Abstract
Protein phosphatase PP4C has been implicated in the DNA damage response (DDR), but its substrates in DDR remain largely unknown. We devised a novel proteomic strategy for systematic identification of proteins dephosphorylated by PP4C and identified KRAB-domain-associated protein 1 (KAP-1) as a substrate. Ionizing radiation leads to phosphorylation of KAP-1 at S824 (via ATM) and at S473 (via CHK2). A PP4C/R3β complex interacts with KAP-1 and silencing this complex leads to persistence of phospho-S824 and phospho-S473. We identify a new role for KAP-1 in DDR by showing that phosphorylation of S473 impacts the G2/M checkpoint. Depletion of PP4R3β or expression of the phosphomimetic KAP-1 S473 mutant (S473D) leads to a prolonged G2/M checkpoint. Phosphorylation of S824 is necessary for repair of heterochromatic DNA lesions and similar to cells expressing phosphomimetic KAP-1 S824 mutant (S824D), or PP4R3β-silenced cells, display prolonged relaxation of chromatin with release of chromatin remodelling protein CHD3. Our results define a new role for PP4-mediated dephosphorylation in the DDR, including the regulation of a previously undescribed function of KAP-1 in checkpoint response.
Keywords: checkpoint, DNA damage signalling, double-strand DNA break, phosphatase
Introduction
Genotoxic conditions leading to double-stranded DNA breaks (DSBs) or replication stress are hazardous by virtue of their ability to stimulate both genomic instability and cellular transformation. To prevent such detrimental consequences, organisms have evolved a complex response to genotoxic stress, generally initiated by the phosphatidylinositol-3 (PI3) kinase-like family of protein kinases. A vast network of ∼700 proteins, including DNA repair and replication proteins, are phosphorylated by these kinases in response to DSBs or replication stress (Matsuoka et al, 2007). The phosphorylated proteins include factors involved in DNA replication and repair, apoptosis and/or cell cycle progression. The functional consequences of phosphorylation have been studied for only a small subset of these factors, and in many cases these modifications impact upon the DNA damage response (DDR). There is increasing evidence that protein phosphatases play an important role in the DSB-induced signalling cascade (Lee and Chowdhury, 2011). Two recent studies investigating the dynamics of phosphorylation, following induction of DSBs, show that over one-third of the captured phospho-peptides were dephosphorylated within minutes of DNA damage (Bennetzen et al, 2010; Bensimon et al, 2010). These data suggest that phosphatases not only play a role in counteracting the DSB-induced phosphorylation later in the damage response but also play a primary role in initiating the repair process. Interestingly, this suggestion is consistent with our recent study showing that PP4 dephosphorylates the essential replication protein A (RPA) on the RPA2 subunit, immediately after DNA damage and that this dephosphorylation event is critical for efficient repair of DSBs (Lee et al, 2010).
In recent years, we and others have identified a role for the PP2A-like phosphatases (PP2AC, PP4C and PP6C) in the DNA damage response DDR (Chowdhury et al, 2005, 2008; Nakada et al, 2008; Mi et al, 2009; Douglas et al, 2010). The catalytic components of these enzymes are typically contained in dimeric or trimeric complexes with tissue/cell type-specific regulatory subunits conferring substrate specificity (Virshup, 2000; Shi, 2009). The catalytic subunit PP4C is significantly overexpressed in malignant lung and breast tissue, and PP4 deficiency impacts repair of DNA replication-mediated damage (Chowdhury et al, 2008; Wang et al, 2008). The role of PP4C in the DDR is broadly conserved in budding yeast, and its homolog, Pph3, impacts DNA repair and the cellular checkpoint response (Hastie et al, 2006; Keogh et al, 2006; O’Neill et al, 2007; Kim et al, 2010). Several putative PP4C-containing complexes have been identified in mammalian cells but their biological functions remain unclear (Gingras et al, 2005; Chen et al, 2008), and so far there are only a few bonafide substrates of PP4 (Zhang et al, 2005; Cha et al, 2008; Toyo-oka et al, 2008; Falk et al, 2010; Zhang and Durocher, 2010). Genetic deletion of the PP4 catalytic subunit PP4C in mice results in early embryonic lethality, underlining its importance in development and maintaining cell health (Shui et al, 2007). Therefore, systematic identification of PP4 substrates is necessary to elucidate its role in the DDR, and during development.
Due to the lack of consensus targeting motifs, identifying substrates of Ser/Thr phosphatases has been a major challenge(Lee and Chowdhury, 2011). In recent times, interaction-based approaches, that is, tandem affinity purification/mass spectrometry, have been the only successful comprehensive strategy enabling the identification of substrates for Ser/Thr phosphatases (Wakula et al, 2003; Gingras et al, 2005; Arroyo et al, 2008). Here, we devised a proteomic method to identify proteins de-phosphorylated by PP4 based on the rationale that phosphoproteins enriched in the absence of a phosphatase are putative substrates. Quantitative phosphoproteomics in the context of PP4 depletion revealed that KRAB-domain-associated protein 1 (KAP-1) is a putative substrate of PP4. KAP-1 (also known as TRIM28, KRIP-1 and TIF1β) is a transcriptional corepressor, which recruits several components of the gene silencing machinery, including heterochromatin protein 1 (HP1) and the chromodomain-helicase-DNA-binding protein 3 (CHD3), to specific genomic loci (Lechner et al, 2000; Schultz et al, 2001, 2002). KAP1 and HP1β interaction is compromised by phosphorylation of KAP1 at S473 (Chang et al, 2008), and this modification occurs in the mitotic phase of the cell cycle (Beausoleil et al, 2004; Chang et al, 2008). In response to DSBs there is rapid, but transient, ATM-mediated phosphorylation of KAP-1 at serine 824 (S824) both at DNA repair foci and throughout the nucleus (Ziv et al, 2006; Goodarzi et al, 2008; Noon et al, 2010). Whereas pan-nuclear pS824-KAP1 dissipates rapidly, pS824-KAP1 foci can persist for longer times. Phosphorylation of KAP-1 at S824 specifically impacts repair of DSBs in heterochromatin (Goodarzi et al, 2008, 2010, 2011; Noon et al, 2010).
The justification for pursuing a PP4 substrate would be provided by any data showing that removal of the phosphorylated form of the protein is necessary for restoration to a ‘normal’, pre-phosphorylation state. Interestingly, constitutive expression of phosphomimetic (S824D) KAP-1 has a distinct cellular phenotype to cells expressing wild-type (WT) KAP-1, with de-repression of several stress-response genes (Li et al, 2007, 2010) and global chromatin relaxation (Ziv et al, 2006). The short half-life of DSB-induced phospho-S824 KAP-1 and the functional consequences of expressing constitutively phosphorylated (S824D) KAP-1 suggested that regulated de-phosphorylation of KAP-1 may be necessary for restoring the ‘normal’ cellular state following DNA damage. Therefore, we investigated the link between PP4C and KAP-1. Biochemical and cytological studies confirmed that PP4C dephosphorylates KAP-1 at S824 and regulates its role in chromatin compaction and gene expression. Furthermore, we observe that CHK2-mediated phosphorylation of another KAP-1 residue, S473, plays a role in enforcing the G2/M checkpoint after ionizing radiation (IR). A PP4C/R3β complex dephosphorylates KAP-1 at S473 to facilitate cell cycle progress.
