Abstract
RNA interference (RNAi) silences gene expression by acting both at the transcriptional and post-transcriptional levels in a broad range of eukaryotes. In the fission yeast Schizosaccharomyces pombe the RNA-Induced Transcriptional Silencing (RITS) RNAi complex mediates heterochromatin formation at non-coding and repetitive DNA. However, the targeting and role of RITS at other genomic regions, including protein-coding genes, remain unknown. Here we show that RITS localizes to specific meiotic genes and mRNAs. Remarkably, RITS is guided to these meiotic targets by the RNA-binding protein Mmi1 and its associated RNA surveillance machinery that together degrade selective meiotic mRNAs during vegetative growth. Upon sexual differentiation, RITS localization to the meiotic genes and mRNAs is lost. Large-scale identification of Mmi1 RNA targets reveals that RITS subunit Chp1 associates with the vast majority of them. In addition, loss of RNAi affects the effective repression of sexual differentiation mediated by the Mmi1 RNA surveillance machinery. These findings uncover a new mechanism for recruiting RNAi to specific meiotic genes and suggest that RNAi participates in the control of sexual differentiation in fission yeast.
Keywords: Mmi1, RITS, RNAi, sexual differentiation, S. pombe
Introduction
RNAi is an RNA-based gene silencing process widespread among eukaryotes with functions in the regulation of cell fate and differentiation, the neutralization of viruses and the maintenance of genome stability (Ghildiyal and Zamore, 2009; Malone and Hannon, 2009; Lau, 2010; Ketting, 2011). RNAi exerts these functions by acting in the cytoplasm as well as in the nucleus in the vicinity of, or directly on, chromatin. For example, RNAi-based processes targeting chromatin contribute to the silencing of transposons during gametogenesis in animals and plants (Klenov et al, 2007; Aravin et al, 2008; Bourc'his and Voinnet, 2010; Lau, 2010), DNA elimination in Tetrahymena thermophila (Mochizuki et al, 2002), DNA methylation in plants (Chan et al, 2004; Matzke et al, 2009), and epigenetic modification of chromatin in animals and the fission yeast Schizosaccharomyces pombe (Volpe et al, 2002; Pal-Bhadra et al, 2004; Fagegaltier et al, 2009; Verdel et al, 2009; Burkhart et al, 2011; Lejeune and Allshire, 2011).
In S. pombe, RNAi mediates formation of heterochromatin and gene silencing at the non-coding and repeated DNA found at the pericentromeric, subtelomeric and mating type regions (Volpe et al, 2002; Noma et al, 2004; Cam et al, 2005). Double-stranded RNA (dsRNA) produced from pericentromeric DNA repeats activates RNAi (Verdel and Moazed, 2005; Buhler and Moazed, 2007; Grewal and Jia, 2007). The ribonuclease III Dicer (Dcr1) digests the dsRNAs into small interfering RNA (siRNA) molecules of 21–23 nucleotides that load onto the Ago1-containing RNAi complex, called RNA-Induced Transcriptional Silencing (RITS) complex (Reinhart and Bartel, 2002; Verdel et al, 2004). RITS is composed of Ago1, the unique member of the Argonaute protein family in S. pombe and onto which the siRNA binds, Chp1, a chromodomain protein, and Tas3, a WG/GW containing protein, that bridges Ago1 to Chp1 (Verdel et al, 2004; Partridge et al, 2007). Transcription of the repeats by RNA polymerase II has been proposed to produce a nascent transcript that serves as a recruiting platform onto which RITS binds thanks to the base-pairing of an Ago1-bound siRNA with the nascent transcript platform (Motamedi et al, 2004; Djupedal et al, 2005; Kato et al, 2005; Buhler and Moazed, 2007). In addition, Chp1 chromodomain specifically recognizes the methylated lysine 9 of histone H3 (H3K9me) residue (Partridge et al, 2002), and anchors RITS to chromatin (Noma et al, 2004; Partridge et al, 2007). H3K9 methylation is catalysed by the histone methyltransferase Clr4/Suv39h (Nakayama et al, 2001). RITS and Clr4 complex (CLRC) physically interact (Zhang et al, 2008; Bayne et al, 2010; Gerace et al, 2010), and both complexes are engaged in a positive self-reinforcing loop, whereby RITS RNA-mediated association with chromatin would stimulate Clr4-dependent H3K9 methylation, which in turn would promote siRNA production that enables more RITS to bind the nascent transcript platform and to participate in heterochromatin formation (Verdel and Moazed, 2005; Buhler and Moazed, 2007; Grewal and Jia, 2007).
Genome-wide mappings have localized several RNAi components to a few genomic sites outside of the major heterochromatin regions in fission yeast. Dcr1 localizes and participates in the silencing of transposon long terminal repeats (Woolcock et al, 2011). In addition, Ago1 together with the H3K9me repressive mark were detected at two meiotic genes, mei4 and ssm4 (Cam et al, 2005). Noticeably, mei4 encodes a master transcription factor that is required for the transcriptional activation of several hundred meiotic genes during sexual differentiation (Horie et al, 1998; Mata et al, 2007). Proper silencing of mei4 is critical to avoid untimely and deleterious expression of these meiotic genes (Harigaya et al, 2006; Mata et al, 2007; Nakase et al, 2009). However, whether Ago1 association with the meiotic genes mei4 and ssm4 reflects a direct role of RNAi in sexual differentiation control is unclear. Moreover, the mechanism recruiting RNAi to mei4 and ssm4 is unknown. During vegetative growth mei4 and ssm4 genes are transcribed, but an RNA surveillance machinery guided by the protein Mmi1 silences their expression by degrading their mRNAs (Harigaya et al, 2006; St-Andre et al, 2010; Yamanaka et al, 2010; Cremona et al, 2011). Mmi1 is an RNA-binding protein that belongs to the YTH protein family (Stoilov et al, 2002). The nuclear exosome component Rrp6, the poly(A)-binding protein Pab2 and the zinc finger protein Red1 are components of the Mmi1 RNA surveillance machinery and important for Mmi1-mediated silencing (Harigaya et al, 2006; St-Andre et al, 2010; Yamanaka et al, 2010; Sugiyama and Sugioka-Sugiyama, 2011).
In this study, we provide evidence that Mmi1 RNA surveillance machinery recruits RITS to mei4 and ssm4 chromatin and mRNAs, during mitotic growth. In addition, upon sexual differentiation induction, RITS localization to mei4 and ssm4 is lost while these genes become active. Thanks to a large-scale identification of Mmi1 RNA targets we also show that Chp1 interacts with the vast majority of them and that these targets accumulate preferentially before meiosis I. Furthermore, we find that inactivation of RNAi alleviates the efficient repression of sexual differentiation mediated by Mmi1. These findings reveal that Mmi1 RNA surveillance machinery directs RNAi to selective meiotic genes. They also suggest that RNAi directly participates in sexual differentiation regulation in fission yeast.
