppargc1a promoter in BAT.
FCoR acts as a ‘repressor’ of Foxo1 and Foxo3a but not of Foxo4. The failure to identify homologous genes in C. elegans, along with the repression of FCoR expression in states of food deprivation, indicates that FCoR may act as an anti-thrifty gene. FCoR regulates insulin sensitivity by altering the size of white adipocytes in WAT and induces cold intolerance by inhibiting Ppargc1a expression. FCoR is thus an attractive therapeutic target for treatment of obesity and type 2 diabetes.
Materials and methods
Antibodies and cell cultures
Anti-FCoR antisera were raised by immunizing rabbits with a peptide corresponding to amino acids 2–16 of the mouse FCoR sequence (GGPTRRHQEEGSAEC). Phospho-threonine 93 rabbit polyclonal antibodies (anti-pT93) were produced by immunizing rabbits with a synthetic phosphorylated peptide coupled to KLH that corresponded to amino acids 85–94 surrounding mouse FCoR Threonine 93 (LDLNSQRS-T (PO3H2)-C). Antibodies were purified using Activated Thiol-Sepharose 4B (Pharmacia) Peptide Columns and peptide affinity chromatography (TAKARA BIO, Inc.). We purchased anti-FLAG (M2) and anti-tubulin from Sigma; anti-cMyc (9E10), anti-FOXO1 (H128), anti-PGC-1 (H300), and anti-GST (Z-5) from Santa Cruz Biotechnology; and anti-phospho-FOXO1 (pT24), and anti-acetylated lysine polyclonal antibodies from Cell Signaling Technology. HEK293 cells and T37i cells were cultured as described previously (Nakae et al, 1999). Preadipocytes cell line 3T3-F442A cells were cultured and induced differentiation into mature adipocytes as described previously (Nakae et al, 2003). Staining 3T3-F442A cells with oil red O was described previously (Nakae et al, 2003).
Yeast two-hybrid screen
Amino acids 1–154 of murine Foxo1 were cloned in-frame into the GAL4 DNA-binding domain plasmid pGBKt7 (Clontech). The GAL4 activation domain 3T3-L1 cDNA library of 3T3-L1 was the kind gift of Dr Alan R Saltiel (University of Michigan Medical Center, Michigan, USA) (Printen et al, 1997). The AH109 yeast strain was used for the library search. The transformation was performed as described in the Clontech Matchmaker two-hybrid system 3 protocol. The transformants were plated on SD/-Ade/-His/-Leu/-Tyr plates in the presence of galactose and then were incubated at 30°C for 3–4 days. Positive interaction was identified by strong β-galactosidase activity. Individual positive clones were isolated using the Yeastmaker yeast plasmid isolation kit (BD Bioscience), sequenced by an ABI310 automated DNA sequencer, and analysed for homology with sequences in the GenBank database using the BLAST algorithm.
Rapid amplification of cDNA ends
In order to determine the complete 5′ and 3′ sequence of Fcor mRNA, we performed both 5′- and 3′-RACE using the SMART RACE cDNA Amplification Kit (Clontech) according to manufacturer’s protocol. In brief, we isolated total RNA from the WAT and BAT of wild-type mice and synthesized first-strand cDNA. After that, we performed 5′- and 3′-RACE PCR using the following gene specific primers: 5′-CACCACTACCATCACCACACTGCACCGG-3′ for 5′-RACE PCR and 5′-CCCCGGGTTTGCAAGGATGGATGGGAATG-3′ for 3′-RACE PCR. The PCR products were separated by electrophoresis in a 1.5% agarose gel (Supplementary Figure 1B). After purification using the QIAquickR Gel Extraction Kit (QIAGEN), PCR products were subcloned into the pCRIIR vector (Invitrogen) using the TA Cloning KitR (Invitrogen) and sequenced.