Results
Differential phosphoproteomics in PP4C-silenced cells
In an effort to broadly profile potential PP4C substrates, we utilized the proteomic strategy outlined in Figure 1A. Duplicate samples of nuclear proteins isolated from cells treated with control (scrambled siRNA) and siRNA targeting PPC4 were digested with trypsin and the resulting peptides were encoded with isobaric iTRAQ tags (Ross et al, 2004). Phosphopeptides were purified with iron-NTA IMAC (Ficarro et al, 2009a) and analyzed by LC-MS/MS (Ficarro et al, 2009b). Data from replicate samples were processed within our multiplierz software framework (Askenazi et al, 2009) and used to derive a 95% acceptance region (Zhang et al, 2010) for iTRAQ intensity ratios (Figure 1B). This approach allowed us to capture experimental variance at the level of enzymatic digestion. Based on this analysis, we identified significant hyperphosphorylation (P-value ?0.05; 197 peptides) in the context of PP4C depletion (Figure 1B). Interestingly, several of the corresponding proteins have well-established roles in the DDR, including KAP-1, CHD4 and 53BP1. In order to validate our phosphoproteomics approach and further explore the potential links between the DDR and subsequent phosphatase activity, we utilized a pan-phosphoserine antibody to immunoprecipate phosphoproteins from PP4C-silenced cells subjected to DNA damage. The immunoprecipitate was probed for KAP-1, 53BP1 and CHD4, along with RPA2, a known PP4 substrate, as a control. All these proteins showed a distinct enrichment in the immunoprecipitates from PP4C-silenced cells exposed to IR (Figure 1C). The enrichment in undamaged cells was detectable but moderate. To verify these results, we examined substrate phosphorylation using the mobility shift of phosphorylated proteins. To enhance the analysis, we used a phosphate-binding metal complex (phos–tag), which enhances the mobility shift of phosphorylated proteins during resolution by SDS–PAGE (Kinoshita et al, 2009). Control and PP4C-silenced cell lysates were analyzed using phos–tag. Consistent with Figure 1C, there was a detectable increase in the phosphorylated form of KAP-1, 53BP1 and CHD4 in the absence of PP4C (Figure 1D). Collectively, these findings verify the potential importance of our screening method and show that several DDR proteins are putative substrates of PP4C. Next, we focused on substantiating direct dephosphorylation and in vivo significance for a single substrate.
Figure 1.
PP4C influences the phosphorylation status of multiple DDR proteins. (A) Schematic for phosphoproteomics-based identification of putative PP4C substrates. (B) Identification of regulated phosphorylation in response to PP4C depletion. The geometric mean of iTRAQ spectral peak height ratios for replicate samples was plotted as a function of the sum of the geometric means of all iTRAQ spectral peak heights. Maximum approximate conditional likelihood (MACL) was used to determine an intensity-based variance function from which the 95% acceptance region was calculated (grey curve and Supplementary Table 1). Several DDR proteins, including KAP1, CHD4 and TP53BP1, exhibited hyperphosphorylation in response to deplection of PP4C. (C) Validation of the PP4 targets. (Left panel) HeLa S3 cells transfected with PP4C or scrambled siRNAs were exposed to IR, lysed after 2 h and immunoprecipitated (IP) using a pan-phosphoSer antibody and probed for DDR proteins as indicated. The relative band intensities are provided below each immunoblot. (Right panel) Proteins identified as putative PP4 substrates were confirmed using Phos–tag. HeLa S3 cells were transfected with PP4C siRNA and irradiated with 10-Gy IR. Lysates were subjected to SDS–PAGE containing 20 μM Phos–tag and immunobloted with indicated antibodies. Lysates treated with λ protein phosphatase (λPP) served as control for the Phos–tag-induced mobility shift.
KAP-1 is a substrate of a PP4/R3β complex
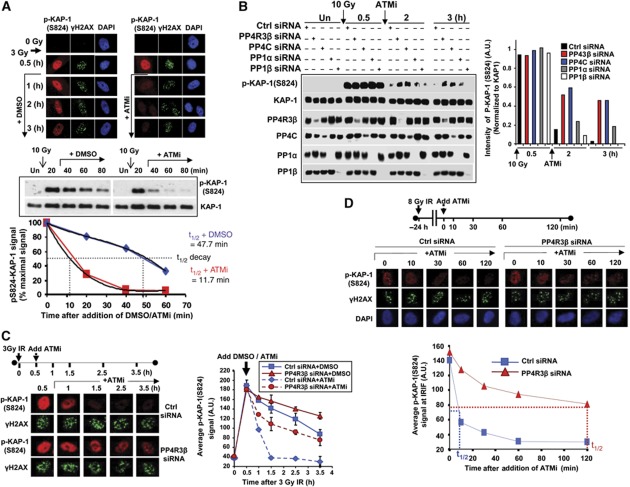
To investigate whether a PP4 complex impacts the phosphorylation of KAP-1 at S824, we silenced the regulatory subunits of PP4, and evaluated basal levels of phosphorylated KAP-1 using a phospho-S824-specific antibody. Although detectable by mass spectrometry, the basal pS824-KAP-1 level in cells without exogenous DNA damage is not visible by immunoblotting using 20–100 μg of cell lysate (Figure 2A). We used 200 μg of cell lysate to observe a pS824-KAP-1 signal in PP4-deficient cells. The increase in p S824-KAP-1 obtained by silencing PP4C was also observed by silencing PP4R3β (Figure 2A). Knocking down the other subunits, PP4R1, PP4R2 or PP4R3α, had a relatively modest effect on pS824-KAP-1 (Supplementary Figure 1). In response to IR there is a rapid increase in pS824-KAP-1, which peaks within 1 h, and significantly drops by 3 h. In the absence of either PP4C or PP4R3β there is a significantly higher amount of pS824-KAP-1 in cells 3 h after IR (Figure 2B). Similar results were obtained using other DNA damaging agents such as bleomycin and doxorubicin (Supplementary Figure 2). Furthermore, there is also persistence of focal pS824-KAP-1 in PP4R3β-silenced primary human fibroblasts most evident between 4–16 h post IR (Figure 2C).
Figure 2.
KAP-1 phosphorylation on S824 is regulated by a PP4C–R3β complex. (A) In unperturbed cells, phosphorylation of KAP-1 on S824 is elevated in PP4-depleted cells. HeLa cells transfected with siRNAs against indicated PP4 subunits were harvested after 72 h and immunoblotting was performed with the indicated antibodies. (B) HeLa cells transfected with siRNAs against PP4C or PP4R3β were irradiated and harvested at the indicated times and p-KAP-1 was assessed by immunoblot using phospho-KAP-1 antibody (phosphoSerine 824). The kinetics of pS824-KAP-1 formation was monitored after irradiation by loading 20 μg of cell lysate per lane. (C) PP4R3β depletion attenuates pS824-KAP-1 turnover after IR. Primary human fibroblasts were transfected with control or PP4R3β siRNAs. After 72 h, cells were irradiated, fixed at the indicated times and immunostained for pS824-KAP1 (red), γH2AX (green) and DAPI (blue). Right Panel: The average pS824-KAP-1 signal intensity per nucleus was quantified using ImageJ software. Data represent average and s.d. of three independent experiments. (D) PP4C/PP4R3β interacts with KAP-1. HeLa S3 cells stably expressing empty vector (FH-Ctrl), FH-tagged KAP-1 (top and bottom panel), and PP4C or PP4R3β (middle panel) were subjected to immunoprecipitation using anti-FLAG beads at indicated time points before or after IR. PP4R3β or control siRNAs were transfected in cells 72 h prior to immunoprecipitation (right panel, bottom). The immunoprecipitate was probed with antibodies against endogenous KAP-1, PP4C and PP4R3β as indicated. Figure source data can be found with the Supplementary data.
R3β mediates the interaction of KAP-1 and PP4C
We reasoned that a PP4C/R3β complex-mediated dephosphorylation of KAP-1 would require interaction of these proteins. We analyzed the association of these proteins by reciprocal immunoprecipitation/immunoblot assays using lysates from HeLa cells expressing FLAG and HA (FH)-tagged KAP-1, PP4C or PP4R3β. KAP-1 associates with PP4C and PP4R3β, and the interaction is enhanced by DNA damage (Figure 2D, upper and middle panel). To examine whether PP4C and PP4R3β independently associate with KAP-1, we silenced PP4R3β and observed that the interaction of PP4C with KAP-1 is dramatically reduced in the absence of PP4R3β (Figure 2D, lower panel). Together, these results suggest that a PP4C/R3β complex interacts with KAP-1 and regulates its phosphorylation status.
