Abstract
Adaptation to endoplasmic reticulum (ER) stress depends on the activation of the unfolded protein response (UPR) stress sensor inositol-requiring enzyme 1α (IRE1α), which functions as an endoribonuclease that splices the mRNA of the transcription factor XBP-1 (X-box-binding protein-1). Through a global proteomic approach we identified the BCL-2 family member PUMA as a novel IRE1α interactor. Immun oprecipitation experiments confirmed this interaction and further detected the association of IRE1α with BIM, another BH3-only protein. BIM and PUMA double-knockout cells failed to maintain sustained XBP-1 mRNA splicing after prolonged ER stress, resulting in early inactivation. Mutation in the BH3 domain of BIM abrogated the physical interaction with IRE1α, inhibiting its effects on XBP-1 mRNA splicing. Unexpectedly, this regulation required BCL-2 and was antagonized by BAD or the BH3 domain mimetic ABT-737. The modulation of IRE1α RNAse activity by BH3-only proteins was recapitulated in a cell-free system suggesting a direct regulation. Moreover, BH3-only proteins controlled XBP-1 mRNA splicing in vivo and affected the ER stress-regulated secretion of antibodies by primary B cells. We conclude that a subset of BCL-2 family members participates in a new UPR-regulatory network, thus assuming apoptosis-unrelated functions.
Keywords: BH3-only proteins, BIM, ER stress, IRE1α modulation, PUMA
Introduction
Endoplasmic reticulum (ER) stress is a hallmark of secretory cells, and many diseases including cancer, neurodegeneration and diabetes, a process that involves the accumulation of misfolded proteins at the ER lumen (Hetz, 2012). Adaptation to ER stress depends on the engagement of the unfolded protein response (UPR) (Ron and Walter, 2007), a signal transduction pathway initiated by the activation of several sensors including inositol-requiring enzyme 1α (IRE1α). IRE1α is an ER-resident Ser/Thr protein kinase and endoribonuclease that upon activation, processes the mRNA of X-box-binding protein-1 (XBP-1). This shifts its codon reading frame resulting in the expression of the transcription factor XBP-1s (spliced XBP-1s) (Calfon et al, 2002; Lee et al, 2002). XBP-1s controls the expression of genes involved in folding, protein quality control, ER translocation and ER/Golgi biogenesis (Hetz and Glimcher, 2009), facilitating adaptation to protein folding stress. Besides, active IRE1α degrades mRNAs encoding certain ER proteins that are predicted to be difficult to fold, thus alleviating ER stress (Hollien and Weissman, 2006; Han et al, 2009; Hollien et al, 2009).
Chronic or irreversible ER stress results in apoptosis, which is regulated by members of the BCL-2 family of proteins (Tabas and Ron, 2011). Apoptosis signals converge at mitochondria, leading to the local activation of the pro-apoptotic multidomain BCL-2 family proteins BAX and BAK (Danial and Korsmeyer, 2004), which are activated by upstream ‘BH3-only’ proteins (Youle and Strasser, 2008). BH3-only proteins can be separated into two subtypes, activators that directly engage BAX and BAK to trigger cytochrome c release and apoptosis (i.e., BID, BIM and PUMA), but are sequestered by anti-apoptotic BCL-2 molecules; and sensitizers or inactivators (i.e., BAD and NOXA) that antagonize specific anti-apoptotic BCL-2 members, releasing activator BH3-only proteins (Kim et al, 2006; Youle and Strasser, 2008; Brunelle and Letai, 2009; Ren et al, 2010). Among these BH3-only proteins, BIM and PUMA are key regulators of ER stress-induced apoptosis (Reimertz et al, 2003; Li et al, 2006; Puthalakath et al, 2007; Kim et al, 2009) (reviewed in Woehlbier and Hetz, 2011).
Several components specifically regulate IRE1α function possibly due to a physical interaction (Gu et al, 2004; Luo et al, 2008; Gupta et al, 2010; Qiu et al, 2010) (reviewed in Hetz, 2012). For example, a novel function of BAX and BAK has been described at the ER where they modulate the amplitude of IRE1α signalling possibly through a physical association with the cytosolic domain of IRE1α (Hetz et al, 2006). Similarly, AIP1 and HSP72 instigate IRE1α signalling possibly due to an interaction (Luo et al, 2008; Gupta et al, 2010). All these findings indicate that IRE1α forms a macromolecular complex in which different signalling and regulatory components assemble around a scaffold that we have referred to as the UPRosome (Hetz and Glimcher, 2008; Hetz 2012). Upon prolonged ER stress, IRE1α activity is turned off (Yoshida et al, 2001; Lin et al, 2007), while PERK (PERK, double-stranded RNA-activated protein kinase (PKR)-like ER kinase) remains active, sensitizing chronically damaged cells to apoptosis (Lin et al, 2009). The ER-located anti-apoptotic protein BAX inhibitor-1 (BI-1) is involved in the inactivation of IRE1α (Bailly-Maitre et al, 2006; Lisbona et al, 2009; Bailly-Maitre et al, 2010), likely due to the direct binding to the UPRosome.