Results
Localization of RITS to mei4 gene and mRNA
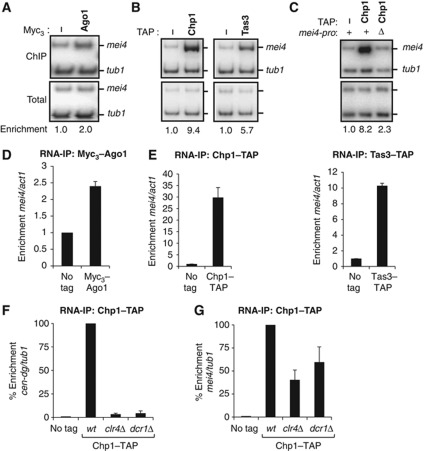
Following on a genome-wide chromatin immunoprecipitation (ChIP) analysis that detected Ago1 at the meiotic gene mei4 (Cam et al, 2005), we investigated whether mei4 might represent a new genomic target for the Ago1-containing complex RITS. As shown in Figure 1A and B (and Supplementary Figure 1A and B), we found that RITS subunits, Ago1, Chp1 and Tas3 specifically localized to mei4, using ChIP experiments. Ago1 immunoprecipitation consistently gave lower enrichment but, as reported previously (Motamedi et al, 2004; Verdel et al, 2004), this is likely due to the less efficient immunoprecipitation using the Myc3 tag, which is imposed by the fact that Ago1 function is disrupted by its fusion to larger tags. Given that at centromeres RITS association with chromatin involves the transcription of pericentromeric DNA by RNA polymerase II (Djupedal et al, 2005; Kato et al, 2005) and its binding to pericentromeric RNA (Motamedi et al, 2004), we checked whether the process of transcription and mei4 mRNA could participate in recruiting RITS to mei4 chromatin. Deleting mei4 promoter led to a considerable loss of Chp1 binding to mei4 (Figure 1C and Supplementary Figure 1C). Moreover, we found that all subunits of RITS interacted with mei4 mRNA, using RNA-IP experiments (Figure 1D and E). Since the histone methyltransferase Clr4 and the ribonuclease Dcr1 are both essential for Chp1 binding to pericentromeric RNA (Figure 1F; Motamedi et al, 2004), we next tested whether they were also necessary for Chp1 interaction with mei4 mRNA. Quite unexpectedly, Chp1 could bind mei4 mRNA in clr4Δ or dcr1Δ cells, although in a less pronounced manner (Figure 1G; ∼40 and ∼60% of the enrichment was kept at mei4 in clr4Δ or dcr1Δ cells, respectively). This binding was still observed in clr4Δ dcr1Δ double mutant cells (Supplementary Figure 1D), thus ruling out the possibility that a Clr4-dependent mechanism was responsible for the Chp1 binding to mei4 observed in dcr1Δ cells and vice versa. Hence, these findings show that RITS localizes to both mei4 gene and mRNA. Furthermore, they reveal that RITS association with mei4 mRNA presents similarities but also clear distinctions compared to its binding to pericentromeric RNA, suggesting the possibility of a distinct mechanism for recruiting RITS to mei4.
Figure 1.
Localization of RITS to mei4 gene and mRNA. (A, B) ChIP analyses of Myc3-Ago1, Chp1-TAP and Tas3-TAP association with the mei4 gene, relative to the control gene tubulin (tub1). α32 P-labelled PCR fragments were run on polyacrylamide gel and quantified with a phosphorimager. (C) ChIP analysis of Chp1-TAP association with mei4 wild type gene or mei4 deleted of its promoter (deletion of nucleotides -307 to -7), relative to tub1. Panels A, B and C show each time the result of one ChIP experiment. The average enrichments obtained from three independent experiments are shown in Supplementary Figures 1A, B and C. (D, E) RNA-IP analyses of Myc3-Ago1, Chp1-TAP and Tas3-TAP association with mei4 mRNA, relative to actin (act1) control mRNA, normalized to an untagged strain. Quantification was done using qRT–PCR. (F, G) RNA-IP analysis of Chp1-TAP association with pericentromeric RNA (cen-dg) or mei4 mRNA, relative to tub1 mRNA. Enrichments are expressed as a percentage of the relative enrichment monitored in wild type. Error bars represent standard deviations (s.d.) from at least three independent biological replicates. Figure source data can be found with the Supplementary data.
Mmi1 protein is required for RITS localization to mei4 and ssm4 mRNAs
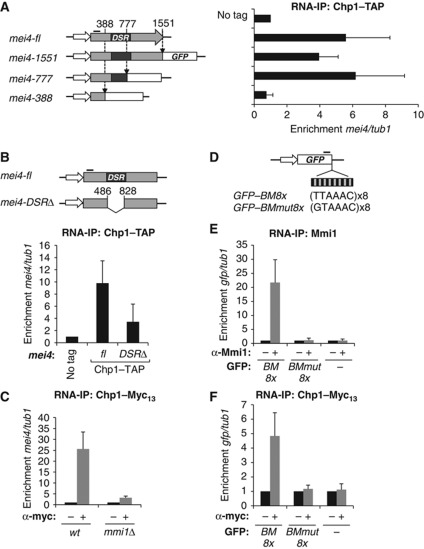
The Clr4- and Dcr1-independent binding of RITS to mei4 transcript led us to investigate if a specific sequence in mei4 mRNA could play a role in recruiting RITS. For this purpose, we made increasing truncations of mei4 open reading frame, starting from its 3′ end (Figure 2A, left panel). Truncations were made by introducing a GFP cassette in frame with the remaining 5′ part of mei4 coding sequence. Using RNA-IP experiments, we examined Chp1 binding to a region of mei4 mRNA present in each deletion mutant, at the 5′ end of its coding sequence. Deletions of the 3′ untranslated region (UTR) or of the 3′ half of mei4 transcript did not impact on Chp1 binding (Figure 2A, right panel, compare histograms mei4-fl to mei4-1551 and mei4-777). However, deletion of an additional 389 nucleotide region led to a near complete loss of Chp1 association with mei4 mRNA (Figure 2A, right panel, compare histograms mei4-777 and mei4-388). Importantly, this region overlapped with most of mei4 determinant of selective removal (DSR) sequence (Figure 2A), the region responsible for mei4 mRNA degradation in mitotic cells (Harigaya et al, 2006). A deletion of only the DSR sequence also led to a considerable loss of Chp1 binding (Figure 2B), thereby demonstrating the critical role of this sequence in recruiting RITS to mei4 mRNA.
Figure 2.
RITS association with mei4 mRNA requires mei4 DSR sequence and the RNA-binding protein Mmi1. (A) Diagram (left panel and not to scale) depicting the three insertion sites made in mei4 gene to gradually delete mei4 open reading frame (at nucleotides +1551, +777 and +388, relative to mei4 start codon). Deletions were made by inserting a GFP cassette (schematized by the white box) composed of the GFP coding sequence fused to the terminator of adh1 followed by the kanamycin resistance gene. RNA-IP analysis (right panel) of Chp1-TAP binding to mei4 mRNA full-length (mei4-fl) or to the three truncated mei4 mRNAs (mei4-1551, mei4-777 and mei4-388), relative to tub1 mRNA. Chp1-TAP binding was checked at a region located in the 5′ part of mei4 mRNA, represented by a thick black line on the diagram. Black box highlights the position of the determinant of selective removal (DSR) sequence. (B) Diagram (upper panel and not to scale) of mei4 gene deleted of just its DSR sequence with no foreign DNA sequence left and RNA-IP experiment (lower panel) of Chp1-TAP binding to the 5′ part of mei4-DSRΔ mRNA, as in (A). Relative enrichments were measured by qRT–PCR and compared to an untagged control strain. (C) RNA-IP analysis of Chp1-Myc13 binding to mei4 mRNA in wild type and mmi1Δ, relative to tub1. (D) Diagram of the GFP coding sequence fused to 8 repeats of the binding motif (BM) TTAAAC (GFP-BM8x) or to 8 repeats of the mutated binding motif GTAAAC (GFP-BMmut8x). The thick black line highlights the region of the GFP mRNA where the association with Mmi1 and Chp1 was examined. (E, F) RNA-IP analyses of Mmi1 and Chp1-Myc13 association with GFP mRNA or the recombinant GFP mRNAs described in (D), relative to tub1. All error bars represent s.d. from at least three independent replicates.