RNA isolation, real-time PCR, and northern blotting
Isolation of total RNA from tissues and cells was performed using the SV Total RNA Isolation System (Promega) according to manufacturer’s protocol. Real-time PCR was performed as described previously (Nakae et al, 2008a). The primers used in this study are described in Table S1. We used the following primers for amplification of Fcor: 5′-ATGGGCGGTCCTACACGCCGCCAT-3′ (sense) and 5′-CTAGCACATGCCTTTAGTCCC-3′ (antisense). Northern blotting was performed using standard techniques. To prepare the probe for Fcor, we performed PCR using pCMV5/cMyc-FCoR plasmid as a template and the same primers as in real-time PCR. The probe used here for β-actin was described previously (Nakae et al, 2008a).
Construction of expression vectors
To construct pCMV5/cMyc-FCoR, we reverse transcribed total RNA from mouse BATs using the GeneAmpR RNA PCR Core Kit (Applied Biosystems) and performed PCR using the following primers: 5′-GG TCTAGA (XbaI) ATGGGCGGTCCTACACGCCGC-3′ (FCoR-S) and 5′-GG TCTAGA (XbaI) CTAGCACATGCCTTTAGTCCC-3′ (FCoR-AS). After treatment with XbaI, PCR products were subcloned into the XbaI-treated pCMV5/cMyc vector (Nakae et al, 1999). To construct pFLAG-CMV2-FCoR, we performed PCR using pCMV5/cMyc-FCoR as a template and FCoR-S and FCoR-AS as primers. XbaI-treated PCR products were subcloned into XbaI-treated pFLAG-CMV2 vector. To construct pFLAG-CMV2-WT FoxO1, XbaI-treated WT-FoxO1 fragments from pCMV5/cMyc-WT FoxO1 vector were subcloned into XbaI-treated pFLAG-CMV2 vector. Construction of pCMV5/cMyc-WT FoxO3a was described previously (Takaishi et al, 1999; Nakae et al, 2001). To construct pCMV5/cMyc-WT FOXO4, human FOXO4 cDNA was amplified by PCR using PFN21AB7772 (Kazusa DNA Research Institute, Chiba, Japan) and the following primers: 5′-GGG ATCGAT (ClaI) ATGGATCCGGGGAATGAGAAT-3′ (sense) and 5′-GGG AAGCTT (HindIII) TCAGGGATCTGGCTCAAAGTT-3′ (antisense). After treatment with ClaI and HindIII, PCR products were subcloned into ClaI/HindIII-treated pCMV5/cMyc vector. All vectors produced for this study were sequenced to confirm that they had the intended sequences.
Site-directed mutagenesis
Site-directed mutagenesis was carried out using the QuickChangeRII Site-Directed Mutagenesis Kit (Stratagene). We used the pCMV5/cMyc-FCoR expression vector as a template. To construct S92A-FCoR, we used the following mutagenic primers: S92A-S 5′-gaactcacaaagaGccacctgctgctgcct-3′ and S92A-AS 5′-aggcagcagcaggtggCtctttgtgagttc-3′. To construct T93A-FCoR, we used T93A-S 5′-gaactcacaaagatccGcctgctgctgcct-3′ and T93A-AS 5′-aggcag cagcaggCggatctttgtgagttc-3′. To construct T93D-FCoR, we used T93D-S 5′-gaactcacaaagatccGActgctgctgcct-3′ and T93D-AS 5′-aggcagcagcagTCggatctttgtgagttc-3′. To construct S92A/T93D-FCoR, we used S92A/T93A-S 5′-gaactcacaaagaGccGcctgctgctgcct-3′ and S92A/T93A-AS 5′-aggcagcagcaggCggCtctttgtgagttc-3′. To construct I78A-FCoR, we used I78A-S 5′-ctacagagccctggctctGCtggaactctatatatcagg-3′ and I78A-AS 5′-cctgatatatagagttccaGCagagccagggctctgtag-3′. To construct T80A-FCoR, we used T80A-S 5′-ccctggctctattggagctctatatatcaggctgg-3′ and T80A-AS 5′-ccagcctgatatatagagctccaatagagccaggg-3′. To construct L81A-FCoR, we used L81A-S 5′-cctggctctattggaactGCatatatcaggctggacttg-3′ and L81A-AS5′-caagtccagcctgatatatGCagttccaatagagccagg-3′. To construct L85A-FCoR, we used L85A-S 5′-ggaactctatatatcaggGCggacttgaactcacaaaga-3′ and L85A-AS5′-tctttgtgagttcaagtccGCcctgatatatagagttcc-3′. To construct L87A-FCoR, we used L87A-S 5′-ctatatatcaggctggacGCgaactcacaaagatccacc-3′ and L87A-AS5′-ggtggatctttgtgagttcGCgtccagcctgatatatag-3′.