In order to identify new IRE1α interacting proteins, we performed a proteomic study and detected the association between PUMA and IRE1α in cells undergoing ER stress. Here, we show that BH3-only proteins modulate the kinetics of IRE1α signalling. The simultaneous deficiency of BIM and PUMA specifically disrupted the maintenance of sustained XBP-1 mRNA splicing over time, yet did not alter the kinetics of its activation. This resulted in a drastic decrease of XBP-1s expression and attenuated the upregulation of XBP-1s-target genes. In vitro studies demonstrated direct binding between BH3-only proteins and IRE1α, associated with a modulation of its RNAse activity. This effect was dependent on the BH3 domain of BIM. Furthermore, we demonstrated a crucial role of several BH3-only proteins in the control of immunoglobulin secretion by primary B cells, a physiological process that requires XBP-1 activity. Finally, BH3-only proteins modulated IRE1α signalling on an animal model of ER stress in the kidney and liver. Our results reveal an additional regulatory checkpoint in IRE1α signalling and suggest a novel biological function of BH3-only proteins at the ER membrane where they determine the kinetics and amplitude of IRE1α signalling.
Results
Physical interaction between BH3-only proteins and IRE1α
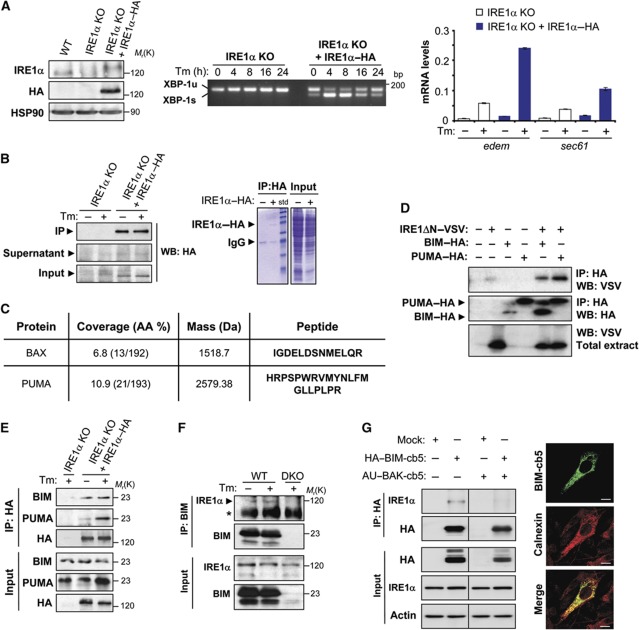
To screen for new potential IRE1α interacting proteins, we stably transduced IRE1α-deficient mouse embryonic fibroblast (MEFs) with retroviruses expressing the HA (human influenza hemagglutinin)-tagged version of full-length human IRE1α (IRE1α–HA). In conditions in which IRE1α–HA expression resembled that of endogenous IRE1α from wild-type (WT) MEFs, the activation and kinetics of XBP-1 mRNA splicing under conditions of ER stress were restored in addition to the upregulation of the target genes edem1 and sec61 (Figure 1A). Then, cells were exposed to the ER stress agent tunicamycin (Tm) for 6 h or left untreated, and IRE1α–HA immunoprecipitated using an anti-HA antibody conjugated to agarose (Figure 1B). To search for the possible association of new BCL-2 family members with IRE1α, we used two-dimensional liquid chromatography together with tandem mass spectrometry, followed by bioinformatic analyses. This approach led to the identification of 40 proteins that interacted with IRE1α exclusively in ER stress conditions. In addition to the known IRE1α interactor, BAX, another BCL-2 family member, PUMA, was discovered to bind to IRE1α (Figure 1C).
Figure 1.
IRE1α interacts with the BH3-only proteins BIM and PUMA. (A) IRE1α-deficient (IRE1α KO) cells were stably transduced with retroviral expression vectors for IRE1α–HA or empty vector. Left panel: the expression levels of IRE1α were analysed by western blot using an anti-IRE1α or anti-HA antibody. HSP90 levels were monitored as loading control. Middle panel: cells were treated or not with 100 ng/ml Tm for the indicated periods, and the levels of XBP-1 mRNA splicing monitored by RT–PCR. PCR fragments corresponding to the XBP-1u or XBP-1s forms of XBP-1 mRNA are indicated. Right panel: IRE1α KO and IRE1α–HA MEFs were treated for 24 h with 100 ng/ml of Tm and the levels of edem and sec61 mRNA were quantified by real-time PCR. Data represent the average and standard error of triplicates representing three independent experiments. (B) Cells described in (A) were treated or not with 100 ng/ml Tm for 6 h and then protein extracts were prepared in 1% CHAPS buffer. IRE1α–HA was immunoprecipitated (IP) and analysed by western blot (left panel). Total extracts, IP and supernatants after IP are shown. In addition, Coomassie blue staining is presented (right panel). (C) IPs presented in (B) were processed as described in Materials and methods and analysed by two-dimensional liquid chromatography together with tandem mass spectrometry. Peptides identified for BAX and PUMA in the analysis are indicated. (D) HEK293T cells were co-transfected with expression vectors for VSV-tagged cytosolic portion of IRE1α (IRE1ΔN–VSV) and HA-tagged PUMA or BIM (PUMA–HA and BIM–HA). After 48 h of transfection, HA-tagged proteins were immunoprecipitated and the possible interaction of BIM and PUMA with IRE1–VSV was analysed by western blot. As control, the expression levels of BIM–HA or PUMA–HA are presented in the IP, in addition to the expression levels of IRE1ΔN–VSV in total extracts. (E) The interaction of BIM and PUMA with IRE1α KO cells reconstituted using IP and western blot analysis. In addition, cells were treated or not with 100 ng/ml Tm for the indicated time points. (F) Endogenous interaction between BIM and IRE1α was analysed after the IP of BIM. BIM/PUMA DKO MEFs cells were used as control. (*) indicates non-specific band. (G) BAX and BAK DKO cells were reconstituted with an AU-BAK-cb5 expression vector or mock, and then transduced with retroviruses expressing HA–BIM-cb5 in the presence of 7.5 μM zVAD-fmk to prevent cell death. Left panel: After 20 h, HA was immunoprecipitated and the association with IRE1α was detected by western blot. Right panel: Immunofluorescence of BAX/BAK DKO cells transduced with HA–BIM-cb5 vectors using the HA tag (green). Calnexin was used as an ER marker (red). Scale bar: 10 μM. Figure source data can be found with the Supplementary data.