Impact of PP4C/R3β on pS824-KAP-1 is not due to ectopic activation of ATM
KAP-1 is a bonafide ATM substrate, and moderate (0–20 Gy) doses of IR do not induce the formation of pS824-KAP1 in ATM-deficient cells (Ziv et al, 2006; Noon et al, 2010). However, ectopic activation of ATM can potentially cause enhanced pS824-KAP-1 levels. PP4 dephosphorylates multiple proteins (H2AX and RPA2) (Chowdhury et al, 2008; Lee et al, 2010) involved in the DDR and it is feasible that PP4 deficiency indirectly activates ATM-induced DNA damage signalling, which in turn leads to the persistence of pS824-KAP-1. In that scenario, continued ATM activity would be necessary for maintaining the pS824-KAP-1 signal in PP4-deficient cells. To evaluate this possibility, we inhibited ATM immediately after IR-induced formation of pS824-KAP-1. The ATM inhibitor (ATMi) has an immediate impact on ATM activity (Shibata et al, 2011), and pS824-KAP-1 levels diminish rapidly after treatment with ATMi (Figure 3A). Depletion of PP4C, or PP4R3β, causes persistence of pS824-KAP-1 several hours after IR even in the presence of ATMi (Figure 3B). A recent study suggested that PP1β de-phosphorylated DNA damage-induced pS824-KAP-1, and PP1α de-phosphorylated basal pS824-KAP-1 (Li et al, 2010). Relative to control cells, there was a small increase in pS824-KAP-1 in PP1α- and PP1β-silenced cells, in the presence of ATMi (Figure 3B). This would suggest that the impact of PP1 on cellular levels of pS824-KAP-1 may be primarily due to inappropriate activation of ATM.
Figure 3.
Hyperphosphorylation of S824-KAP-1 in PP4-silenced cells is not due to ectopic activation of ATM. (A) pS824-KAP-1 is lost rapidly in the absence of ongoing ATM activity. (Upper panel) 1BR3 primary fibroblasts were irradiated with 3-Gy IR and, 0.5 h later, were incubated with either DMSO or 10 μM of ATMi and harvested at the indicated times. Cells were fixed and immunostained for pS824-KAP-1 (red), γH2AX (green) and DAPI (blue). (Lowerpanel) GM02188 lymphoblastoid cells were irradiated with 10-Gy IR and, 20 min later, were incubated with DMSO or ATMi. Cells were harvested at the indicated times, processed and immunoblotted for pS824-KAP-1 and total KAP-1 (top panel). Immunoblot signal was quantified and plotted (lower panel). The half-lives of signal decay were calculated. (B) PP4C/PP4R3β depletion attenuates pS824-KAP-1 turnover after IR independent of ATM activity. HeLa cells were transfected with siRNAs. After 72 h, cells were irradiated with 10-Gy IR. At 0.5 h post IR, DMSO or ATMi was added. Cells were harvested at the indicated times and immunoblotting was performed and band intensities quantified using the Odyssey Infrared Imaging System. The intensity of pS824-KAP-1 was normalized relative to total KAP-1 and represented graphically. (C) PP4R3β depletion attenuates pS824-KAP-1 turnover after IR independent of ATM activity. Primary human fibroblasts were transfected with siRNAs. After 72 h, cells were irradiated with 3-Gy IR. At 0.5 h post IR, DMSO or ATMi was added. Cells were fixed at the indicated times and immunostained for pS824-KAP1 (red), γH2AX (green) and DAPI (not shown). The data were quantified as in Figure 2C. (D) PP4R3β regulates pS824-KAP-1 at late-repairing IRIF. Primary human fibroblasts were transfected with control or PP4R3β siRNA. After 72 h, cells were irradiated with 8-Gy IR. After 24 h, ATMi was added (t=0 min) and cells were fixed at the indicated times up to 120 min. Fixed cells were then immunostained for pS824-KAP-1 (red), γH2AX (green) and DAPI (blue). The average pS824KAP-1 signal intensity (from (A)) overlapping with γH2AX foci (i.e., ‘at IRIF’) was quantified using ImageJ software. Data represent average and s.d. of three independent experiments. The approximate half-life (t1/2) of pS824-KAP-1 signal following the addition of ATMi is indicated. Data represent average and s.d. of three independent experiments. Figure source data can be found with the Supplementary data.
Cytological detection of pS824-KAP-1 shows a very distinct localization pattern (Goodarzi et al, 2010). Shortly after DNA damage, pS824-KAP-1 is detected throughout the nucleus, and the pS824-KAP-1 foci at DSBs are not distinctly visible. After several hours, pan-nuclear pS824-KAP-1 dissipates, and intense pS824-KAP-1 foci at late-repairing IR-induced foci (IRIF) are observable at sites marked by γ-H2AX foci and heterochromatic markers (Noon et al, 2010). Addition of ATMi shortly after IR leads to a rapid decrease in nuclear pS824-KAP1 in control cells. In agreement with the immunoblots shown in Figure 3B, PP4R3β-silenced cells show persistence of nuclear pS824-KAP-1 even in the presence of ATMi (Figure 3C). Importantly, pS824-KAP1 at late-repairing foci is critical for HC-DSB repair whereas pan-nuclear pS824-KAP1 appears to be dispensable (Noon et al, 2010). To study the impact of PP4R3β depletion on late pS824-KAP-1 foci, ATMi was added 24 h after a higher dose of IR (8 Gy). Under these conditions only late-repairing IRIF with pS824-KAP-1 foci are detectable. Consistent with previous results, pS824-KAP1 foci at late-repairing IRIF disappeared rapidly following the addition of an ATM inhibitor in control cells, while persisting selectively in the PP4R3β-depleted cells (Figure 3D). In all these experiments, the depletion of PP4R3β was observed only in cells transfected with the PP4R3β siRNA. These results strongly suggest that the PP4C–R3β complex directly dephosphorylates KAP-1, and that the persistence of pS824-KAP1 in PP4-depleted cells is not due to prolonged activation of ATM.
Functional impact of PP4-mediated dephosphorylation of SS824-KAP-1
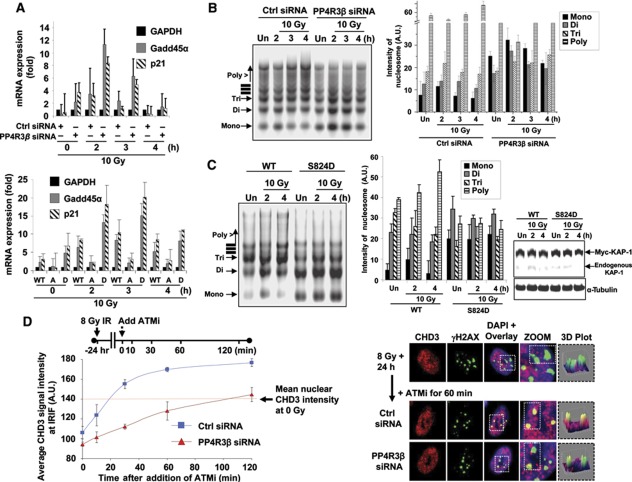
So far, two major functional consequences of expressing the phosphomimetic (S824D) KAP-1 mutant have been reported; one is the global de-condensation of chromatin, which is reflected by increased susceptibility to micrococcal nuclease (MNase) (Ziv et al, 2006) and the second is the ectopic de-repression of specific stress-response genes (Li et al, 2007, 2010). The molecular details of pS824-KAP-1-induced chromatin relaxation in the context of DSB repair has been recently elucidated. KAP-1 phosphorylation triggers the release of the nucleosome remodeler, CHD3, from DSB sites allowing localized de-condensation of chromatin (Goodarzi et al, 2011). We hypothesize that PP4 deficiency is functionally equivalent to expressing the phosphomimetic (S824D) KAP-1 mutant. To test this idea, we compared the expression level of KAP-1 target genes, p21 and Gadd45α, in PP4R3β-silenced cells, and cells where the endogenous KAP-1 had been replaced with the S824D- or S824A-KAP-1 mutant. IR-induced expression of p21 and Gadd45α is significantly enhanced by the depletion of PP4R3β or by the expression of phosphomimetic (S824D) KAP-1 mutant (Figure 4A). Conversely in cells expressing the phosphonull (S824A) KAP-1 mutant, these genes are not responsive to DNA damage. We also conducted chromatin relaxation assays using partial MNase digestion in the same set of cells. The depletion of PP4R3β, or expression of the S824D-KAP-1 mutant, caused a constitutive increase in cellular chromatin accessibility (Figure 4B and C). Consistent with previous data (Ziv et al, 2006), there is no additional de-condensation of chromatin after IR in cells expressing the S824D-KAP-1 mutant (Figure 4C). Continued ATM activity is required for the pS824-KAP-1-mediated dispersal of CHD3 from late-repairing IRIF (Goodarzi et al, 2011). We assessed the level of CHD3 at late IRIFs in PP4R3β-silenced cells (Figure 4D). As anticipated based on the previously observed persistence of pS824-KAP-1 following PP4R3β depletion (Figure 3D), silencing PP4R3β impedes the relocalization of CHD3 to late-repairing DSBs following the addition of ATMi. Together, these results strongly suggest that PP4-mediated dephosphorylation of KAP-1 S824 has a significant impact on its cellular function, and is necessary for an optimal DDR.