Construction of adenoviral vectors and adenoviral transduction
To construct an adenoviral vector encoding FCoR, we amplified the cMyc-FCoR cDNA fragment using the pCMV5/cMyc-FCoR expression vector as a template and the following primers: 5′- GG GCTAGC (NheI) ATGGAGCAGAAGCTGATCAGC-3′ (sense) and 5′-GG GCTAGC (NheI) CTAGCACATGCCTTTAGTCCC-3′ (antisense). After treatment with NheI, the PCR fragment was subcloned into NheI-treated pShuttle2 vector (Clontech). After sequencing the vectors to confirm that they had the intended sequences, and confirming protein expression in HEK293 by transient transfection, an I-CeuI- and PI-SceI-treated fragment was subcloned into Adeno-X viral DNA (pAdeno-X-cMyc-FCoR) (Clontech). The adenovirus vector was generated by transfecting HEK293 cells with the pAdeno-X-cMyc-FCoR plasmid.
For knockdown of FCoR in 3T3-F442A cells, we used DNA-based adenoviral vector-mediated technology (Knockout RNAi Systems; Clontech Laboratories, Inc.), with 5′-GCATGTGCTAGCATGCATAGC-3′ and 5′-GCTAGCATGCATAGCCTAATG-3′ as the targeted sequences of shRNA1 and shRNA2, respectively. The sequences of shRNA1 and shRNA2 are 5′-GATCCG GCATGTGCTAGCATGCATAGC TTCAAGAGA GCTATGCATGCTAGCACATGC CTTTTTT TCTAGA G-3′ and 5′-GATCCG GCTAGCATGCATAGCCTAATG TTCAAGAGA CATTAGGCTATGCATGCTAGC CTTTTTT TCTAGA G-3′, respectively. We selected an RNAi target sequence for FCoR using the Block-iT RNAi Designer (Invitrogen). We used the BD Adeno-X Expression System 1 (BD Biosciences) to generate a recombinant adenovirus encoding FCoR-shRNA. The generation of adenovirus encoding the cCN Foxo1 (T24A/S253A/S316A) was described previously (Nakae et al, 2006). For induction of endogenous Igfbp1, SV40-transformed hepatocytes were transduced with adenovirus encoding CN Foxo1 as described previously (Nakae et al, 2006). For transduction of 3T3-F442A cells with adenovirus encoding FCoR-shRNA, 3T3-F442A cells were transduced with adenovirus as described previously (Nakae et al, 2003).
Western blotting and fractionation of cytoplasmic and nuclear proteins
We homogenized tissues and lysed cells in buffer containing 50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors (Roche Diagnostics). After centrifugation to remove insoluble material, the proteins in 30 μg of lysate were separated using 8 or 14% SDS–PAGE, and western blotting was performed using the indicated antibodies. Immunoprecipitation was performed as described previously (Cao et al, 2006). Fractionation of cytoplasmic and nuclear extracts was performed using NE-PER extraction reagents (Pierce). Protein concentrations in the cytoplasmic and nuclear extracts were determined using the Micro BSA protein assay kit (Pierce), and the proteins in aliquots (30 μg) were resolved by 14% SDS–PAGE followed by western blotting using the indicated antibody.