To confirm the physical association between PUMA and IRE1α, we first transiently transfected HEK293T cells with HA-tagged PUMA, as well as a VSV-tagged version of the cysotolic domain of IRE1α containing both the kinase and endoribonuclase activities. As additional controls we included expression vectors for other BH3-only proteins such as BIM, BNip3, BMF, BLK, NIX and DP5, all of which were HA tagged (Kim et al, 2006). After immunoprecipitation (IP) of the HA-tagged BH3-only proteins, we detected the specific co-precipitation of VSV–IRE1α with PUMA, and further identified an association with BIM and BNip3, but not with any of the other BH3-only proteins tested (Figure 1D; Supplementary S1). We then validated the interaction of IRE1α with endogenous BH3-only proteins in IRE1α KO cells reconstituted with physiological levels of IRE1α–HA (Figure 1A). We were able to confirm an association of BIM and PUMA with IRE1α when the HA tag was pulled-down (Figure 1E). Tm treatment enhanced the association of PUMA with IRE1α consistent with our proteomic analysis, whereas the interaction with BIM was not affected by ER stress (Figure 1E). We also corroborated the presence of endogenous BIM/IRE1α complexes when BIM was immunoprecipitated and then the association with IRE1α was determined (Figure 1F). Finally, we examined the possible contribution of the ER-located pool of BIM in the interaction with IRE1α and its relation to BAX/BAK. Using the reticular targeting signal of cytochrome b5, BAX/BAK double-knockout (DKO) cells were engineered to express an ER-located version of BAK (Supplementary Figure S1B) (Klee et al, 2009). We then enforced the expression of an ER-targeted version of HA-BIM using a retroviral system as previously described (Figure 1G, right panel) (Klee et al, 2009). IP of ER-located BIM demonstrated an interaction with endogenous IRE1α, which was dramatically reduced by the expression of BAK (Figure 1G). This interaction was not modulated by Tm treatment (Supplementary Figure S1B). Taken together, these results indicate that BIM and PUMA form a protein complex with IRE1α.
Expression of BIM and PUMA regulate the maintenance of XBP-1 mRNA splicing
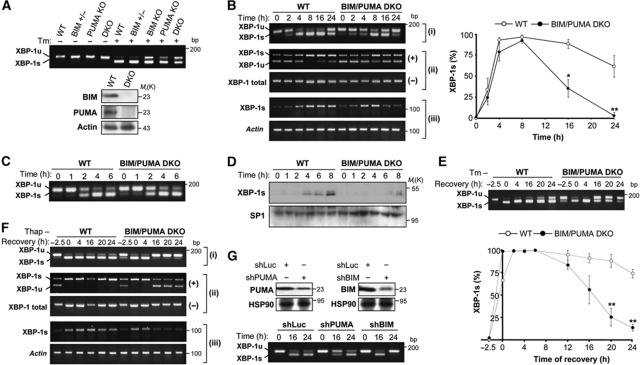
To explore the putative role of BH3-only proteins in UPR signalling, we generated MEFs lacking BIM or PUMA or both BIM and PUMA (DKO) (Kim et al, 2009). Cells were treated with 100 ng/ml Tm, and XBP-1 mRNA splicing was assessed by reverse transcription polymerase chain reaction (RT–PCR). As compared with the corresponding WT or BIM heterozygous MEFs, BIM or PUMA KO cells presented a reduction in the levels of Tm-inducible XBP-1 mRNA splicing. This effect was even stronger in BIM/PUMA DKO cells (Figure 2A). BNip3 was not studied in MEFs because its expression was not detectable at basal levels or after induction of ER stress (Supplementary Figure S1C).
Figure 2.