Since mei4 DSR sequence is bound by the RNA-binding protein Mmi1 (Harigaya et al, 2006), we next evaluated the importance of Mmi1 for RITS association with mei4 mRNA. Because the deletion of mmi1 strongly affects cell growth and viability, due to the deleterious untimely expression of Mei4 transcription factor (Harigaya et al, 2006), we combined mmi1 deletion with a mutation in mei4 that disrupts its function but preserves the DSR sequence, which we called mei4-828 (see experimental procedure for further details). Using RNA-IP experiments, we showed that Chp1 binding to mei4-828 mRNA was reduced to near background level in mmi1Δ cells (Figure 2C). This loss was not the result of a destabilization of Chp1 protein in mmi1Δ cells as shown by a western blot analysis (Supplementary Figure 2A), indicating that Mmi1 is required for recruiting RITS to mei4 mRNA.
In addition to mei4, Ago1 had been detected at a second meiotic gene, ssm4 (Cam et al, 2005). Interestingly, ssm4 mRNA had also been independently identified as a direct target of Mmi1 (Harigaya et al, 2006). RNA-IP experiments showed that RITS subunits associated with ssm4 mRNA (Supplementary Figure 2B and C). Moreover, Chp1 binding to ssm4 mRNA was dependent on Mmi1 (Supplementary Figure 2D). Based on these results, we conclude that Mmi1 is required for recruiting RITS to at least two of its targets, mei4 and ssm4 mRNAs.
Targeting Mmi1 to a GFP mRNA is sufficient to induce the recruitment of Chp1
Because Mmi1 protein binds directly to mei4 and ssm4 mRNA (Harigaya et al, 2006), we hypothesized that Mmi1 may act as a specificity factor guiding RITS to these meiotic mRNAs. To test this hypothesis, we assessed Chp1 association with a GFP mRNA fused to Mmi1 RNA-binding motifs. Initially, the identity of Mmi1 RNA-binding motif was unknown but a bioinformatic approach (described in the experimental procedure) led us to identify the UNAAAC RNA sequence as a potential Mmi1-binding site, with the UUAAAC motif being the most represented motif (Supplementary Figure 3). Mmi1 specific binding to the UNAAAC motif was demonstrated and analysed in detail in another study (Yamashita et al, 2012). As anticipated, RNA-IP experiments showed that Mmi1 associated in vivo with a GFP mRNA containing 8 repeats of the UUAAAC binding motif (GFP-BM8x), but not with the GFP mRNA alone or fused to the mutated binding motif GUAAAC repeated 8 times (GFP-BMmut8x; Figure 2D and E). We next examined Chp1 binding to the same GFP mRNA derivatives. Importantly, Chp1 interacted with the GFP-BM8x mRNA, but not with the other two GFP mRNAs (Figure 2F). These results strongly support the possibility that Mmi1 binding to a target RNA is sufficient to promote the recruitment of RITS to this RNA.
RITS associates with mei4 and ssm4 genes in a Mmi1- and Clr4-dependent fashion
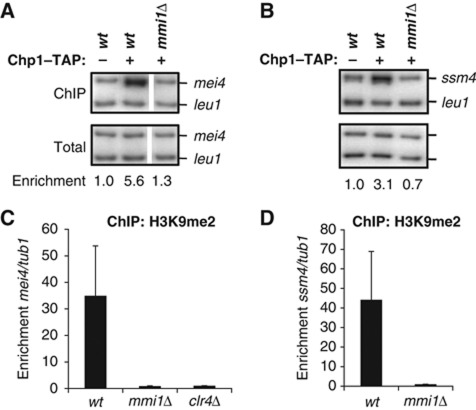
Given that RITS localized to mei4 chromatin (Figure 1A and B), we next investigated whether Mmi1 was also important for recruiting RITS to chromatin. Using ChIP experiments, we observed a dramatic reduction of Chp1 localization to mei4 in mmi1Δ cells, indicating that Mmi1 is critical for RITS association with the mei4 gene (Figure 3A and Supplementary Figure 4A). Similarly, Chp1 associated with the ssm4 gene in a Mmi1-dependent manner (Figure 3B and Supplementary Figure 4B). Because Chp1 stable binding to chromatin requires its interaction with the H3K9me mark (Partridge et al, 2007) and this mark had been detected at the mei4 and ssm4 genes (Cam et al, 2005), we examined whether H3K9 methylation was affected in mmi1Δ cells. Indeed, H3K9 methylation was completely lost in mmi1Δ and clr4Δ cells (Figure 3C and D), suggesting that Mmi1 mediates the Clr4-dependent H3K9 methylation, which is then recognized by the chromodomain of Chp1 to stably anchor RITS to the chromatin of mei4 and ssm4.
Figure 3.
RITS associates with mei4 and ssm4 genes in a Mmi1-dependent fashion. (A, B) ChIP analysis of Chp1-TAP binding to mei4 and ssm4 chromatin in wild type or mmi1Δ, relative to the control gene leu1. Panel A and B are the results of one experiment. The average enrichments obtained from three independent experiments are shown in Supplementary Figure 4A and B. Note that in panel A the white separation between the second and third lane is to indicate that the lanes come from the same gel but were non-adjacent and that the gel was cropped to juxtapose them. (C) ChIP analysis of H3K9me2 deposition on mei4 gene in wild type, mmi1Δ and clr4Δ, relative to tub1. (D) ChIP analysis of H3K9me2 deposition on ssm4 gene in mmi1Δ, relative to tub1. Error bars represent s.d. from three independent experiments. Figure source data can be found with the Supplementary data.
In parallel, we analysed the possible implication of Mmi1 in recruiting RITS to the major heterochromatic sites. However, in contrast to the mei4 and ssm4 genes, Chp1 localization to all heterochromatin regions tested in mmi1Δ cells was similar to wild type cells (Supplementary Figure 4C). Moreover, pericentromeric transcriptional gene silencing of a ura4 reporter gene inserted at the inner most repeat of centromere 1 (imr1) was not impaired in mmi1Δ cells (Supplementary Figure 4D), indicating that Mmi1 is not essential for gene silencing at constitutive heterochromatin regions.
Mmi1-mediated repression of sexual differentiation is affected in RNAi deficient cells
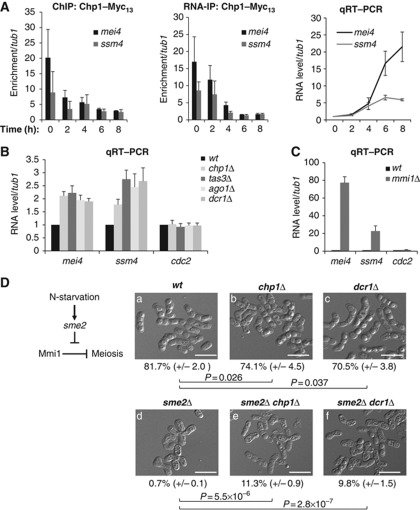
The fact that RITS was recruited to mei4 and ssm4 chromatin and mRNAs by a Mmi1-dependent mechanism suggested that RNAi may directly participate in Mmi1-mediated gene silencing. In support of this possibility, we observed that upon sexual differentiation induced in diploid cells by nitrogen starvation, Chp1 was gradually lost from both the chromatin and mRNAs of mei4 and ssm4, while both genes became activated (Figure 4A). To test the contribution of RNAi to Mmi1-mediated gene silencing more directly we analysed mei4 and ssm4 mRNA levels in different RNAi deficient cells, by quantitative reverse transcription PCR (qRT–PCR). We observed a reproducible ∼1.5 to 2.5 fold increase in mei4 and ssm4 mRNA levels in chp1Δ, tas3Δ, ago1Δ and dcr1Δ cells (Figure 4B). This increase appeared specific as the levels of cdc2 and tub1 control RNAs did not vary significantly under the same conditions. However, when compared to mei4 and ssm4 mRNA accumulation in mmi1Δ cells, the accumulation observed in RNAi deficient cells appeared relatively weak (Figure 4C). This result may reflect important redundancy between RNAi and other RNA degradation activities acting in Mmi1 RNA surveillance machinery (see further below).