Detection of phosphorylated FCoR protein
For detection of phosphorylated FCoR, HEK293 cells were transfected with the indicated vectors encoding cMyc-FCoR. Cells were stimulated with forskolin (20 μM) for the indicated time and harvested using lysis buffer containing phosphatase inhibitor as described previously (Nakae et al, 2000). The proteins in the cell lysates were separated by 14% SDS–PAGE and transferred onto Immobilon-P Transfer Membrane (Millipore Corporation). After blocking, the membrane was incubated with Phos-tagR BTL-104 (NARD Institute Ltd.) according to manufacturer’s protocol followed by detection with streptavidin-horseradish peroxidase conjugate (ECL; Amersham, Buckinghamshire, UK). The membranes were reprobed as per manufacturer’s protocol.
In-vitro translation and glutathione S-transferase fusion protein pull-down assay
The wild-type Fcor cDNA fragment was generated by PCR using pCMV5/cMyc-FCoR expression vector as a template and the following primers: 5′-GG GAATTC (EcoRI) GGATGGGCGGTCCTACACGCCGCCAT-3′ (sense) and 5′-GG GAATTC (EcoRI) CTAGCACATGCCTTTAGTCCC-3′ (antisense). The fragment was cloned in-frame into the EcoRI site of pGEX-4T-2. In-vitro translation of Foxo1 and the pull-down assay were performed as described previously (Cao et al, 2006).
In-vitro acetylation assay
GST-Foxo1-C1 fusion protein (20 μg) was incubated at 30°C for 1 h with 4 μg of GST, GST-FCoR fusion protein, or 0.5 μg of recombinant p300 protein (ACTIVE MOTIF) and 25 nCi of [14C] acetyl-CoA (Perkin-Elmer) in the reaction buffer (50 mM Tris–HCl (pH 8.0), 100 mM NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 5 mM sodium butyrate). Reaction products were separated by 14% SDS–PAGE, stained with Coomassie brilliant blue, dried, exposed to a BAS 5000 imaging plate (FujiFilm, Japan) and analysed.
In-vitro phosphorylation assay
Five micrograms of GST, GST-WT FCoR, GST-T93A, GST-T93D, and GST-RRAS as a positive control (GST-RRAS is a short sequence of salt-inducible kinase (SIK) 2 and a substrate of PKA) were incubated at 30°C for 10 min with 0.5 units of the catalytic subunit of cAMP-dependent protein kinase (Promega) in the reaction buffer (50 mM Tris–HCl (pH 7.4), 0.1 mM DTT, 5 mM MgCl2, 0.5 mM ATP). Reaction products were separated by 14% SDS–PAGE and transferred onto a nylon membrane. Phosphorylated products were detected using Phos-tagR BTL-104 as described above and then blotted with anti-GST antibody.
Generation of adipose tissue-specific FCoR transgenic mice
We cloned wild-type Fcor cDNA into the SmaI site of plasmid pCMV5-aP2, into which the ClaI- and SmaI-treated 5.4-kb promoter-enhancer fragment of the mouse aP2 gene was subcloned into the ClaI/SmaI-treated pCMV5/cMyc vector (Nakae et al, 2008a). We excised the transgene with ClaI and XhoI, gel-purified it and injected it into fertilized eggs from BDF1 X C57BL/6 mice. The resulting embryos were implanted into CD-1 foster mothers. We screened the offspring for transgene transmission by PCR. Of 11 independent transgenic lines obtained, 9 founders transmitted the transgene through the germ line. We crossed each founder with C57BL/6 mice, obtained F1 mice, sacrificed one of the mice in each line in which transgene was transmitted and examined transgene expression in adipose tissues. We obtained three independent Fcor transgenic lines (Acc. No. CDB0446T: http://www.cdb.riken.jp/arg/TG%20mutant%20mice%20list.html) in which the transgene was expressed in adipose tissues. Primers for genotypying are 5′-ATG GAG CAG AAG CTG ATC AGC-3′ and 5′-CAT CCT TTC TCT GTG ATA CTG-3′.