BIM and PUMA double deficiency precludes the maintenance of sustained XBP-1 mRNA splicing under ER stress conditions. (A) WT, BIM−/−, PUMA+/−, PUMA−/− or BIM−/− PUMA−/− DKO MEFs were incubated for 3 h in the absence or presence of 100 ng/ml Tm and XBP-1 mRNA splicing was monitored by RT–PCR. PCR fragments corresponding to the XBP-1u or XBP-1s forms of XBP-1 mRNA are indicated in the upper panel. BIM and PUMA deficiency was confirmed by immunoblot (bottom panel). (B) WT and BIM/PUMA DKO cells were treated with 100 ng/ml of Tm for indicated time points and then xbp-1 mRNA splicing was monitored by RT–PCR. (i) For regular splicing assay, the percentage of XBP-1 mRNA splicing was calculated after densitometric analysis of the XBP-1u and XBP-1s-related PCR products to quantify the percentage of splicing in each time point (left panel). A representation of the quantification all single experiments is presented in Supplementary Figure S2A. (ii) PstI-based XBP-1 mRNA splicing assay. PCR products were incubated in the absence (−) or presence of PstI (+) for further electrophoresis analysis. The splicing excises the PstI site from the amplicon. (iii) Specific primers to amplify XBP-1s were employed using semi-quantitative PCR. Actin was used as loading control. Bottom panel: Data represent the average and standard error of three independent experiments analysed with regular splicing assay. Statistically significant differences are indicated (*P<0.01; **P<0.001). (C) Cells were treated as indicated in (B) at indicated time points and XBP-1 mRNA splicing was monitored by RT–PCR. (D) WT and BIM/PUMA DKO cells were treated as in (B), and the expression of XBP-1s was monitored by immunoblot of nuclear extracts. SP1 levels were monitored as loading control. (E) WT and BIM/PUMA DKO cells were incubated with 0.5 μg/ml Tm for 2.5 h. Then, cells were washed three times with PBS to remove Tm, and fresh culture media was added. The levels XBP-1 mRNA splicing was monitored by RT–PCR as described in (B). Upper panel: A representative experiment is presented. Bottom panel: The percentage of XBP-1 mRNA splicing was calculated and data represent the average and standard error of three independent experiments. Statistically significant differences are indicated (**P<0.001). (F) A similar experiment was performed as described in (E) after treatment with a pulse of 500 nM Thap for 2.5 h and analysed with all XBP-1 mRNA assays presented in (B). (G) WT MEFs cells were stably transduced with lentiviruses expressing shRNA constructs against Luciferase (Luc), BIM (shBIM) or PUMA (shPUMA). Cells were incubated for indicated time points with 100 ng/ml of Tm. Decreased expression of BIM and PUMA was confirmed by western blot analysis (upper panels). The levels XBP-1 mRNA splicing was monitored by RT–PCR as described in (B) (lower panel). Figure source data can be found with the Supplementary data.
After persistent ER stress, the levels of XBP-1 mRNA splicing progressively decline due to IRE1α inactivation (Yoshida et al, 2001; Lin et al, 2007; Lisbona et al, 2009). To evaluate the effects of BIM/PUMA on the sequential activation and inactivation of IRE1α, we quantified XBP-1 mRNA splicing by RT–PCR in time-course experiments. The inactivation of XBP-1 mRNA splicing was markedly accelerated in BIM/PUMA DKO cells, yielding full recovery of the unspliced XBP-1u (XBP-1u) form to baseline levels after 24 h of Tm treatment. In the same conditions, at 24 h post-ER stress, WT control cells exhibited still ∼70% XBP-1 mRNA splicing (Figure 2B; Supplementary Figure S2A). In sharp contrast, the kinetics of activation of XBP-1 mRNA splicing were not drastically different in WT and BIM/PUMA DKO cells, showing a similar slope of increase that reached 100% of splicing after 6 h of treatment with Tm. Only a slight, yet statistically insignificant (ANOVA test) delay in the activation phase of XBP-1 mRNA splicing (between 4 and 8 h) was observed in BIM/PUMA DKO cells when compared with WT control. A similar pattern was observed when ER stress was induced by means of dithiothreitol (DTT) (Supplementary Figure S2B). Consistent with this result, careful kinetic analysis of early activation time points reveal no effects on XBP-1 mRNA splicing in BIM and PUMA DKO cells (Figure 2C). We confirmed the effects of BIM and PUMA deficiency on the attenuation of XBP-1 mRNA splicing using two alternative assays based on PstI digestion of PCR products or the selective amplification of the XBP-1s form using specific primers (Figure 2B; see Materials and methods). Finally, dose response and kinetic experiments demonstrated that the effects of BIM and PUMA on XBP-1 mRNA splicing were only observed in conditions of mild ER stress (Supplementary Figure S2C).
Analysis of XBP-1s protein expression revealed a strong reduction in ER-stressed BIM and PUMA DKO cells as compared with WT control cells (Figure 2D), suggesting the requirement of sustained IRE1α signalling for the expression of a detectable pool of XBP-1s. To address the possible effects of BIM and PUMA on the inactivation of XBP-1 mRNA splicing in a different setting, we exposed WT and BIM/PUMA DKO cells to a pulse of high concentrations of Tm (500 ng/ml) for 2.5 h to induce a rapid induction of full XBP-1 mRNA splicing. Then, Tm was washed out to monitor the recovery of the XBP-1u mRNA form over time. Using this approach, drastic differences were observed in the inactivation of XBP-1 mRNA splicing. Sustained and maximal splicing was observed in WT cells up to 24 h post-Tm treatment (Figure 2E), whereas BIM and PUMA DKO cells recovered significant levels of XBP-1u mRNA (Figure 2E). Similar results were observed when cells were exposed to a pulse of thapsigargin (Thap) using three splicing assays (Figure 2F), confirming that BIM and PUMA influenced the inactivation (but not the activation) phase of XBP-1 mRNA splicing. As a control, we assessed the maintenance of the activity of the ER stressors Tm or Thap after prolonged incubation times (Supplementary Figure S3A). To corroborate the role of BIM and PUMA on the regulation of XBP-1 mRNA splicing, we knocked down BIM or PUMA with the stable delivery of shRNA constructs using lentiviral vectors. As shown in Figure 2G, reducing the expression of these two BH3-only proteins attenuated the levels of XBP-1 mRNA splicing in cells undergoing prolonged ER stress. As an additional control, we monitored the mRNA stability of XBP-1 mRNA, observing no differences in the decay of the XBP-1 mRNA in WT and BIM/PUMA DKO cells (Supplementary Figure S3B). Taken together, our results suggest that the expression of BIM and PUMA is essential for sustaining XBP-1 mRNA splicing upon ER stress.