Figure 4.
Effective Mmi1-mediated silencing activity is impaired in RNAi deficient cells. (A) Time course analysis from diploid cells induced to undergo sexual differentiation by nitrogen starvation. ChIP (left panel) and RNA-IP (middle panel) analyses of Chp1-Myc13 association with respectively the chromatin and mRNAs of mei4 and ssm4, relative to tub1. qRT–PCR analysis (right panel) of mei4 and ssm4 RNA levels, relative to tub1 RNA, during the time course analysis. (B) qRT–PCR analysis of mei4, ssm4 and cdc2 RNA levels in chp1Δ, tas3Δ, ago1Δ or dcr1Δ, relative to tub1 RNA and normalized to the wild type. (C) qRT–PCR analysis of mei4, ssm4 and cdc2 RNA levels in mmi1Δ, relative to tub1 RNA and normalized to the respective RNA levels in wild type. All error bars represent s.d. from three independent biological replicates. (D) Diagram depicting the sme2-dependent inhibition of Mmi1 upon sexual differentiation (left panel). Representative images of cells counted after sexual differentiation induction to measure sporulation frequency of the wild type (a), chp1Δ (b), dcr1Δ (c) and sme2Δ (d) single mutants, and sme2Δ chp1Δ (e) and sme2Δ dcr1Δ (f) double mutants (right panel). Sporulation frequency was determined by counting ascospores and the total number of cells under a microscope. An ascospore was counted as the product of two cells. Sporulation frequency expressed in percentage is represented as a mean (±s.d.) calculated from three independent biological replicates. P values were calculated using the Student’s t-test. White scale bars=10 μm. Figure source data can be found with the Supplementary data.
In parallel, we assessed the role of RNAi on the overall Mmi1-mediated repression of sexual differentiation by measuring sporulation efficiency. For this purpose, we used an established read out assay based on the rescue of the sme2Δ-dependent meiotic arrest (Yamashita et al, 1998; Harigaya et al, 2006; St-Andre et al, 2010; Yamanaka et al, 2010). Upon sexual differentiation, sme2 non-coding locus participates in Mmi1 inhibition by sequestering it (Figure 4D, left panel; Harigaya et al, 2006; Yamashita et al, 2012). In sme2Δ cells induced to differentiate, Mmi1 is not sequestered and not inhibited, Mmi1 target genes are thus kept silenced, and sexual differentiation eventually arrests before meiosis I. Measure of the sporulation efficiency in chp1Δ or dcr1Δ cells showed that it was consistently lower than in wild type cells (Figure 4D, right panel, compare images b and c to a), probably because of chromosome segregation defects caused by poor heterochromatin formation at pericentromeric repeats, as suggested previously (Hall et al, 2003). In contrast, we observed a higher sporulation frequency in sme2Δ chp1Δ or sme2Δ dcr1Δ cells compared to sme2Δ cells (Figure 4D, right panel, compare images e and f to d, respectively). Collectively, these results support a role for RNAi in Mmi1-mediated repression of sexual differentiation.
Differential contribution of Mmi1 RNA surveillance factors to RITS localization to mei4
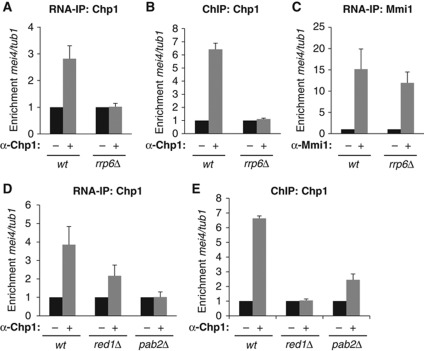
The 3′ to 5′ exonuclease Rrp6 plays a major role in Mmi1-mediated degradation of mei4 and ssm4 mRNAs (Harigaya et al, 2006; St-Andre et al, 2010; Yamanaka et al, 2010; Sugiyama and Sugioka-Sugiyama, 2011). To further dissect the mechanism by which Mmi1 RNA surveillance machinery recruits RNAi to mei4 and ssm4, we analysed the contribution of Rrp6 to this recruitment. RNA-IP and ChIP experiments showed that, like Mmi1, Rrp6 is critical for recruiting Chp1 to mei4 mRNA and gene, respectively (Figure 5A and B). H3K9 methylation at mei4 was also completely dependent on Rrp6 (Supplementary Figure 5A). In contrast, Mmi1 still bound mei4 mRNA in rrp6Δ cells (Figure 5C). Furthermore, loss of Chp1 binding to mei4 mRNA and chromatin was not caused by a destabilization of Chp1 protein, as shown by western blot analysis (Supplementary Figure 5B). From these results we conclude that Rrp6 acts downstream of Mmi1 albeit it is also required for RITS association with mei4 mRNA and gene.
Figure 5.
Mmi1 RNA surveillance factors, Rrp6, Red1 and Pab2 show distinct contributions to RITS association with mei4. (A) RNA-IP analysis of Chp1 association with mei4 mRNA in wild type or rrp6Δ, relative to tub1 mRNA. Relative enrichment was normalized to IP done without antibody. (B) ChIP analysis of Chp1 association with mei4 gene in wild type or rrp6Δ, relative to tub1. (C) RNA-IP analysis of Mmi1 binding to mei4 mRNA in wild type or rrp6Δ, relative to tub1 mRNA. (D) RNA-IP analysis of Chp1 association with mei4 mRNA in wild type, red1Δ or pab2Δ, relative to tub1 mRNA. (E) ChIP analysis of Chp1 association with mei4 gene in wild type, red1Δ or pab2Δ, relative to tub1. All error bars represent s.d. from three independent experiments.
Red1 and Pab2 are two other proteins recently found to be part of Mmi1 mRNA surveillance machinery (St-Andre et al, 2010; Yamanaka et al, 2010; Sugiyama and Sugioka-Sugiyama, 2011). RNA-IP experiments showed that while Chp1 association with mei4 mRNA remained relatively high in red1Δ cells, with approximately 50% of the enrichment observed in wild type cells, it was completely lost in pab2Δ cells (Figure 5D). At the chromatin level, ChIP experiments showed the opposite tendency, with Chp1 localization to mei4 completely lost in red1Δ cells but only partially lost in pab2Δ cells (Figure 5E). We hypothesized that these opposite effects of deleting red1 and pab2 might come from the fact that Mmi1 RNA surveillance machinery recruits RITS by two non-exclusive mechanisms. The first one would act at the RNA level and preferentially involve Pab2 whereas the second one acting at the chromatin level would rather implicate Red1. Consistent with this possibility, H3K9 methylation at mei4 was only reduced in pab2Δ cells whereas it was completely lost in red1Δ cells (Supplementary Figure 5A). In agreement with Pab2 playing an important role in recruiting RITS to Mmi1 RNA targets, no significant effect on mei4 mRNA accumulation was observed in ago1Δ pab2Δ double mutant cells, compared to the pab2Δ cells (Supplementary Figure 5C). Altogether, these results show that factors of the Mmi1 mRNA surveillance machinery contribute in distinct manners and degrees to the recruitment of RITS to Mmi1 target genes or mRNAs.