Generation of FcorKO
The mouse genomic DNA clone containing exons 1 and 2 of the Fcor gene was obtained from a genomic DNA library derived from the 129/Sv mouse strain. The Fcor targeting construct consisted of 5.5 kb of genomic sequence that was immediately 5′ of the first exon, followed by a 4.5-kb LacZ-cassette, a Neo-resistance gene, and 3.5 kb of genomic sequence from intron 1, exon 2, and intron 2. The SpeI-linearized targeting vector was electroporated into TT2 embryonic stem (ES) cells (Yagi et al, 1993) as described previously. G418-resistant clones were isolated and screened by PCR and Southern blotting. In all, 30 out of 115 G418-resistant clones had undergone the desired homologous recombination. Positive clones were injected into CD-1 8-cell stage embryos, and the chimeric male offspring was mated to C57BL/6 females. Mice carrying the mutation in the heterozygous state were intercrossed to produce homozygous Fcor mutants (Acc. No. CDB0612K: http://www.cdb.riken.jp/arg/mutant%20mice%20list.html), and 20- to 24-week-old mice were used for analysis. Primers for detection of wild-type allele (204 bp) are 5′-ATGGGCGGTCCTACACGCCGC-3′ and 5′-CGGTGCAGTTCCCAGCCCTAC-3′. Primers for detection of knockout allele (234 bp) are 5′-AGCCCTTCCCCGCTGTGCACG-3′ and 5′-GGGTTTTCCCAGTCACGACGT-3′.
Animal studies, analytical procedures, IPGTT, and ITT
We used only male mice for the following experiments, as they are more susceptible to insulin resistance and diabetes. Animals were fed a standard chow diet and water ad libitum in sterile cages in a barrier animal facility with a 12-h/12-h light/dark cycle. Wild-type littermates were used as controls. All experimental protocols using mice were approved by the animal ethics committee of the Keio University School of Medicine and Kobe University Graduate School of Medicine. A HFD was started at weaning (4 weeks of age) and continued for 20 weeks. We used the same HFD and measured the blood glucose and insulin levels as described previously (Nakae et al, 2008a). We carried out all assays in duplicate. Each value represents the mean of two independent determinations. Glucose, ITT, and CT scanning were performed as described previously (Nakae et al, 2008a). The rectal temperature of mice was measured using Thermal SensorR (Shibaura Electronics Co. Ltd).
Luciferase assay
The IGFBP-1 or G6Pase luciferase assay was performed as described previously (Nakae et al, 2006). To construct a mouse Ppargc1a promoter-luciferase vector, mouse genomic DNA was amplified by PCR using the following primers: 5′-GG GCTAGC (NheI) TCATTGACTCAGGAACGACA-3′ (forward primer) and 5′-GG GCTAGC (NheI) CCAGTCACATGACAAAGCTA-3′ (antisense primer). After digestion with NheI, the PCR product was subcloned into NheI-treated pGL3-Basic vector (Promega) and sequenced to rule out the presence of mutations. For the mouse Ppargc1a promoter luciferase assay, HEK 293 cells were plated onto 12-well culture dishes. Transfections were carried out on cells at a 70–80% stage of confluence using 1.5 μg of pGL3/Basic-Ppargc1a reporter vector, and/or 0.6 μg of pCMV5/cMyc empty vector or pCMV5/cMyc-FCoR expression vectors. Synthetic Renilla luciferase reporter vector (phRL-SV40; Promega) (10 ng) was used as an internal control of transfection efficiency. After transfection, cells were cultured in DMEM containing 10% fetal calf serum and incubation was continued for 36 h. After that, cells were stimulated with forskolin (20 μM) for 6 h and the cells were harvested for luciferase assay.