Deficient upregulation of XBP-1s-target genes and decreased IRE1α-dependent mRNA decay in BIM and PUMA DKO cells
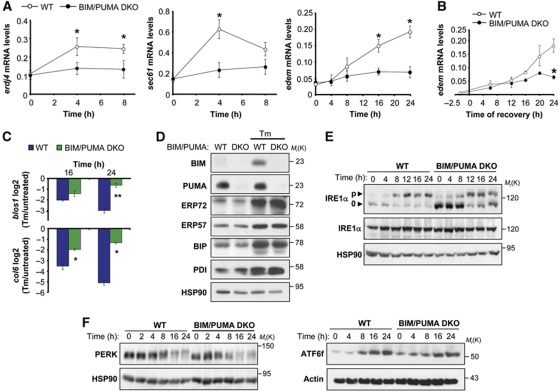
We explored the impact of BIM/PUMA DKO on UPR downstream responses. Treatment of WT cells with Tm led to a more pronounced upregulation of the XBP-1s-target genes erdj4, sec61 and edem (Lee et al, 2003b) at the mRNA level when compared with BIM/PUMA DKO cells (Figure 3A and B), consistent with the drastic decrease of XBP-1s expression in BIM/PUMA DKO cells. bip mRNA or protein levels (a UPR-target gene independent of XBP-1s) were not significantly affected (Figure 3D and Supplementary Figure S2D). These results are according with the hypothesis that sustained XBP-1 mRNA splicing is required to generate active XBP-1s-dependent responses.
Figure 3.
Effects of the BIM/PUMA DKO on IRE1α-dependent downstream responses. (A) WT and BIM/PUMA DKO MEFs were treated with 100 ng/ml of Tm and the levels of erdj4, sec61 and edem mRNA were quantified by real-time PCR. (B) WT and BIM/PUMA DKO cells were incubated with 0.5 μg/ml Tm for 2.5 h. Then, cells were washed three times with PBS and fresh culture media was added and the levels edem mRNA were quantified at the indicated time points by real-time PCR as shown in (A). (C) WT and BIM/PUMA DKO cells were treated with 100 ng/ml Tm for 16 and 24 h, and the mRNA decay of the IRE1α targets blos1 and col6 was monitored by real-time PCR and normalized with respect to the levels of rpl19 as a housekeeping gene. Values were normalized with the mRNA levels under untreated conditions. (D) BIM and PUMA WT and DKO cells were treated with 100 ng/ml Tm for 16 h, and the expression levels of indicated proteins were monitored by western blot. (E) Phosphorylation levels of IRE1α upon Tm-induced ER stress were analysed in WT and BIM/PUMA DKO cells after treatment of cells with 100 ng/ml Tm for indicated time points. Followed by a Phostag assay and western blot analysis (p, indicates phosphorylated and 0, indicates non or hypophosphorylated band). Total IRE1α were also analysed using conventional electrophoresis and western blot analysis (bottom panel). Of note, differences in the total mass of IRE1α is regularly observed with the PhosTag assay possibly due to inefficient transfer in the western blot analysis. Data represent the analysis of four independent experiments. (F) PERK phosphorylation shift was analysed in cells described in (E) on an 8% polyacrylamide gel (left panel). In parallel, the active fragment of ATF6 was monitored in WT and BIM/PUMA DKO cells undergoing ER stress. In (A–C), all data represent the average and standard error of three independent experiments. Statistically significant differences are indicated (*P<0.01; **P<0.001). Figure source data can be found with the Supplementary data.
Recent reports indicate that the RNAse activity of IRE1α mediates the rapid degradation of a subset of mRNAs that either encode ER proteins with transmembrane domains or secreted proteins that may be difficult to fold (Hollien and Weissman, 2006). Targets for IRE1α-dependent mRNA decay were defined in MEFs and included genes such as blos1, and col6 (Hollien et al, 2009). While WT cells exposed to mild ER stress (100 ng/ml Tm) exhibited a marked decay of the blos1 and col6 mRNAs, little decay was observable in BIM/PUMA DKO cells cultured in similar conditions (Figure 3C). When cells were exposed to higher doses of Tm (3 μg/ml), the DKO did not influence the decay of blos1 or col6 mRNA (Supplementary Figure S2F).
BIM and PUMA specifically regulate the IRE1α signalling branch
We performed several control experiments to exclude unrelated effects of BIM and PUMA on ER homeostasis. A variety of BCL-2 protein family members regulate ER calcium homeostasis (Rodriguez et al, 2011), which could affect the folding capacity of the ER and the susceptibility of cells to ER stress. Thus, we monitored the release of ER calcium in BIM/PUMA DKO cells after stimulation with Thap or ionomycin in the absence of extracellular calcium. No differences in the kinetics and amplitude of ER calcium release were observed between BIM/PUMA DKO cells and WT controls (Supplementary Figure S5A). In contrast, BAX/BAK DKO cells exhibited drastic changes in ER calcium fluxes (Supplementary Figure S5B), as previously described (Scorrano et al, 2003; Zong et al, 2003). In agreement with these results, no alterations were observed in BIM/PUMA DKO cells in the basal or inducible expression of a variety of essential ER chaperones and foldases that are not regulated by XBP-1 (Lee et al, 2003b), including BiP, ERp57, PDI, ERp72, calreticulin and calnexin (Figure 3D and data not shown). These data suggest that calcium homeostasis and protein folding at the ER is not altered by BIM and PUMA double deficiency, ruling out possible non-specific effects on the basal physiology of the ER.