Widespread binding of Chp1 to Mmi1 RNA targets
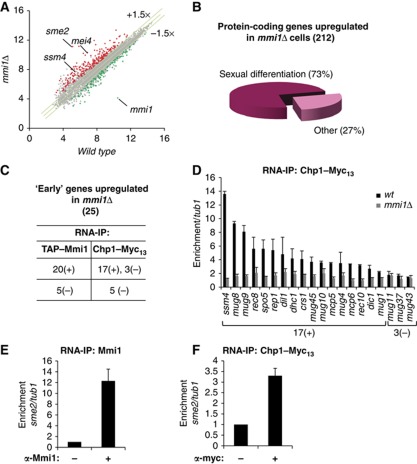
Finally, in order to investigate RITS binding to Mmi1 target mRNAs in a comprehensive manner, we conducted a transcriptome analysis of mmi1Δ cells to first identify Mmi1 RNA targets, and then to examine Chp1 binding to these targets. As indicated before (and further described in the experimental procedure section) to alleviate the severe growth defects associated with mmi1 deletion we conducted the transcriptome analysis on the mmi1Δ mei4-828 double mutant cells. From the genome-wide transcriptomic analysis, we found a total of 212 protein-coding genes whose mRNA level was upregulated at least 1.5 fold in mmi1Δ cells, compared to wild type or to mei4-828 cells (Figure 6A and Supplementary Figure 6A and B). Noticeably, more than 70% of these mRNAs accumulate during sexual differentiation (Figure 6B), according to the classification described by Mata and co-workers (Mata et al, 2002). This result confirmed that Mmi1 likely recognizes and triggers degradation of sexual differentiation-specific mRNAs, as proposed in the initial characterization of Mmi1 (Harigaya et al, 2006). 90 mRNAs were also found to be less abundant in mmi1Δ cells compared to wild type or mei4-828 cells, but gene ontology analysis showed no enrichment for a given regulatory function or a specific cellular process, suggesting that their down-regulation is probably an indirect effect of mmi1Δ deletion.
Figure 6.
Transcriptome of mmi1Δ cells and large-scale identification of RNAs bound by Mmi1 and Chp1. (A) Scatter plot of S. pombe genome-wide gene expression measured from hybridization signals of RNAs isolated from mmi1Δ cells (Y axis) and from wild type cells (X axis). Red and green dots represent genes up and down regulated, respectively. Grey dots represent genes with no change in their RNA level in mmi1Δ compared to wild type and mei4-828 cells. Each hybridization signal is an average of two independent biological replicates and is plotted using a log2 scale. Note that the mmi1Δ cells used for the transcriptome analysis also contain the mei4-828 mutation (further described in the experimental procedure section). Transcriptome of mei4-828 cells is highly similar to wild type cells as shown in Supplementary Figure 6A. (B) Pie chart representing the distribution of the protein-coding genes upregulated in mmi1Δ cells according to their expression during sexual differentiation (Mata et al, 2002). (C) Table summarizing the RNA-IP analyses of TAP-Mmi1 and Chp1-Myc13 association with 25 mRNAs specific of the ‘early’ phase of sexual differentiation and accumulating in mmi1Δ, relative to tub1 mRNA control. An RNA was considered significantly associated with Mmi1 or Chp1 when enriched at least two fold after RNA-IP. Enriched and non-enriched mRNAs are classified as (+) and (−), respectively. (D) RNA-IP analysis of Chp1-Myc13 association with Mmi1 mRNA targets in wild type or mmi1Δ, relative to tub1. 17(+) and 3(−) denote respectively RNAs that are and are not significantly enriched in Chp1-Myc13 RNA-IP in (D). (E, F) RNA-IP analyses of Mmi1 or Chp1-Myc13 association with sme2 RNA, relative to tub1. Error bars represent s.d. from at least three independent experiments.
S. pombe sexual differentiation gene expression programme is composed of four successive waves of transcription, called N-starvation, early, middle and late phases (Mata et al, 2002). A search for the overrepresentation of Mmi1 RNA-binding motifs among the mRNAs accumulating in mmi1Δ cells revealed a strong bias towards mRNAs accumulating in the phase ‘early’ of sexual differentiation (Supplementary Table I). Using RNA-IP experiments, we undertook to test Mmi1 binding to all ‘early’ mRNAs accumulating in mmi1Δ cells. Due to the low abundance of some of these mRNAs we were able to test Mmi1 binding to 25 of the 29 ‘early’ mRNAs accumulating in mmi1Δ cells. We found that 20 of them showed at least 4 fold enrichments over background after Mmi1 immunoprecipitation, whereas the other 5 mRNAs did not show any significant enrichment (Figure 6C and Supplementary Table II). Strikingly, Chp1 bound to 17 out of the 20 Mmi1 RNA targets but not to any of the other 5 ‘early’ mRNAs not targeted by Mmi1 (Figure 6C). Furthermore, Chp1 binding to all 17 mRNAs was lost in mmi1Δ cells (Figure 6D). In parallel, we examined Chp1 and H3K9me localization to the 20 genes expressing the Mmi1 RNA targets, by ChIP experiments. Remarkably, in contrast to the widespread association of Chp1 with Mmi1 RNA targets, Chp1 and H3K9 methylation were detected respectively at only 3 and 4 of the corresponding genes (Supplementary Figure 6C and D). From these results, we conclude that Chp1 binding to Mmi1 RNA targets is rather systematic, whereas its binding to the chromatin of the corresponding genes may be more restricted.
Since our transcriptome analysis was performed with a tiling array, which includes the intergenic regions, we also examined the level of non-coding RNAs in mmi1Δ cells. Surprisingly, the most upregulated RNA in the whole transcriptome analysis was expressed from sme2, the non-coding locus that sequesters Mmi1 during sexual differentiation. RNA-IP experiments showed that Mmi1 and Chp1 bound sme2 non-coding RNA in vivo (Figure 6E and F). 26 other non-coding RNAs accumulated in mmi1Δ cells (Supplementary Table III). However, none of them showed an overrepresentation of Mmi1 RNA-binding motifs, supporting the idea that they were not direct targets of Mmi1. Thus, the transcriptome analysis of mmi1Δ cells combined with RNA-IP experiments revealed a widespread association of Chp1 with Mmi1 RNA targets, including a non-coding RNA with a key function in the control of sexual differentiation.
Discussion
Our findings provide evidence that the Mmi1 RNA surveillance machinery guides RNAi to specific meiotic mRNAs and genes. They also suggest for the first time a direct implication of RNAi in the control of sexual differentiation in yeast. The major findings are as follow: first, RITS localizes to the chromatin and mRNAs of two meiotic genes, mei4 and ssm4, during vegetative growth, and this association is lost upon sexual differentiation when both genes are activated. Second, localization of RITS to mei4 and ssm4 genes and their respective mRNAs requires the Mmi1 RNA surveillance machinery. Furthermore, Mmi1 binding to a recombinant mRNA containing Mmi1 hexameric RNA-binding motifs is sufficient to induce the recruitment of Chp1, consistent with Mmi1 being the specificity factor for recruiting RITS to selective meiotic genes and mRNAs. Third, a large-scale identification of Mmi1 targets reveals that Chp1 binds to the vast majority of them in a Mmi1-dependent manner. Fourth, in agreement with a role of RNAi in Mmi1-mediated repression of sexual differentiation, a loss of RNAi partially suppresses the sexual differentiation block caused by a constitutive activation of Mmi1. Below we discuss the implications of these findings for the targeting of RITS and for the role of RNAi in sexual differentiation.