To construct GAL4-Foxo1 expression vectors (PM-WT, -6KQ, and -6KR FoxO1), mouse Foxo1 cDNA was amplified by PCR using the pCMV5/cMyc-WT, -6KQ, or -6KR Foxo1 expression vectors as templates (Kitamura et al, 2005) and the following primers: 5′-GG GGATCC (BamHI) GT ATG GCC GAG GCG CCC CAG GTG GTG-3′ (sense) and 5′-GG GGATCC (BamHI) TTA GCC TGA CAC CCA GCT GTG TGT-3′ (antisense). After treatment with BamHI, PCR products were subcloned into a BamHI-treated pM vector (Clontech). To construct GAL4-Foxo3a, the Foxo3a cDNA fragment from EcoRI/HindII-treated PCMV5/cMyc/Foxo3a vector (Nakae et al, 2001) was ligated into an EcoRI/HindII-treated PM vector. To construct GAL4-FOXO4, human FOXO4 cDNA was amplified by PCR using PFN21AB7772 (Kazusa DNA Research Institute, Chiba, Japan) and the following primers: 5′-GG GTCGAC (SalI) GCATGGATCCGGGGAATGAGAATTCA-3′ (sense) and 5′-GG AAGCTT (HindIII) TCAGGGATCTGGCTCAAAGTTGAA-3′ (antisense). After treatment with SalI and HindIII, PCR products were subcloned into the SalI/HindIII-treated pM vector. Plasmids were sequenced to confirm that they had the intended sequences. The 5XGAL4-luciferase reporter plasmid (pTAL-5XGAL4) was described previously (Katoh et al, 2006). For the 5XGAL4-luciferase assay, HEK293 cells were plated onto 12-well dishes. When the cells showed 70–80% confluence, transfections were carried out using 1.5 μg of pTAL-5XGAL4, 0.3 μg of several kinds of pM vector, and 0.6 μg of pCMV5/cMyc empty vector or pCMV5/cMyc-FCoR expression vectors. The synthetic Renilla luciferase reporter vector (phRL-SV40; Promega) (10 ng) was used as an internal control for transfection efficiency.
For luciferase assays using the J3-tk-Luc vector, HEK 293 cells were plated onto 12-well culture dishes. When the cells showed 70–80% confluence, transfections were carried out using 0.5 μg of the J3-tk-Luc reporter vector, 0.1 μg of pcDNA3-hPPARγ, 0.1 μg of pcDNA3-hRXRα, 0.1 μg of pFLAG-CNFoxo1 and/or 1.0 μg of pCMV5/cMyc empty vector or pCMV5/cMyc-FCoR expression vectors. The synthetic Renilla luciferase reporter vector (phRL-SV40; Promega) (10 ng) was used as an internal control for transfection efficiency. All vectors have been described previously (Vu-Dac et al, 1995; Tachibana et al, 2005). After transfection, cells were cultured in DMEM containing 10% fetal calf serum and incubation was continued for 36 h with 10 μM of rosiglitazone for 24 h.
Chip assay
3T3-F442A cells were seeded onto 15 cm-culture dishes and induced to differentiate into mature adipocytes as described previously (Nakae et al, 2003). On day 4 (clonal expansion) and day 6 (end of clonal expansion) after induction, cells were trypsinized and fixed with 1% formaldehyde for 1 h at 37°C. The DNA solution for Chip PCR was prepared according to the protocol in the Chip Assay Kit (Upstate). We performed immunoprecipitation with anti-FCoR serum, anti-FOXO1 (H128; Santa Cruz Biotechnology, Inc.), or an equal amount of normal rabbit IgG (Santa Cruz Biotechnology, Inc.). We subjected the samples to PCR using the following primers: for mouse Cdkn1a, 5′-aggaggaagactgggcatgtc-3′ and 5′-gagttggatccctagtaaggc-3′. For a Chip assay of Ppargc1a promoter, T37i cells infected with adenovirus encoding cMyc-FCoR were fixed with 1% formaldehyde for 1 h at 37°C. The DNA solution for the Chip assay was prepared as described previously (Nakae et al, 2006). We used an anti-cMyc mouse monoclonal antibody (9E10; Santa Cruz Biotechnology, Inc.) for immunoprecipitation of exogenous cMyc-FCoR. We subjected the samples to PCR using the following primers for mouse Ppargc1a, S3: 5′-cctatgagatccacggaaag-3′ (sense) and 5′-cgctcctcatgtgatacatt-3′ (antisense), and S4: 5′-aatgtatcacatgaggagcg-3′ (sense) and 5′-ccagtcacatgacaaagcta-3′ (antisense).