We then monitored the specificity of BIM and PUMA on the regulation of distinct UPR stress sensors. The phosphorylation state of IRE1α was analysed using a Phostag™ protocol (Yang et al, 2010). Time-course experiments indicated early dephosphorylation of IRE1α in BIM and PUMA DKO cells in addition to a slight decrease in the amplitude of phosphorylation (Figure 3E). Analysis of total IRE1α levels with conventional western blot indicated no changes in BIM and PUMA DKO cells (Figure 3E). Consistent with these results, accumulation of large IRE1α oligomers was reduced after prolonged ER stress as measured with non-denaturing gels (Supplementary Figure S4A). Moreover, BIM and PUMA deficiency did not alter the subcellular distribution of IRE1α (Supplementary Figure S4B). IRE1α is known to control the activation of JNK through binding to TRAF2 (Urano et al, 2000). The transient phosphorylation of JNK observed in WT cells exposed to a low concentration of Tm (100 ng/ml, see 2 h) was absent in BIM/PUMA DKO cells (Supplementary Figure S4C). In contrast, activation of PERK was not drastically affected in DKO cells as monitored by a shift in its molecular weight by western blot analysis (Figure 2F, left panel). Similarly, the generation of ATF6 active fragment was similar in both WT and BIM and PUMA DKO cells (Figure 2F, right panel). Altogether, our data indicate that BIM and PUMA double deficiency leads to early inactivation of IRE1α signalling, resulting in decreased XBP-1s expression, reduced upregulation of its target genes, attenuated JNK activation, and deficient IRE1α-mediated decay of specific ER stress-relevant mRNA species.
A BH3 mimetic or the expression of BAD modulate the levels XBP-1 mRNA splicing
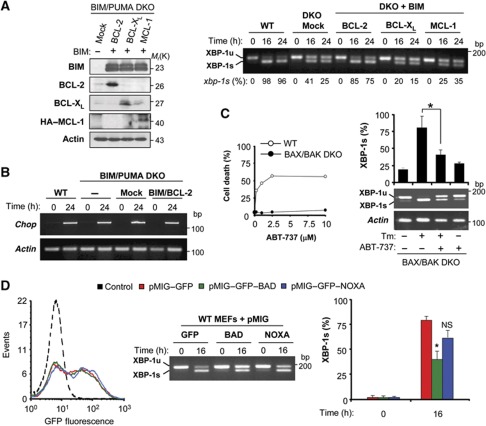
Under resting conditions, BIM and PUMA are kept in check by inhibitory interactions with anti-apoptotic proteins from the BCL-2 family including BCL-2 itself, BCL-XL and MCL-1 (Kim et al, 2006; Youle and Strasser, 2008; Brunelle and Letai, 2009). We investigated whether a pool of BIM/PUMA protein that interacts with anti-apoptotic BCL-2 family proteins might participate in the regulation of IRE1α. BIM and PUMA DKO cells were transduced with bicistronic retroviruses co-expressing BIM in combination with BCL-2, BCL-XL or MCL-1 (Kim et al, 2006) (Figure 4A, left panel). Surprisingly, relatively normal levels of XBP-1 mRNA splicing were only restored in BIM/PUMA DKO cells expressing BIM/BCL-2, not in those expressing BIM/MCL-1 or BIM/BCL-XL (Figure 4A, right panel).
Figure 4.
Effects of BAD or the BH3 mimemtic ABT-737 on XBP-1 mRNA splicing. (A) BIM and PUMA DKO cells were stably transduced with bicistronic retroviral vectors expressing BIM in combination with BCL-2, BCL-XL or MCL-1 (see Materials and methods for details). As control, cells were transduced with an empty vector (Mock). The expression of the aforementioned proteins was confirmed by immunoblot. Actin levels were monitored as loading control (left panel). Then, cells were exposed to 100 ng/ml Tm, and XBP-1 mRNA splicing levels determined by RT–PCR over time (right panel). The percentage of XBP-1 mRNA splicing (XBP-1s %) is presented calculated after densitometric analysis (bottom of gel). (B) WT or DKO cells transduced with the empty vector (DKO+Mock) or BIM/BCL-2 expression retroviruses were incubated with Tm (100 ng/ml) for 24 h. After treatment, chop mRNA was monitored by RT–PCR using actin mRNA as loading control. (C) WT or BAX and BAK DKO cells were treated with indicated concentrations of ABT-737 and after 24 h cell viability was analysed after PI staining and FACS analysis (left panel). Then BAX and BAK DKO cells were treated with 100 ng/ml of Tm in the absence or presence of ABT-737 (10 μM). At indicated time points, levels of XBP-1 mRNA splicing were monitored by RT–PCR. Data represent the analysis of three independent experiments. Statistically significant differences are indicated (*P<0.01). (D) BIM and PUMA WT cells were transiently transduced with pMIG–GFP retroviral expression vectors expressing BAD, NOXA or empty vector. Left panel: Efficiency of retroviral transduction was monitored by quantifying GFP fluorescence using FACS. Right panels: After 72 h of transduction, cells were treated with 100 ng/ml Tm, and the levels of XBP-1 mRNA splicing were monitored by RT–PCR. Bars represent the average and standard error of three independent experiments. Statistically significant differences are indicated (*P<0.01).