Figure 4.
PP4-mediated dephosphorylation of KAP-1 on S824 impacts its role in the DNA damage response. (A) PP4-mediated regulation of KAP-1 impacts the expression of Gadd45α and p21. HeLa cells were transfected with PP4R3β siRNA, or endogenous KAP-1 was replaced with WT, mutant forms A (KAP1-S824A) or D (KAP1-S824D). After IR, cells were harvested and RNA purified at indicated times and quantitative real-time PCR (qRT–PCR) was performed. Data represent average and s.d. of four independent experiments. (B, C) PP4R3β depletion and KAP1 phosphomimetic mutant (S824D) prolong damage-induced chromatin relaxation. HeLa cells were transfected with control or PP4R3β siRNA, or endogenous KAP-1 was replaced with WT or KAP1-S824D mutant (D). Cells were irradiated and harvested at indicated times. Nuclei were purified, treated with micrococcal nuclease; DNA isolated and analyzed as described in experimental procedures. The relative intensity of each nucleosomic form (mono, bi, tri or poly) is expressed as the percentage of the total signal (for a given lane). Bar graphs represent average and s.d. of three independent experiments. The KAP-1 replacement has been shown in the immunoblot with endogenous and Myc-tagged KAP-1 indicated. (D) The ‘return’ of CHD3 to sites of slow-repairing DSBs following ATMi addition is significantly attenuated in PP4R3β-depleted cells. Primary human fibroblasts were transfected with either control or PP4R3β siRNA. After 72 h, cells were irradiated with 8-Gy IR. At 24 h post IR, cells were treated with ATMi and harvested 10, 30, 60 and 120 min later. To examine the retention of CHD3 at sites of late-repairing damage, cells were pre-extracted with PBS containing 0.1% (v/v) Triton × 100 for 30 s before being fixed and immunostained for CHD3, γH2AX and DAPI (left panel). The signal intensity of CHD3 at regions of γH2AX foci (as determined by computer analysis) was measured (∼200 foci per sample). The data represent the mean and s.d. of multiple experiments. siRNA-mediated knock-down efficiency was independently verified for each experiment and was >80% (of cells with good knock-down) (right panel). It shows representative images at 60 min time point after ATMi addition from quantified data. In the zoomed-in and three-dimensional plotted images, note the relative difference in red–green overlap (yellow signal), indicative of changes in CHD3 abundance at sites of ongoing DSB repair, following the addition of ATMi for 1 h.
CHK2 phosphorylates KAP-1 at S473 in response to IR
The S473 residue on KAP-1 has been recently reported to be phosphorylated in response to DNA damage (Bennetzen et al, 2010; Blasius et al, 2011). In our proteomic analysis with PP4-depleted cells a phosphopeptide with the S473 residue was enriched (Supplementary Table 1). The comparison of KAP1 orthologues showed that the S473 residue is highly evolutionarily conserved, and this sequence fits the full consensus sequence for the checkpoint kinases (Figure 5A, upper panel). Inhibition of ATM impacts the phosphorylation of KAP-1 at S473 and S824 (Supplementary Figure 3). Since ATM activates CHK2 (Ahn et al, 2000; Cai et al, 2009), it is very likely that the impact of ATM on S473 is via CHK2. This is validated by the observation that silencing CHK2 (Figure 5A, lower panel) or treating cells with CHK2 inhibitors (Supplementary Figure 3) abrogates the IR-induced phosphorylation of S473 but does not impact S824.
Figure 5.
IR-induced phosphorylation of KAP1 on S473 is regulated by a combination of ATM/CHK2 and PP4C/R3β. (A) Alignment of region surrounding S473 of KAP1 across different organisms. Sequence alignment was performed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2) and consensus sequence is shown in color-shaded boxes (Upper panel). Phosphorylation of KAP-1 on S473 after IR is CHK2-mediated. HeLa cells after transfection with CHK1 or CHK2 siRNAs were irradiated after 3 days and harvested at indicated times (lower panel). Immunoblotting was performed with the indicated antibodies. (B) HeLa cells transfected with siRNAs for PP4C or PP4R3β were irradiated and harvested at the indicated times and pS473-KAP-1 was assessed by immunoblot using phospho-KAP-1 antibody (phosphoSerine 473). (C) PP4R3β depletion attenuates pS473-KAP-1 turnover after IR independent of Chk2 or ATM activity. HeLa cells transfected with siRNAs were irradiated after 3 days. At 0.5 h post IR, Chk2i (left panel) and ATMi (right panel) were added. Cells were harvested at the indicated times and immunoblotting was performed with p-KAP-1 antibody (S473). Figure source data can be found with the Supplementary data.
PP4C/R3β complex dephosphorylates pS473-KAP1
The kinetics of IR-induced phosphorylation of S473 is markedly different from the S824 residue but at all indicated time points there is elevated pS473-KAP-1 in the absence of either PP4C or PP4R3β (Figure 5B). As described for S824 (Figure 3B), we ruled out the possibility that continued S473 phosphorylation in PP4R3β-silenced cells is due to ectopic activation of ATM (Figure 5C, right panel) or CHK2 (Figure 5C, left panel) by treating cells with ATMi or CHK2 inhibitor immediately after the phosphorylation of S473.
Direct in vitro dephosphorylation of KAP-1 at S824 and S473
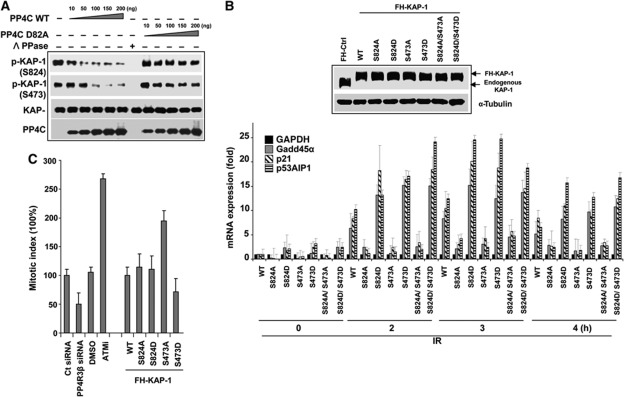
To determine whether PP4C can dephosphorylate pS824-KAP-1 and p-S473-KAP-1 directly, we immunopurified endogenous phospho-KAP-1 from IR-treated cells and performed dephosphorylation assays (Figure 6A). PP4C dephosphorylates both pS824-KAP-1 and p-S473-KAP-1 in a dose-dependent manner. Catalytically inactive PP4C mutant and λ phosphatase served as controls. Together, these results strongly suggest that PP4C directly dephosphorylates both KAP-1 phosphoresidues, S473 and S824, with comparable efficacy.