Immunohistochemistry, immunofluorescence, and histological analysis
For histological analysis, we removed epididymal fat tissues from 20-week-old mice, fixed the specimens in 10% paraformaldehyde, and embedded them in paraffin. We mounted consecutive 10 μm sections on slides and stained them with haematoxylin and eosin. Measurement of WAT adipocyte size was performed using FLVFS-LS software (Flovel, Tokyo) by manually tracing at least 500 adipocytes for each genotype. Immunofluorescence using SV40-transformed hepatocytes or HEK293 cells was performed as described previously (Nakae et al, 2006). After transduction with adenoviruses encoding cMyc-FCoR and FLAG-3A FoxO1, cMyc-tagged FCoR was visualized in SV40-transformed hepatocytes with anti-cMyc monoclonal antibody and fluorescein isothiocyanate-conjugated anti-mouse IgG. FLAG-tagged CN FoxO1 was visualized with OctA-Probe (D-8) (sc-807, Santa Cruz Biotechnology, Inc.) and a Cy3-conjugated anti-rabbit IgG.
Statistics
We calculated descriptive statistics using ANOVA followed by Fisher’s test (Statview; SAS Institute, Inc.). P-values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr Alan R Saltiel (University of Michigan, Ann Arbour, Michigan, USA) for providing the 3T3-L1 cDNA library, Dr Bruce M Spiegelman (Dana-Farber Cancer Institute, Boston, MA, USA) for providing promoter-enhancer fragment of the mouse aP2 gene, Dr Tadahiro Kitamura (Laboratory of Metabolic Signal, Metabolic Signal Research Center, Gunma University, Japan) for providing the pCMV5-cMyc-6KQ and -6KQ FoxO1 expression vectors, and Dr Masao Doi and Dr Hitoshi Okamura (Department of System Biology, Kyoto University Graduate School of Pharmaceutical Science, Kyoto, Japan) for technical advice about the in-vitro acetylation assay. We also thank Dr Masato Kasuga (Research Institute, International Medical Centre of Japan, Tokyo, Japan) for helpful discussion. This work was supported by a grant for the 21st Century COE Program ‘CenteR of Excellence for Signal Transduction Disease: Diabetes Mellitus as a Model’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Grants-in-Aid for Scientific Research for Priority Areas No. 18052013 from the Ministry of Education, Science, Sports, and Culture in Japan to JN; by a grant from the Takeda Science Foundation to JN; by a grant from the Astellas Foundation for Research on Metabolic Disorders to JN; by a grant from Novo Nordisk Insulin Research to JN; and by a grant from Nippon Boehringer Ingelheim Co. Ltd. to HI.
Author contributions: JN, HT, and ST designed the experiments. JN, YC, FH, YK, RS, TA, and HK carried out the experiments. TT and JS prepared vectors for the J3-tk-luciferase assay. JN and ST analysed all data. HI gave detailed comments of the paper. JN wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB (2001) Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409: 729–733 [DOI] [PubMed] [Google Scholar]
- Accili D, Arden KC (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117: 421–426 [DOI] [PubMed] [Google Scholar]
- Armoni M, Harel C, Karni S, Chen H, Bar-Yoseph F, Ver MR, Quon MJ, Karnieli E (2006) FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem 281: 19881–19891 [DOI] [PubMed] [Google Scholar]
- Banks AS, Kim-Muller JY, Mastracci TL, Kofler NM, Qiang L, Haeusler RA, Jurczak MJ, Laznik D, Heinrich G, Samuel VT, Shulman GI, Papaioannou VE, Accili D (2011) Dissociation of the Glucose and Lipid Regulatory Functions of FoxO1 by Targeted Knockin of Acetylation-Defective Alleles in Mice. Cell Metab 14: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L (2006) C. elegans SIR-2.1 Interacts with 14-3-3 Proteins to Activate DAF-16 and Extend Life Span. Cell 125: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Cannon B, Houstek J, Nedergaard J (1998) Brown adipose tissue. More than an effector of thermogenesis? Ann NY Acad Sci 856: 171–187 [DOI] [PubMed] [Google Scholar]
- Cao Y, Kamioka Y, Yokoi N, Kobayashi T, Hino O, Onodera M, Mochizuki N, Nakae J (2006) Interaction of FoxO1 and TSC2 induces insulin resistance through activation of the mammalian target of rapamycin/p70 S6K pathway. J Biol Chem 281: 40242–40251 [DOI] [PubMed] [Google Scholar]
- Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A (2003) Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52: 642–649 [DOI] [PubMed] [Google Scholar]
- Dowell P, Otto TC, Adi S, Lane MD (2003) Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem 278: 45485–45491 [DOI] [PubMed] [Google Scholar]