To study the possible requirement of the BH3 domain in the regulation of XBP-1 mRNA splicing, we employed the well-characterized BH3 domain mimetic, ABT-737, which disables the interaction between BCL-2 and BH3-only proteins through a competitive inhibition (Oltersdorf et al, 2005). Since ABT-737 induces apoptosis on a BAX/BAK-dependent manner (Figure 4C, left panel) and the interaction of BIM with IRE1α is independent of BAX and BAK (Figure 1G), we exposed BAX and BAK DKO MEFs to Tm in the presence or absence of ABT-737, and then XBP-1 mRNA splicing was monitored. ABT-737 treatment significantly attenuated the induction of XBP-1 mRNA splicing by Tm (Figure 4C, right panel). Similar results were obtained when these experiments were performed in WT cells treated in the presence of zVAD-fmk (Supplementary Figure S6A and B). To complement this pharmacological approach, we transiently expressed the inactivator BH3-only proteins BAD or NOXA using equal titters of retroviruses to selectively disrupt the association of BIM/PUMA with BCL-2/BCL-XL or MCL-1, respectively (Kim et al, 2006). Consistent with the effects of BCL-2 on XBP-1 mRNA splicing, only expression of BAD (but not that of NOXA) accelerated the inactivation of XBP-1 mRNA splicing in WT cells undergoing ER stress (Figure 4D).
BH3-only proteins interact with IRE1α through the BH3 domain and regulate its RNAse activity on a cell-free system
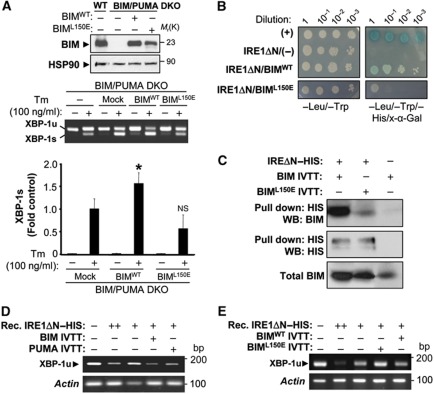
The BH3 domain of BH3-only proteins is essential for their apoptosis-regulatory activities. We performed site-directed mutagenesis on the BH3 domain of BIM and tested its impact on the levels of XBP-1 mRNA splicing. We transiently transfected expression vectors for BIMWT, BIML150E or empty vector (Figure 5A) into BIM and PUMA DKO cells (∼30% efficiency of transfection), and then exposed cells to 100 ng/ml Tm. An enhancement of XBP-1 mRNA splicing was observed after enforced expression of BIMWT, but not BIML150E mutant (Figure 5A).
Figure 5.
BIM controls XBP-1 mRNA splicing possibly through a direct interaction involving the BH3 domain. (A) BIM and PUMA DKO were transiently transfected with expression vectors for BIMWT, BIML150E or empty vector in the presence of 50 μM zVAD-fmk. After 20 h, cells were stimulated with Tm for 16 h and XBP-1 mRNA splicing monitored by RT–PCR. Percentage of XBP-1 mRNA is indicated. Upper panel: As control, the expression of BIM was monitored by western blot. Middle panel: XBP-1 mRNA splicing was monitored over time and quantified. Of note, transient transfection leads to an enhancement of XBP-1 mRNA splicing after Tm treatment. This control was used to normalize the levels of XBP-1 mRNA splicing and calculate the contribution due to BIM expression as a fold induction from Mock control (lower panel). Bars represent the average and standard error of three independent experiments. Statistically significant differences are indicated (*P<0.01). Efficiency of transfection was measured by FACS for all experiments (∼30% efficiency). (B) The possible interactions between BIMWT, BIML150E with the cytosolic domain of IRE1α (IRE1ΔN) were tested using a two-hybrid system (see Materials and methods). Different serial dilutions of yeast cultures are presented in control (−Leu/−Trp) and selection media (−Leu/−Trp/−His/X-α-Gal). (+): Indicate a positive control for the assay. (C) Recombinant IRE1ΔN–HIS (1 μg) was incubated with IVTT BIMWT or BIML150E. Their interactions were tested after pulling down IRE1ΔN–HIS and performing a western blot analysis (see Materials and methods). (D) The endoribonuclease activity of 0.1 μg of IRE1ΔN–HIS (+) was monitored in vitro using the conditions described in Supplementary data in the presence of IVTT BIM-EL, PUMA or a mock preparation. Substrate was added (total mRNA) and after 1 h, mRNA was re-extracted and endoribonuclease activity of IRE1α was analysed by RT–PCR. Actin levels were monitored as loading control. As positive control, mRNA was treated with 1 μg IRE1ΔN–HIS (++). (E) In parallel, using the conditions described in (h), the effects on the IRE1α RNAse activity of with IVTT BIMWT or BIML150E were tested. Figure source data can be found with the Supplementary data.
Additionally, we investigated the possible direct binding of BIM with IRE1α. Using a yeast two-hybrid system, we were able to confirm the interaction of BIMWT and IRE1α-ΔN in living cells (Figure 5B). In contrast, BIML150E considerably lost the interaction with IRE1α-ΔN under the same experimental conditions (Figure 5B). We also tested the possible physical association of BH3-only proteins with IRE1α using purified components. The cytosolic domain of human IRE1α was purified from insect cells using a HIS tag (IRE1ΔN–HIS), and then incubated with in vitro transcribed and translated (IVTT) BIM. Pull-down experiments then revealed an interaction of IRE1ΔN–HIS and BIMWT (Figure 5C). Remarkably, a reduction of the binding between IRE1ΔN–HIS and BIML150E protein was observed (Figure 5C).