Figure 6.
PP4 directly dephosphorylates KAP-1 and impacts expression of stress-reponse genes and G2/M checkpoint. (A) PP4 dephosphorylates KAP1 in vitro. Wild-type PP4C and mutant PP4C (D82A) were purified using the baculoviral system and were serially diluted in the phosphatase reaction. λ phosphatase served as positive control for the reaction. PP4C dephosphorylates phospho-KAP1 on both S824 and S473 in a dose-dependent manner. Phosphatase reactions were probed with indicated antibodies. (B) PP4-mediated regulation of KAP-1 impacts the expression of Gadd45α, p21 and p53AIP1. In HeLa cells, endogenous KAP-1 was replaced with WT, mutant forms A (S824A or/and S473A) and D (S824D or/and S473D). After IR, cells were harvested and RNA purified at indicated times and quantitative real-time PCR (qRT–PCR) was performed. Data represent average and s.d. of four independent experiments. (C) Hyperphosphorylation of KAP1 on S473 impacts G2/M checkpoint. U2OS cells after depleting PP4R3β or replacing endogenous KAP-1 with wild-type, phosphonull mutants, or phosphomimetic mutants were exposed to irradiation (5 Gy) and then released in medium for 3 h and fixed. Mitotic cells were stained with anti-phospho-S10-Histone H3 (p-H3) antibody and quantified by flow cytometry. Data were analyzed with FlowJo software. The results from three independent experiments are graphically represented (S473A, P<0.029; S473D, P<0.011; PP4R3β, P<0.0321; ATMi, P<0.041). Figure source data can be found with the Supplementary data.
Dynamics of S473 phosphorylation impacts expression of stress-response genes and cell viability after IR
Phosphorylation of S473 was originally described in the context of cell cycle progression (Beausoleil et al, 2004; Chang et al, 2008) and correlated with diminished interaction of KAP-1 with HP1 and de-repression of the cyclin A2 gene (Chang et al, 2008). However, it was shown recently that damage-induced phosphorylation of S473 does not alter association of KAP-1 with HP1 (Blasius et al, 2011), and interestingly the phosphorylation of S824 and S473 occurs independent of each other (Blasius et al, 2011). Therefore, the impact of S473 on heterochromatin remains a contentious issue. Based on the function of KAP-1 as a transcriptional repressor and our observation with S824 phosphomutants (Figure 4A), we reasoned that phosphorylation of S473 may allow the de-repression of KAP-1 target genes involved in the stress response. The expression level of KAP-1 target genes, p21 Gadd45α and p53AIPI, was assessed in cells where the endogenous KAP-1 had been replaced with the single KAP-1 mutants, S824D, S473D, S824A or S473A, or the double KAP-1 mutants, S473D–S824D or S473A–S824A (Figure 6B). IR-induced expression of KAP-1 target genes were significantly and comparably enhanced by the expression of phosphomimetic S824D or S473D-KAP-1 mutant (Figure 6B). However, mutating both residues did not have an additive impact on gene expression. Expressing the phosphonull S824A or S473A-KAP-1 mutant, individually or in combination, represses the DNA damage-induced transcription of these genes. This result suggests that the expression of these stress response genes is tightly regulated by KAP-1 phosphorylation, where phosphorylation of both S473 and S824 is necessary for de-repression, and PP4C/R3β-mediated dephosphorylation of these residues restores the repressed state. We assessed the impact of both S824 and S473 on cell survival in response to IR using clonogenic assays. Mutation of each residue, individually or in combination, had a significant impact on viability after IR (Supplementary Figure 4).
CHK2-mediated phosphorylation of S473 influences the G2/M checkpoint
CHK2 exerts checkpoint control by phosphorylating its substrates, therefore it is fair to hypothesize that CHK2-mediated phosphorylation of KAP-1 may also contribute to the cellular checkpoint response. To evaluate whether phosphorylation of KAP-1 influences the G1/S checkpoint, cells expressing the KAP-1 phosphomutants (S824D, S824A, S473D and S473A) were exposed to IR and transition to S-phase monitored by BrdU incorporation. Although there was a significant drop in BrdU staining after IR (indicating an effective G1/S checkpoint), there was no difference between control cells and cells expressing the KAP-1 phosphomutants (Supplementary Figure 5). The ATM/CHK2 signalling pathway is the key enforcer of the G2/M checkpoint (Smith et al, 2010) and PP4C depletion delays the transition of cells to mitosis (Nakada et al, 2008; Lee et al, 2010). To assess the impact of CHK2- and PP4-mediated regulation of KAP-1 phosphorylation on the G2/M checkpoint, PP4R3β-silenced cells or cells expressing the KAP-1 phosphomutants were exposed to IR, and the mitotic index determined by analyzing expression of phospho-H3 after a 3-h incubation. Cells expressing KAP-1-wt or the KAP-1- S824D or S824A mutants had a G2/M checkpoint similar to control cells, whereas the significant proportion of cells expressing the KAP-1-S473D mutant had a prolonged G2/M checkpoint. Conversely, the KAP-1-S473A mutant appears to have a ‘leaky’ checkpoint with a significantly higher percentage of cells in mitosis (Figure 6C). Consistent with the phenotype of S473D mutant, we found that PP4R3β-silenced cells had an extended checkpoint with ∼50% reduction of cells in mitosis after IR (Figure 6C). Cells pre-treated with ATMi served as a control for this experiment. These results strongly suggest that CHK2-mediated phosphorylation of KAP-1 at S473 has a functional impact on DDR, which is distinct from the ATM-mediated phosphorylation of S824. Importantly, a PP4C/R3β complex dephosphorylates both these residues to broadly regulate the function of KAP-1 in the DDR.
The phenotype induced by silencing PP4R3β is mediated by KAP-1
Depletion of PP4C/R3β has an impact on DDR that is comparable to the phenotype induced by the expression of phosphomimetic (S824D or S473D) KAP-1 mutants (Figures 4 and 6). However, it is not clear that the impact of PP4R3β silencing on KAP-1 target genes, or on the G2/M checkpoint, is mediated directly by KAP-1. Theoretically if indeed the function of PP4R3β was mediated by KAP-1, then the phenotype induced by silencing PP4R3β would be ‘rescued’ by expressing the KAP-1 phosphonull mutants (S824A, S473A or both). Consistent with this notion, the ectopic induction of KAP-1 target genes in PP4R3β-silenced cells was completely blocked when the WT KAP-1 was replaced with S824A, S473A or S824A/S473A KAP-1 mutants (Figure 7A). Typically, PP4R3β-silenced cells have a prolonged G2/M checkpoint with fewer cells entering mitosis after IR. However, this phenotype is selectively reversed by the expression of the S473A KAP-1 mutant and not by the S824A KAP-1 mutant (Figure 7B). In combination, these results strongly suggest that a PP4C–R3β complex impacts the DDR by directly dephosphorylating KAP-1 at S824 and S473.
Figure 7.
KAP-1 phosphonull mutants reverse the phenotype induced by silencing PP4R3β. (A) In cells expressing phosphonull KAP-1 mutants, PP4 depletion does not affect the expression of Gadd45α, p21. In HeLa cells, endogenous KAP-1 was replaced with WT, mutant forms A (S824A or/and S473A), and either control siRNA or PP4R3β siRNA was transfected. After IR, cells were harvested and RNA purified at indicated times and quantitative real-time PCR (qRT–PCR) was performed. Data represent average and s.d. of four independent experiments. (B) KAP-1 S473A mutant, not S824A, reverses G2/M checkpoint phenotype induced by PP4R3β depletion. The G2/M checkpoint assay was performed as described in Figure 6D after silencing PP4R3β with U2OS cells expressing wild-type or phosphonull KAP-1 mutants. The results from three independent experiments are graphically represented (WT/PP4R3β siRNA, P<0.041; S824A/PP4R3β siRNA, P<0.011; S473A/Ctrl siRNA, P<0.019; S473A/PP4R3β siRNA, P<0.009). (C) Working model. In response to IR, ATM directly phosphorylates KAP-1 at S824 and indirectly induces the phosphorylation at S473 via CHK2. Phosphorylation of both S824 and S473 regulates chromatin structure and the expression of KAP-1 target genes, whereas only S473 is involved in the checkpoint response. A PP4 complex dephosphorylates KAP-1 at S824 and at S473 impacting the role of KAP-1 in DDR.