RITS association with meiotic genes and (m)RNAs
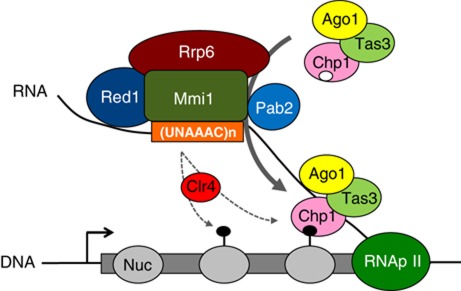
In S. pombe the association of RITS with major heterochromatic regions has been extensively studied. The mechanism of RITS recruitment to chromatin is best understood at the pericentromeric heterochromatin. At this genomic region, RITS binding to pericentromeric chromatin involves an RNA platform produced by transcription of the DNA repeats by RNA polymerase II (Verdel and Moazed, 2005; Buhler and Moazed, 2007; Grewal and Jia, 2007). RITS binding to pericentromeric RNA requires Clr4 and Dcr1, which respectively methylates H3K9 and produces siRNAs that load onto RITS (Motamedi et al, 2004). Based on this Clr4 and Dcr1 dependency and other results, it has been proposed that the binding of Chp1 chromodomain to H3K9me and of Ago1-loaded siRNA to a nascent transcript are key events in recruiting RITS to pericentromeric heterochromatin. In this study, we identify a new mechanism that involves the Mmi1 RNA-binding protein and its associated RNA degradation machinery in recruiting RITS to specific meiotic genes and mRNAs. From our findings, we propose a model for the recruitment of RITS to these meiotic mRNAs and genes (see Figure 7 for further details). Mmi1 RNA-binding protein recognizes the hexameric motif UNAAAC that we and others have identified (Chen et al, 2011; Yamashita et al, 2012). Thanks to its sequence-specific binding Mmi1 acts as a specificity factor for recruiting RITS to selective meiotic mRNAs and chromatin during vegetative growth. Once sexual differentiation is induced, Mmi1 is inhibited and RITS loses its localization to the meiotic targets. Several factors of the Mmi1 RNA surveillance machinery differentially promote RITS binding to chromatin or mRNAs. Like Mmi1, the Rrp6 protein is required for RITS binding to both the mRNA and gene of mei4. In contrast, Pab2 and Red1 proteins are probably part of two non-exclusive mechanisms recruiting RITS preferentially at the mRNA and chromatin, respectively. By using a combination of RNA-IP and ChIP experiments, we show that Chp1 associates with most Mmi1 RNA targets whereas it is only detected in a small proportion of their corresponding genes. H3K9 methylation is also detected in only a limited fraction of Mmi1-silenced genes. Very recently, it was similarly reported that H3K9 methylation at mei4, ssm4 and other meiotic genes is dependent on Mmi1 RNA surveillance machinery, which interacts with Clr4 H3K9 methyltransferase (Zofall et al, 2012). In agreement with our findings, H3K9 methylation was detected at only 6 out of the 22 Mmi1-silenced genes identified by Harigaya and co-workers (Harigaya et al, 2006) and in our study. From these data, we postulate that the more restricted localization of Chp1 and H3K9me to chromatin may be because Red1-dependent mechanism acts on chromatin of only a subset of genes silenced by Mmi1, conversely to the Pab2-dependent mechanism, which would act on most, if not all, Mmi1 RNA targets. It will be important to address this possibility in future studies. However, it should be noted that testing the specific contribution of Pab2- and Red1-dependent mechanisms is challenging as these mechanisms are overlapping and reinforce RITS recruitment to both chromatin and RNA targets. Also, at this point we cannot completely exclude that another possibility such as a poorer detection limit for the ChIP compared to the RNA-IP experiment is responsible for this difference. Importantly, regardless of the mechanisms, our findings provide evidence that an RNA degradation machinery plays a critical role in recruiting the RNAi effector complex RITS to mRNAs and chromatin of cell fate-regulated genes.
Figure 7.
Model for the Mmi1 RNA surveillance-driven recruitment of RITS to meiotic (m)RNAs and genes. Mmi1 binds to a target meiotic RNA through the recognition of its hexameric RNA-binding motif (UNAAAC) and recruits the RNA surveillance machinery. Rrp6, Pab2 and Red1 proteins are factors important for Mmi1 RNA surveillance machinery. Recruitment of this machinery enables or stabilizes the binding of RITS to the target RNA. RITS can bind both meiotic RNAs and their corresponding genes. Pab2 protein assists mostly RITS binding to the RNA, whereas Red1 would favour the recruitment of RITS to chromatin by mediating Clr4-dependent H3K9 methylation. These two mechanisms working side-by-side would further reinforce the recruitment of RITS to chromatin and mRNAs. Furthermore, a selective regulation of only one of the two mechanisms may be a mean to modulate RITS binding to the RNA versus chromatin of the target gene. Nuc: nucleosome; Black lollipop: H3K9me repressive histone mark; White circle on Chp1 depicts its chromodomain; RNApII: RNA polymerase II; (UNAAAC)n: cluster of Mmi1 hexameric binding motifs.
A role for an RNA-binding protein in recruiting an Argonaute-containing complex to selective RNAs has been described in other eukaryotes. The HuR AU-rich RNA-binding protein recruits or stabilizes the binding of a RISC complex to two different mRNAs, in animal cell lines (Kim et al, 2009; Glorian et al, 2011). Similarly, in A. thaliana the RNA-binding protein KTF1, which participates in the RNAi-dependent process called RNA-directed DNA methylation (RdDM), interacts with the Argonaute protein AGO4 and directs or stabilizes AGO4 binding to nascent transcripts (He et al, 2009; Rowley et al, 2011). Together with our results, these findings set a paradigm for the promoted or assisted recruitment of Argonaute-containing complexes to RNA and chromatin targets in a wide range of eukaryotes, including unicellular organisms such as fission yeast.
In addition, our findings reveal that the nuclear exosome component Rrp6 is essential for the targeting of RNAi to chromatin regions and mRNAs. In this regard, it is interesting to note that a genome-wide transcriptome analysis found that RNAs emanating from genomic regions targeted by the RNAi-based RdDM accumulate in several mutants of the nuclear exosome in A. thaliana (Chekanova et al, 2007). Thus, it is tempting to speculate that the connection between exosome-mediated RNA degradation, RNAi and epigenetic modification of chromatin uncovered in fission yeast might also exist in plants and other eukaryotes. Furthermore, the finding that Mmi1 and Chp1 target a non-coding RNA expressed from sme2 locus suggests that Mmi1/exosome-dependent recruitment of RNAi may extend to non-coding RNAs implicated in the control of cellular differentiation (Harigaya et al, 2006; Yamashita et al, 2012). Recently multiple non-coding RNAs identified in several organisms were found to exert important regulatory functions, including the control of cellular differentiation (Pauli et al, 2011). Whether some of these non-coding RNAs could be regulated by a similar mechanism remains to be determined.