- Fan W, Imamura T, Sonoda N, Sears DD, Patsouris D, Kim JJ, Olefsky JM (2009) FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J Biol Chem 284: 12188–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A (2005) FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24: 7410–7425 [DOI] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM (2003) Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 100: 15712–15717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Jiang M, Chambon P, Metzger D (2001) Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc Natl Acad Sci USA 98: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR (2007) SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 6: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, Witters LA, Nakamura H, Okamoto M (2006) Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. Febs J 273: 2730–2748 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Li P, Huntley J, Chang JP, Arden KC, Olefsky JM (2009) FoxO1 haploinsufficiency protects against high-fat diet-induced insulin resistance with enhanced peroxisome proliferator-activated receptor gamma activation in adipose tissue. Diabetes 58: 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D (2005) FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab 2: 153–163 [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Lazar MA (2009) New developments in adipogenesis. Trends Endocrinol Metab 20: 107–114 [DOI] [PubMed] [Google Scholar]
- Lochner A, Moolman JA (2006) The many faces of H89: a review. Cardiovasc Drug Rev 24: 261–274 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Han S, Kitamura T, Accili D (2006) Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest 116: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Barr V, Accili D (2000) Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. Embo J 19: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, Hayashi Y (2006) The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest 116: 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Cao Y, Oki M, Orba Y, Sawa H, Kiyonari H, Iskandar K, Suga K, Lombes M, Hayashi Y (2008a) Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 57: 563–576 [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, Accili D (2003) The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell 4: 119–129 [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Silver DL, Accili D (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest 108: 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Oki M, Cao Y (2008b) The FoxO transcription factors and metabolic regulation. FEBS Lett 582: 54–67 [DOI] [PubMed] [Google Scholar]
- Nakae J, Park BC, Accili D (1999) Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem 274: 15982–15985 [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK (2010) Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 72: 219–246 [DOI] [PubMed] [Google Scholar]
- Printen JA, Brady MJ, Saltiel AR (1997) PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science 275: 1475–1478 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423: 550–555 [DOI] [PubMed] [Google Scholar]
- Qiang L, Banks AS, Accili D (2010) Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem 285: 27396–27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Kobayashi Y, Tanaka T, Tagami M, Sugiyama A, Katayama T, Ueda C, Yamasaki D, Ishimoto K, Sumitomo M, Uchiyama Y, Kohro T, Sakai J, Hamakubo T, Kodama T, Doi T (2005) Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms. Nucl Recept 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y (1999) Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA 96: 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM (2005) Drosophila melted modulates FOXO and TOR activity. Dev Cell 9: 271–281 [DOI] [PubMed] [Google Scholar]
- Vu-Dac N, Schoonjans K, Kosykh V, Dallongeville J, Fruchart JC, Staels B, Auwerx J (1995) Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J Clin Invest 96: 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR 3rd, Fischer WH, Thomas JB, Montminy M (2011) A hormone-dependent module regulating energy balance. Cell 145: 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A (2006) SMK-1, an essential regulator of DAF-16-mediated longevity. Cell 124: 1039–1053 [DOI] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S (1993) A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem 214: 70–76 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hou H, Haller EM, Nicosia SV, Bai W (2005) Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J 24: 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.