Discussion
In the human genome, there are 40 potential Ser/Thr phosphatases to counter the activity of 428 kinases known or predicted to phosphorylate Ser/Thr residues (Douglas et al, 2010). Therefore, it is fair to assume that each Ser/Thr protein phosphatase has a large number of substrates. However, there are no standard methods to comprehensively identify proteins dephosphorylated by a Ser/Thr protein phosphatase. We have devised an unbiased proteomic approach to identify PP4C targets, and this strategy if appropriately optimized may be broadly applicable. We obtained a list of phosphoproteins enriched in the absence of PP4C, which are therefore putative substrates of the phosphatase. The utility of this method will be determined by the proportion of direct targets obtained from the initial list. We are currently in the process of systematically validating the targets, and evaluating the global efficacy of this new approach. We mined the initial analysis to identify KAP-1 as a putative target of PP4C, and demonstrate that PP4-mediated de-phosphorylation of KAP-1 regulates its role in the DDR. Our results now show that the phospho-signalling network centering KAP-1 is initiated by ATM at one end and balanced by a PP4C/R3β complex at the other end. Although it is widely accepted that phosphorylation of KAP-1 is necessary for the efficient repair of DSBs, our data underscore the point that the PP4 complex-mediated de-phosphorylation of KAP-1 may play an equally important role in the DDR.
A mechanistic understanding of DNA repair in heterochromatin remains largely unclear, but the phosphorylation of KAP-1 is a key step in this otherwise opaque process. Concentrated localization of ATM at DNA lesions, and consequent phosphorylation of S824-KAP-1 at DSBs, is essential for heterochromatic DSB repair (Noon et al, 2010). Lack of factors (such as 53BP1, RNF168, etc) that recruit, and/or stabilize, ATM activity at IRIF impede the formation and maintenance of pS824-KAP-1 at these sites (Noon et al, 2010). pS824-KAP-1’s precise function at DSB sites is still emerging, but the chromatin relaxation phenotype of cells expressing the phosphomimetic (S824D) KAP-1 mutant suggests that pS824-KAP1 at DSBs ‘loosens’ the tightly compacted heterochromatin, and potentially enables the function of DNA repair factors at these regions. This idea is further supported by recent data showing the pS824-KAP1-mediated release of CHD3 from DSB sites facilitates DNA repair (Goodarzi et al, 2011). The condensed, transcriptionally inactive status of heterochromatin is tightly regulated, and it is conceivable that DNA damage-induced de-compaction needs to be countered rapidly after the repair is complete. Here, we provide evidence that the PP4C/PP4R3β, phosphatase complex, counteracts heightened ATM activity, and dephosphorylates pS824-KAP1 to restore heterochromatin to its pre-DNA damage state. This is an important step towards understanding how ‘normal’ heterochromatin is restored after DDR.
Although the phosphorylation of KAP-1 at S473 has been recently reported, the functional implications of this modification have not been described. We provide the first evidence that CHK2-mediated phosphorylation of the S473 residue influences the expression of KAP-1 target genes involved in the stress response. More importantly, the phosphorylation of S473 has a significant impact on the G2/M checkpoint. A role in checkpoint control adds a new dimension to the function of KAP-1 in the DDR. Although the ATM/CHK2 signalling pathway is involved in the phosphorylation of both S824 and S473, our results clearly distinguish the impact of these two phosphoresidues on the DDR (Figure 7C).
In the absence of the PP4 complex, p-KAP-1 levels are gradually diminished albeit with slower kinetics than control cells. This would suggest that some proportion of IR-induced p-KAP-1 may be removed independently of PP4 by other mechanisms (such as selective degradation) or by other phosphatases. A recent study has suggested that PP1α dephosphorylates basal pS824-KAP-1 and PP1β dephosphorylates DNA damage-induced pS824-KAP-1 (Li et al, 2010). However, this study observed that depletion of PP1β had no impact on pS824-KAP-1 levels, and silencing PP1α caused a moderate increase in pS824-KAP-1 independent of DNA damage (Li et al, 2010). Contrary to the expectations, PP1α/β interacts with wild-type KAP-1 and the phosphonull (S824A) KAP-1 mutant, but does not interact with the phosphomimetic (S824D) KAP-1 mutant. We find that in the presence of the ATM inhibitor, depletion of PP1α/β has minimal impact on pS824-KAP1. These results would suggest that PP1 may indirectly impact pS824-KAP-1 levels, but it is unlikely to directly de-phosphorylate pS824-KAP-1 generated by DNA damage. Like the PI3-like kinases, the PP2A-like phosphatases have overlapping substrates and roles in the cellular DDR. Therefore, it is feasible that PP2AC and/or PP6C may be involved in dephosphorylating p-KAP-1. Future studies will elucidate the mechanism by which other factors, including phosphatases, contribute to the removal of phosphorylated KAP-1.
Materials and methods
Cell cultures, antibodies and reagents
HeLa S3, HeLa and primary human fibroblasts were grown in DMEM supplemented with 10% (v/v) FBS. Antibodies used were against KAP-1 (BD Transduction Laboratories), pS824-KAP-1 (Bethyl), pS473-KAP-1 (BioLegend), 53BP1 (Cell signaling), CHD3 (Bethyl), CHD4 (Bethyl), RPA2 (Cell Signaling), PP4R1 (Bethyl), PP4R2 (Bethyl), PP4R3α (Bethyl), PP4R3β (Bethyl), PP4C (Bethyl), PP1α (Bethyl), PP1β (Bethyl), CHK1 (Cell Signaling), CHK2 (Cell Signaling), Flag-tag (Sigma), α-tubulin (Sigma) and Phoshphoserine-agarose conjugate (Abcam). CHK1 inhibitor (SB218078) was obtained from Calbiochem. CHK2 inhibitor, Doxorubicin and Bleomycin were obtained from Sigma-Aldrich. Phos–tag was purchased from the Phos-tag Consortium (http://www.phos-tag.com/english/index.html).
Phosphoproteomics
Nuclei were fractionated from control and PP4C knock-down cells following a hypotonic fractionation protocol (Dignam et al, 1983). Nuclear extracts were prepared using four volumes of lysis buffer [150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 0.5% NP40 and 10% glycerol, containing a cocktail of phosphatase (Sigma) and protease inhibitors (Roche)]. For each sample (Ctrl and PP4C knock-down), two 100-μg aliquots of nuclear extracts were denatured using 0.1% RapiGest SF (Waters) and reduced with 10 mM DTT for 45 min at 56°C. Reduced cysteines were alkylated with 20 mM iodoacetamide at room temperature and in the dark for 30 min. Proteins were digested with 5 μg of trypsin overnight at 37°C. RapiGest was removed following the manufacturer’s recommendation. Tryptic peptides were de-salted on a micro-elution plate (Waters), recovered with 100 μl of a 80%/0.1% acetonitrile/TFA solution and dried by vacuum centrifugation. Each of the four samples (replicate control (scrambled siRNA) and replicate PP4C knock-down) was solubilized in 30 μl of 500 mM tri-ethyl ammonium bicarbonate and labelled with an isoform of iTRAQ reagent for 1 h (114 and 115 for the two control samples, 116 and 117 for the two PP4C knock-down samples). The four samples were concentrated by vacuum centrifugation, combined and purified as described above. Phosphopeptides were enriched as described previously (Ficarro et al, 2009a) using Magnetic NTA beads (Qiagen) and analyzed by LC-MS/MS on an LTQ-Orbitrap Velos (Thermo). The data files were processed in our multiplierz software framework (Askenazi et al, 2009; Parikh et al, 2009; Webber et al, 2011) to provide separate peak lists (.mgf files) for the CAD and HCD MS/MS spectra. The two files were submitted to Mascot (v2.3) and searched against all 34048 entries in a concatenated reverse–forward human NCBI RefSeq database (released on 11/08/2010).