RNAi and sexual differentiation
Although RNAi has been clearly involved in sexual differentiation in multiple eukaryotes its implication in yeast sexual differentiation has remained elusive. Recently, numerous antisense RNAs differentially expressed during vegetative growth and sexual differentiation have been identified (Bitton et al, 2011; Chen et al, 2012). These antisense RNAs could potentially produce double stranded RNAs that would activate RNAi upon or during sexual differentiation (Bitton et al, 2011). In this study, we now show that RNAi is linked to the control of sexual differentiation by the Mmi1 mRNA surveillance system. This system imposes a robust gene silencing essential for the proper inhibition of sexual differentiation gene expression programme during mitotic growth in fission yeast but no connection to RNAi had been found initially (Harigaya et al, 2006). We show that the Mmi1 RNA surveillance machinery plays a critical role in recruiting RNAi to meiotic mRNAs and genes, and that RITS may interact with most Mmi1 RNA targets. In addition, the partial rescue of the sme2Δ-induced sexual differentiation arrest in the absence of Dcr1 or Chp1 further supports the implication of RNAi in Mmi1-mediated inhibition of sexual differentiation. Noticeably, the silencing defects observed in RNAi deficient cells are weak compared to mmi1Δ cells. We believe this may be due to redundancy between RNAi and the nuclear exosome RNA degradation activities. In this regard, inactivation of Dis3, another nuclease acting together with Rrp6 in Mmi1-mediated gene silencing leads to only minor silencing defects (St-Andre et al, 2010). The implication of diverse nucleases, including Ago1 slicing and Dcr1 dicing activities, in Mmi1 RNA surveillance system may in fact be essential for the proper regulation of sexual differentiation programme under specific environmental conditions. In addition, an intriguing possibility is that RNAi might have another function at Mmi1 target genes. In C. elegans, similarly to Ago1 in S. pombe, the Argonaute protein CSR1 targets sexual differentiation-specific genes (Claycomb et al, 2009; van Wolfswinkel et al, 2009). However, it does not seem to play a role in silencing their expression. Instead, it has been proposed that CSR1 contributes to efficient chromosome segregation during mitotic and meiotic cell divisions, possibly by recruiting to meiotic genes chromatin modifiers, such as the cohesins. Intriguingly, an RNAi-dependent recruitment of the Rad21 cohesin has been described at mei4 (Gullerova and Proudfoot, 2008). Hence, we cannot exclude that RITS recruitment to Mmi1 silenced genes may have other regulatory functions, such as to contribute to genome stability by loading for example cohesins to specific chromosomal regions.
In animals, RNAi is essential to sexual differentiation and is directly involved in the regulation of this cellular process by acting at several levels (Lau, 2010; Suh and Blelloch, 2011). We present evidence that RNAi is also directly involved in the regulation of sexual differentiation in fission yeast. Thanks to the identification of Mmi1 RNA-binding motif and the transcriptome of mmi1Δ cells, our findings indicate that RITS may bind to a set of mRNAs and genes silenced in mitotic cells and expressed relatively early during sexual differentiation. Importantly, among the mRNAs targeted we identified rep1 and mei4, which code for two key transcription factors of the sexual differentiation gene expression programme. If their silencing is defective during vegetative growth, as when Mmi1 function is impaired, a potentially large proportion of this sexual differentiation programme is expressed at the wrong time, which ultimately provokes severe growth defects and death of the cell (Harigaya et al, 2006). In this regard, it is interesting to note that similarly in several other eukaryotes RNAi is critical for silencing the expression of sexual differentiation-specific genes, which maintains the germline stem cell (GSC) self-renewal and proliferation capacity (Lau, 2010; Suh and Blelloch, 2011). Together these results argue that the implication of RNAi in the control of sexual differentiation and possibly its function in regulating this cellular process may well be conserved in numerous eukaryotes, including fission yeast.
Material and methods
Strain, media and plasmid construction
S. pombe strains used in this study are listed in Supplementary Table IV. Standard procedures were used for growth and manipulations of S. pombe cells (Moreno et al, 1991; Sabatinos and Forsburg, 2010). Gene deletion and epitope tagging were made following the PCR-based methodology (Bahler et al, 1998). The mmi1Δ cells were generated from a parental strain possessing an inactive form of mei4 (named mei4-828), as deletion of mmi1Δ leads to severe growth and viability defects due to the deleterious expression of Mei4. mei4-828 mutant strains were made by inserting at nucleotide 829 of mei4 coding sequence a GFP cassette, composed of the GFP coding sequence fused to adh1 terminator and the kanamycin resistance gene. The recombinant mei4-gfp gene still possesses mei4 DSR sequence but has lost the sequence coding for the activation domain of Mei4. The transcriptome analysis, as all other experiments involving mmi1Δ cells, was conducted with the double mutant mmi1Δ mei4-828 cells. Sporulation of diploid cells was induced by nitrogen starvation and carried out as described (Moreno et al, 1991; Mata et al, 2002). Sporulation efficiency of h90 cells was measured after growing cells on synthetic sporulation agar (SPA) for 2 days at 30 °C, and was calculated as the number of ascospores formed over the total number of cells counted. pRGT1 plasmids (derived from pREP1) express, under the control of the adh promoter, the GFP mRNA alone (pRGT1-GFP), in fusion to 8 repeats of the motif UUAAAC (pRGT1-GFP-TTAAAC8x) or in fusion to 8 repeats of the mutated motif GUAAAC (pRGT1-GTAAAC8x). Construction of the plasmids is detailed elsewhere (Yamashita et al, 2012).
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed mainly as described previously (Motamedi et al, 2004). For ChIP done with antibodies, immunoprecipitation (IP) was carried out for 2 h using 2 μg of anti-H3K9me2 (Abcam, ab1220-100), anti-Myc 9E10 (Covance, MMS-150P) or anti-Chp1 (Abcam, ab18191), followed by 1 h incubation with 20 μl 1/1 slurry protein A-sepharose (GE Healthcare). Quantitative PCR was conducted with 4 μl of a 1/8 dilution of the IP DNA and 4 μl of a 1/50 dilution of the input. Relative enrichment was calculated as the ratio of signal of interest to signal of control in IP over input. Primers used are listed in Supplementary Table V.
RNA-immunoprecipitation (RNA-IP)
RNA-IP experiments were mainly as described (Motamedi et al, 2004). Briefly, cells grown to an OD600 of 1.3 (approximately 2 × 107 cells/ml) were crosslinked with 1% formaldehyde for 15 min. Whole cell extracts were treated with 700 units of DNase I (Sigma) for 1 h at 30 °C in a 50 mM HEPES-NaOH pH7.5, 150 mM NaCl, 1 mM EDTA pH8, 1% Triton X100, 0.1% Na-deoxycholate, 5 mM DTT buffer supplemented with 25 mM MgCl2 and 5 mM CaCl2. IP was performed as described in ChIP section. Anti-Mmi1 IP was carried out using 10 μl of rabbit antibody. Immunoprecipitated RNA was isolated by phenol/chloroform extraction and reverse transcribed according to the manufacturer instructions (Superscript II, Invitrogen). Quantitative analysis was performed using real-time PCR. For RNAs present in low abundance a quantitative preamplification step was made as described (Lao et al, 2006; Smith et al, 2011). 4 μl of a 1/400 final dilution of the preamplification reaction was used for the second quantitative PCR. Primers are listed in Supplementary Table V.
Quantitative PCR (qPCR)
qPCR was performed on the LightCycler480 machine (Roche) using MESA Green qPCR mix (Eurogentec) and the following programme: 15 min incubation at 95 °C, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 70 °C for 30 s. Quantitative preamplification was done under the same conditions but with 14 cycles instead of 40.
Prediction of Mmi1 RNA-binding motif
Search for motif enrichment in mei4, ssm4, rec8 and spo5 DSR sequences was first performed with oligo-analysis programme from RSAT (http://rsat.scmbb.ulb.ac.be/rsat/), which uses an enumerative word discovery approach to detect statistically overrepresented motifs in a set of sequences (van Helden et al, 1998). Default settings and single strand as the oligomer counting mode were used. In addition, based on the detailed analysis of rat Yth521-B protein, the founding member of the YTH family, which demonstrated that its YTH RNA-binding module recognizes a hexameric sequence motif (Zhang et al, 2010), we searched for enrichment of hexameric sequences. The UUAAAC hexameric motif was found overrepresented in the DSR sequences with an E-value for occurrences of 2.4E-3. A second analysis with the prediction algorithms Consensus, MEME, Gibbs and MDscan from melina II (http://melina2.hgc.jp/public/index.html) (Okumura et al, 2007) further validated the overrepresentation of the UUAAAC motif in the four DSR sequences. This analysis also indicated that the UGAAAC, UCAAAC and UAAAAC motifs were potentially overrepresented in these sequences. Using a non-parametric and asymmetric Mann-Whitney-Wilcoxon test, UNAAAC motif was found to be statistically overrepresented with a P-value=3.33E-4 in mRNA sequences of mei4, ssm4, rec8 and spo5, compared to the control set formed by the 7039 annotated genomic regions in S. pombe.
Transcriptome analysis
Cultures of 25 ml YEA S. pombe cells were grown to approximately 7.106 cells/ml. Total RNA was extracted using hot phenol method. 100 μg total RNA was further purified using RNAeasy Qiagen columns, incubated with DNAse I (Roche) digestion for 30 min at 37 °C, and extracted by classical phenol extraction. Microarray hybridization, processing and analysis were performed on the ProfileXpert core facility (Bron, France) following instructions of the manufacturer (Affymetrix, Santa Clara, CA, USA) and using a high-density oligonucleotide array (GeneChip S. pombe Tiling 1.0FR Array). Microarrays were read with a confocal laser (Genechip scanner 3000, Affymetrix). CEL files were generated using the Affymetrix GeneChip Command Console (AGCC) software 3.0. Probe sequences (from Affymetrix BPMAP file) were aligned to the genome sequence of S. pombe from the S. pombe Genome project, using the R/Bioconductor package Starr. Probe sequences were mapped to unique positions on the genome and a new BPMAP file was built (available upon request). RMA background correction and quantile normalization were applied to the data using Robust Multiarray Average (RMA) statistical algorithm (Partek Genomic Suite software 6.5, St. Louis, MO, US). Transcriptome analysis of annotated regions was conducted performing a median summarization of probes covering each gene from Ensembl Fungi Database (EF1). Transcriptome analyses were done in duplicates, each duplicate corresponding to an independent biological replicate. A total of four pair-wise comparisons was carried out, where each annotated region from one duplicate was compared with the corresponding annotated region from the other duplicate. Only annotated genomic regions showing a variation greater than 1.5 fold in all four pair-wise comparisons were retained. A selected region was considered differentially expressed only if the ratio of the fold-changes (FC) [mmi1Δ/wt over mei4-828/wt] was above 1.5 fold. Transcriptome analysis was visualized using Integrated Genome Browser (IGB). The complete set of CEL files is available at the GEO database under accession number GSE31119.
Supplementary Material
Acknowledgments
We thank El Cherif Ibrahim and Fabienne Hans for their critical reading, Sophie Rousseaux for her help in editing the manuscript, Ramesh Pillai for constructive discussions, Danesh Moazed and Saadi Khochbin for their constant support, Sophie Barral for her contribution to the initial phase of this work. We thank Thierry Gautier, Eric Coissac and Laura Gregoire for their help with the bioinformatic analyses, François Bachand, Danesh Moazed and Jean-Paul Javerzat for strains and plasmids, and Séverine Croze for her help with the microarray hybridization. A.V. was first supported by a doctoral fellowship from the French Ministry of Research and later on from l’Association pour la Recherche contre le Cancer (ARC). Research done in Tokyo was supported by Grants-in-Aid for Scientific Research to AY and MY from Japan Society for the Promotion of Science. Research in our laboratory was supported by the international Human Frontier Science Programme Organization (HFSPO), the ARC, the Institut National de la Santé Et de la Recherche Médicale (INSERM) Avenir programme and an European Research Council (ERC) Starting Grant.
Author contributions: EH, AV, LTT and AV designed the experiments; EH, AV and LTT performed most of the experiments; EH, AV, LTT, DP and AV wrote the paper; BG, EL, NN, JL and DP contributed to the transcriptome analyses; CB and JP made specific contributions to the characterisation of Mmi1 and sme2; AY, YS and MY provided plasmid constructions and contributed to the sexual differentiation rescue experiments; All authors discussed the results and the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, Allshire RC (2010) Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Grallert A, Scutt PJ, Yates T, Li Y, Bradford JR, Hey Y, Pepper SD, Hagan IM, Miller CJ (2011) Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Mol Syst Biol 7: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Voinnet O (2010) A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330: 6004617–622 [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D (2007) Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S (2011) A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet 7: e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, Alonso J, Brukhin V, Grossniklaus U, Ecker JR, Belostotsky DA (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J (2011) The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE 6: e26804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Rosebrock AP, Khan SR, Futcher B, Leatherwood JK (2012) Repression of meiotic genes by antisense transcription and by Fkh2 transcription factor in Schizosaccharomyces pombe. PLoS ONE 7: e29917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D Jr., Mello CC (2009) The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona N, Potter K, Wise JA (2011) A meiotic gene regulatory cascade driven by alternative fates for newly synthesized transcripts. Mol Biol Cell 22: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Bouge AL, Berry B, Poisot E, Sismeiro O, Coppee JY, Theodore L, Voinnet O, Antoniewski C (2009) The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci U S A 106: 21258–21263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D (2010) The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell 39: 360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S (2011) HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ 18: 1692–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI (2003) RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK (2009) An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C (1998) The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol 18: 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (2005) RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Ketting RF (2011) The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao K, Xu NL, Yeung V, Chen C, Livak KJ, Straus NA (2006) Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem Biophys Res Commun 343: 85–89 [DOI] [PubMed] [Google Scholar]
- Lau NC (2010) Small RNAs in the animal gonad: guarding genomes and guiding development. Int J Biochem Cell Biol 42: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147 [DOI] [PubMed] [Google Scholar]
- Mata J, Wilbrey A, Bahler J (2007) Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol 8: R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA (2002) Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110: 689–699 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Nakase Y, Hirata A, Shimoda C, Nakamura T (2009) Ectopic overproduction of a sporulation-specific transcription factor induces assembly of prespore-like membranous compartments in vegetative cells of fission yeast. Genetics 183: 1195–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Okumura T, Makiguchi H, Makita Y, Yamashita R, Nakai K (2007) Melina II: a web tool for comparisons among several predictive algorithms to find potential motifs from promoter regions. Nucleic Acids Res 35: (Web Server issue)W227–W231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ (2007) Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell 26: 593–602 [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT (2011) Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet 7: e1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL (2010) Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol 470: 759–795 [DOI] [PubMed] [Google Scholar]
- Smith MJ, Pascal CE, Grauvogel Z, Habicht C, Seeb JE, Seeb LW (2011) Multiplex preamplification PCR and microsatellite validation enables accurate single nucleotide polymorphism genotyping of historical fish scales. Mol Ecol Resour 11: Suppl 1268–277 [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F (2010) Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem 285: 27859–27868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Rafalska I, Stamm S (2002) YTH: a new domain in nuclear proteins. Trends Biochem Sci 27: 495–497 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R (2011) Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. Embo J 30: 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Blelloch R (2011) Small RNAs in early mammalian development: from gametes to gastrulation. Development 138: 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF (2009) CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Moazed D (2005) RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett 579: 5872–5878 [DOI] [PubMed] [Google Scholar]
- Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L (2009) Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol 53: 245–257 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M (2011) Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol 18: 94–99 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M (2010) Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. Embo J 29: 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M (2012) Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biology 2: 120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Watanabe Y, Nukina N, Yamamoto M (1998) RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95: 115–123 [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, Stamm S (2010) The YTH domain is a novel RNA binding domain. J Biol Chem 285: 14701–14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.