The search parameters were set as follows: The mass tolerance values for fragment ions were set at 0.6 Da and 0.02 Da for CAD and HCD spectra, respectively. Other common search parameters were set as follows: maximum of two missed cleavages; fixed modifications for cysteine (carbamidomethyl), peptide N-terminus and lysine residues (iTRAQ4plex); and variable oxidation on methionine and phosphorylation on serine and threonine. The search CAD and HCD results were merged using.mzScripts. We allowed a 5% false discovery rate (FDR) for peptides retrieved with identical sequences from the corresponding CAD and HCD scans. Peptides with discordant sequences retrieved from the CAD and HCD scans were discarded. Peptides identified in only one scan type were retained only if they had an FDR score below 1%.
Pairs of reporter ions corresponding to replicate measurements (114/115 and 116/117) were first corrected to obtain a median ratio of 1:1. For each phosphopeptide, we calculated the mean ratio for each pair of reporter ions. Statistics for these combined phosphopeptide iTRAQ ratios were calculated as described (Zhang et al, 2010). For clarity, we do not display phosphopeptides with missed tryptic cleavages or oxidized methionine residues in Figure 1B.
Phosphoprotein detection with Phos–tag
HeLa S3 cells were transfected with either control or PP4C siRNA. After 72 h, cells were lysed and the lysates were subjected to SDS–PAGE on 3% (w/v) polyacrylamide gel strengthened with 0.5% (w/v) agarose containing 20 μM Phos–tag, followed by immunoblotting with indicated antibodies.
siRNAs and transfection
siRNA duplexes (Invitrogen) were transfected using Lipofectamine 2000 (Invitrogen). The PP4 siRNAs were described previously (Chowdhury et al, 2008; Lee et al, 2010). The other siRNAs were as follows:
PP1α, siRNA #1 sense 5′-UAUUUCUUGGCUUUGGCGGAAUUGC-3′, antisense 5′-GCAAUUCCGCCAAAG CCAAGAAAUA-3′.
PP1β, siRNA #1 sense 5′-UUCUGCUUCAGUC AUCUGCACAAUC-3′, antisense 5′-GAUUGUGCAGAUGACUGAAGCAGAA-3′.
KAP-1, siRNA #1 sense 5′-GAGGACUACAACCUUAUUGUUAUUG-3′, antisense 5′-CAAUAACAAUAAGGUUGUA GUCCUC-3′.
To replace endogenous KAP-1 with WT or mutants, HeLa cells were transfected with siRNA-resistant KAP-1 plasmids using Lipofectamine 2000, and after 30 h, cells were reverse transfected with KAP-1 siRNA by RNAiMAX (Invitrogen) for 24 h.
Plasmids
siRNA-resistant KAP-1 constructs were made by mutating the siRNA target site to 5′GAGGACTACAATTTGATTGTTATTG-3′. KAP-1 mutants were constructed by QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Primers used were the following:
S824A-F, 5′-CTGGTGCTGGCCTGAGTGCCCAGGAGC-3′;
S824A-R, 5′-GCTCCTGGGCACTCAGGCCAGCACCAG-3′;
S824D-F, 5′-GCCTGGTGCTGGCCTGAGTGACCAGGAGCTG-3′;
S824D-R, 5′-CAGCTCCTGGTCACTCAGGCCAGCACCAGGC′-3′;
S473A-F, 5′-AAACGGTCCCGCGCAGGTGAGGGCGA-3′;
S473A-R, 5′-TCGCCCTCACCTGCGCGGGACCGTTT-3′;
S473D-F, 5′-AACGGTCCCGCGATGGTGAGGGCGA-3′;
S473D-R, 5′-TCGCCCTCACCATCGCGGGACCGTT-3′;
KAP-1 siRNA-resistant-F, 5′-CTGAGGACTACAATTTGATTGTTATTGAACGTG-3′;
KAP-1 siRNA-resistant-R, 5′-CACGTTCAATAACAATCAAATTGTAGTCCTCAG-3′.
Immunofluorescence
Cells plated on glass slides were fixed for 10 min with fixative (3% (w/v) PFA, 2% (w/v) sucrose and 1 × PBS) and permeablized for 1 min with 0.2% (v/v) Triton X-100 in PBS. Cells were rinsed with PBS and incubated with primary antibody diluted in PBS+2% (w/v) BSA for 1 h at room temperature (RT). Cells were washed three times, incubated with secondary antibody (diluted in PBS+2% (w/v) BSA) for 30 min at RT in the dark, incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min and washed three times with PBS. Slides were mounted using Vectasheild and visualized using a Zeiss Axioplan microscope and Simple-PCI software. Simple-PCI software analysis was used for experiments quantifying nuclear or sub-nuclear levels of IF signal. The anti-KAP-1 phosphoserine 824 antibody is an affinity-purified, polyclonal rabbit antibody recognizing amino acids 820–830 of human KAP-1 with a phosphoserine at 824. Identical results were achieved using commercial anti-KAP-1 pS824 from Bethyl, USA. Anti-γH2AX is ab18311 from Abcam. Appropriate secondary antibodies were HRP, FITC, Cy3 or TRITC conjugates from Sigma.
Co-immunprecipitation
HeLa S3, expressing FH-PP4C, FH-PP4R3β or FH-KAP-1, were lysed in buffer containing 50 mM Tris–HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% (v/v) NP-40 and protease inhibitor cocktail (Roche). Anti–Flag-agarose (Sigma) or anti–c-Myc antibody (+ protein A/G PLUS agarose (Santa Cruz)) were incubated with lysate at 4°C for 16 h. Immunocomplexes were washed three times with buffer containing 50 mM Tris–HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA and 0.5% (v/v) NP-40. The immunoprecipitated proteins were resolved by SDS–PAGE and analyzed by immunoblot. For immunoprecipitation with anti-phosphoserine-agarose, we used control and PP4C-depleted HeLa S3 cells.
Quantitative PCR
Quantitative PCR was used to analyze RNA isolated from HeLa cells, where the endogenous KAP-1 has been replaced with indicated mutants, and also from PP4R3β-silenced cells, using protocols described previously (Moskwa et al, 2011). Gene-specific primers used are as follows:
p21-F, 5′-TTTCTCTCGGCTCCCCATGT-3′; p21-R, 5′-GCTGTATATTCAGCATTGTGGG-3′; Gadd45α–F, 5′-AGGAAGTGCTCAGCAAAGCC-3′; Gadd45α–R, 5′-GCACAACACCACG TTATCGG-3′.
Chromatin relaxation assay
HeLa cells (PP4R3β-silenced or expressing KAP-1-S824D mutant) were used for this experiment. Cells were treated with NCS, resuspended in 10 V HBSS buffer (340 mM sucrose, 15 mM Tris pH 7.5, 15 mM NaCl, 60 mM KCl, 10 mM DTT, 0.15 mM, Spermine, 0.5 mM Spermidine and protease inhibitors) and incubated at 4°C for 10 min with periodic vortexing before centrifugation at 11 000 r.p.m. for 10 min at 4°C. Discarding supernatant, pellets containing nuclei were resuspended in 5 V HBSS, vortexed, spun as above and supernatants removed. Repeat this wash step two more times. Washed nuclei were resuspended in 5 V 1:1 HBSS+glycerol and stored at −20°C until next step. Nuclei were pelleted as before and resuspended in 250 μl (for ∼50 μl pellets) of 1 × MNase digestion buffer (60 mM KCl, 15 mM NaCl, 15 mM Tris pH 7.5, 250 mM sucrose, 1 mM CaCl2 and 0.5 mM DTT). Digestion reactions were initiated by adding 5 U Nuclease S7 (Roche) and incubating at 25°C for up to 10 min with occasional gentle agitation. Every few minutes, 40 μl of resuspended nuclei was removed to tubes containing 1.5 μl 0.5 M EDTA (pH 8.0) to terminate the reaction (the tubes were prepared in advance). When all samples were collected, 8 μl 5%(w/v) SDS+1 mg/ml Proteinase K was added, mixed and incubated at 37°C for 30 min. Each sample was diluted with 200 μl MNase buffer, 250 μl 25:24:1 phenol:chloroform:isoamyl-alcohol was added, vortex, spinned 14 000 r.p.m. for 2 min and supernatant was transfered to a clean tube. About 1 ml water saturated ether was added (prepared by mixing 10 ml water per 30 ml ether), vortex, spinned 14 000 r.p.m. for 2 min and supernatant was discarded. Ether wash was repeated. About 25 μl 3 M NaAcetate was added (pH 5.8), vortex, 750 μl cold ethanol was added and stored at −20°C for >16 h. Centrifuged at 14 000 r.p.m. for 20 min at 4°C and supernatant was discarded. Pellets were washed with 70% ethanol, dried and resuspended in 30 μl water. [DNA] was measured and 5–10 μg on 1.2% agarose gels in 1 × TAE was resolved.
In vitro dephosphorylation assay
We performed the in vitro dephosphorylation assay as described (Lee et al, 2010). Anti-flag beads were used to isolate p-KAP-1 from HeLa S3 cells stably expressing FH-KAP-1 after irradiation.
G2/M checkpoint assay
U2OS cells (PP4R3β-silenced or expressing KAP-1 mutants) were irradiated (5 Gy). After 3 h recovery in medium, cells were fixed in 4% formaldehyde and permeabilized in cold 90% methanol. Approximately 0.5 × 105 cells were stained with Phospho-Histone H3 (Cell Signaling, 1:100) for 1 h at room temperature and then stained with Alexa488-conjugated donkey anti-rabbit Ig at room temperature, followed by propidium iodide (PI/RNase staining buffer, BD Biosciences) staining. Cellular fluorescence was monitored with flow cytometry. Two-dimensional dot plots were generated by FloJo software.
Supplementary Material
Acknowledgments
DC is supported by R01CA142698 (NCI), JCRT and RSG-12-079-01 (ACS). D-HL was supported by NIH-training grant (CA 009078-34). AAG was supported by a grant from the Association for International Cancer Research to the PAJ laboratory. The PAJ laboratory is additionally supported by a grant from the Medical Research Council.
Author contributions: D-HL performed most of the experiments with help from YP and GOA. Most of the microscopy was done by AAG. JAM, PAJ and DC designed the experiments, and DC wrote the paper with the proteomic analysis provided by JAM. PAJ, AAG and JAM helped with edits.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE (2000) Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 60: 5934–5936 [PubMed] [Google Scholar]
- Arroyo JD, Lee GM, Hahn WC (2008) Liprin alpha1 interacts with PP2A B56gamma. Cell Cycle 7: 525–532 [DOI] [PubMed] [Google Scholar]
- Askenazi M, Parikh JR, Marto JA (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods 6: 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9: 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y (2010) ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3: rs3. [DOI] [PubMed] [Google Scholar]
- Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP (2011) A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol 12: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Chehab NH, Pavletich NP (2009) Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell 35: 818–829 [DOI] [PubMed] [Google Scholar]
- Cha J, Chang SS, Huang G, Cheng P, Liu Y (2008) Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J 27: 3246–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC (2008) Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC Mol Biol 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GI, Tisayakorn S, Jorgensen C, D’Ambrosio LM, Goudreault M, Gingras AC (2008) PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem 283: 29273–29284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J (2005) Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell 20: 801–809 [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J (2008) A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell 31: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP (2010) Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol 30: 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JE, Chan AC, Hoffmann E, Hochwagen A (2010) A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev Cell 19: 599–611 [DOI] [PubMed] [Google Scholar]
- Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA (2009a) Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem 81: 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, Smith ED, Boyer L, Schlaeger TM, Luckey CJ, Marto JA (2009b) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal Chem 81: 3440–3447 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R (2005) A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics 4: 1725–1740 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo P, Lobrich M (2010) The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair (Amst) 9: 1273–1282 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Hastie CJ, Vazquez-Martin C, Philp A, Stark MJ, Cohen PT (2006) The Saccharomyces cerevisiae orthologue of the human protein phosphatase 4 core regulatory subunit R2 confers resistance to the anticancer drug cisplatin. Febs J 273: 3322–3334 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, Lieberman J, Shen X, Buratowski S, Haber JE, Durocher D, Greenblatt JF, Krogan NJ (2006) A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501 [DOI] [PubMed] [Google Scholar]
- Kim JA, Hicks WM, Li J, Tay SY, Haber JE (2010) Protein phosphatases Pph3, Ptc2, and Ptc3 play redundant roles in DNA double strand break repair by homologous recombination. Mol Cell Biol 31: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Ujihara H, Koike T (2009) Mobility shift detection of phosphorylation on large proteins using a Phos-tag SDS-PAGE gel strengthened with agarose. Proteomics 9: 4098–4101 [DOI] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ III (2000) Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol 20: 6449–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Chowdhury D (2011) What goes on must come off: phosphatases gate-crash the DNA damage response. Trends Biochem Sci 36: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D (2010) A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol 17: 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK (2007) Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem 282: 36177–36189 [DOI] [PubMed] [Google Scholar]
- Li X, Lin HH, Chen H, Xu X, Shih HM, Ann DK (2010) SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal 3: ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Mi J, Dziegielewski J, Bolesta E, Brautigan DL, Larner JM (2009) Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS One 4: e4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Chen GI, Gingras AC, Durocher D (2008) PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep 9: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- O’Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates JR III, Aparicio OM, Romesberg FE (2007) Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci USA 104: 9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh JR, Askenazi M, Ficarro SB, Cashorali T, Webber JT, Blank NC, Zhang Y, Marto JA (2009) Multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, Lobrich M, Jeggo PA (2011) Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol Cell Biol 30: 3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui JW, Hu MC, Tan TH (2007) Conditional knockout mice reveal an essential role of protein phosphatase 4 in thymocyte development and pre-T-cell receptor signaling. Mol Cell Biol 27: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108: 73–112 [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, Inagaki M, Hiraiwa N, Muramatsu M, Wynshaw-Boris A, Yoshiki A, Hirotsune S (2008) Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. J Cell Biol 180: 1133–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM (2000) Protein phosphatase 2A: a panoply of enzymes. Curr Opin Cell Biol 12: 180–185 [DOI] [PubMed] [Google Scholar]
- Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M (2003) Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem 278: 18817–18823 [DOI] [PubMed] [Google Scholar]
- Wang B, Zhao A, Sun L, Zhong X, Zhong J, Wang H, Cai M, Li J, Xu Y, Liao J, Sang J, Chowdhury D, Pfeifer GP, Yen Y, Xu X (2008) Protein phosphatase PP4 is overexpressed in human breast and lung tumors. Cell Res 18: 974–977 [DOI] [PubMed] [Google Scholar]
- Webber JT, Askenazi M, Marto JA (2011) mzResults: an interactive viewer for interrogation and distribution of proteomics results. Mol Cell Proteomics 10: M110 003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Durocher D (2010) De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E (2005) Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev 19: 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Askenazi M, Jiang J, Luckey CJ, Griffin JD, Marto JA (2010) A robust error model for iTRAQ quantification reveals divergent signaling between oncogenic FLT3 mutants in acute myeloid leukemia. Mol Cell Proteomics 9: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.