Finally, we monitored the possible effects of BH3-only proteins on the endoribonuclease activity of IRE1α in a recently described cell-free assay (Lisbona et al, 2009; Gupta et al, 2010). Recombinant IRE1ΔN–HIS was incubated for 1 h with IVTT BIM or PUMA. Then, a mixture of total mRNA was added as a substrate for IRE1α endoribonuclease activity in the presence of ATP. After 1 h of incubation, mRNA was re-extracted, and the cleavage of XBP-1u mRNA in the splicing site was monitored by RT–PCR as a sign of IRE1α ribonuclease activity. As an internal control, actin mRNA levels were monitored. The enzymatic activity of recombinant IRE1α was enhanced in the presence of in vitro synthesized BIM or PUMA (Figure 5D; Supplementary S6C). The effects of BIM over IRE1ΔN–HIS RNAse activity were ablated when BIML150E was used in the in vitro splicing assay (Figure 5E). Altogether, these results suggest that a subset of BCL-2 family members participate in a regulatory network that regulates the kinetics of IRE1α signalling possibly through a direct modulation of its RNAse activity.
BH3-only proteins regulate XBP-1 mRNA splicing in vivo
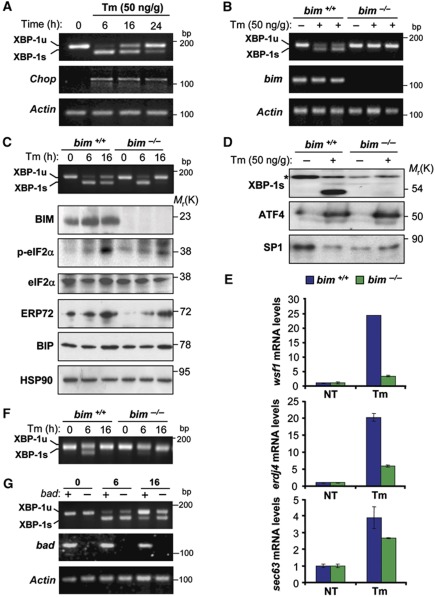
To establish that IRE1α is regulated by BH3-only proteins in vivo, we intraperitoneally injected WT and bim−/− mice with Tm (50 ng/kg). In WT livers, Tm-induced XBP-1 mRNA splicing reached a maximum at 6 h post-injection, and then returned almost completely to the unspliced form at 24 h. In contrast, the upregulation of chop mRNA was sustained 24 h post-Tm injection in WT mice, suggesting that the attenuation of XBP-1 splicing resulted from a specific downregulation rather than, for example, from Tm detoxification (Figure 6A). Remarkably, bim−/− livers presented a strong attenuation of XBP-1 mRNA splicing at 16 h compared with control mice (Figure 6B). In contrast, XBP-1 mRNA splicing levels were similar in bim+/+and bim−/− livers 6 h post-injection, suggesting that BIM modulated the decline of XBP-1 mRNA splicing rather than its activation (Figure 6C). The effects of BIM expression on the kinetics of XBP-1 mRNA splicing translated into a major reduction in XBP-1s protein levels, as detectable in liver nuclear extracts from bim−/− mice (Figure 6D). In agreement with these results, the induction of the XBP-1-target genes wfs1, erdj4 and sec63 were reduced in bim−/− mice (Figure 6E). Remarkably, expression of ATF4 was not affected by bim deficiency (Figure 6D). Similarly, induction of BiP or ERp72 were not drastically altered in BIM-deficient mice (Figure 6C), and only a slight alteration in phosphorylation of eIF2α was observed, further supporting the conclusion that BIM expression specifically regulates the IRE1α/XBP-1 arm of the ER stress response in vivo. Additionally, we monitored the effects of BIM on the regulation of Tm-triggered XBP-1 mRNA splicing in other organs such as the kidney, in which Tm (50 ng Tm/g) caused a mild response in WT mice that is attenuated in bim−/− mice (Figure 6F). Finally, we study the possible involvement of other BH3-only protein in the regulation of XBP-1 mRNA splicing in vivo. Of note, bad−/− livers exhibited a delayed downregulation of XBP-1 mRNA splicing upon Tm injection (Figure 6G), consistent with the results obtained in vitro after BAD overexpression.
Figure 6.
bim deficiency attenuates XBP-1 mRNA splicing in vivo. (A) To monitor the kinetics of XBP-1 mRNA splicing in an animal model of ER stress, bim+/+ mice were injected with a single dose of Tm (50 ng/g, i.p.). After injection, animals were sacrificed at different time points, and the levels of XBP-1 mRNA splicing were monitored by RT–PCR on total liver cDNAs. As a control, chop and actin expression levels were monitored by semi-quantitative RT–PCR. (B) bim+/+ or bim−/− mice were injected with Tm (50 ng/g, i.p.) and XBP-1 mRNA splicing was monitored after 16 h of injection. bim and actin levels were monitored as controls. Each well of the gel represents independent animals. (C) bim+/+ or bim−/− mice were injected with Tm (50 ng/g, i.p.) and XBP-1 mRNA splicing was monitored after indicated time points. In addition, phophorylation of eIF2α, and levels of BiP, ERp72, BIM, total eIF2α and Hsp90 were monitored by western blot in the same samples. (D) The levels of XBP-1s were analysed by western blot of nuclear extracts from liver samples, as described in (B). In addition, ATF4 was monitored in the same samples. A non-specific band was used as loading control (*), as well as SP1 nuclear protein levels. (E) wfs1, erdj4 and sec63 mRNA levels were quantified in samples described in (B) using real-time PCR and normalized with β-actin levels in each sample. Each animal cDNA was analysed in triplicates. Data represent mean and s.d. (F) In parallel, in the same animals presented in (C), the levels of XBP-1 mRNA splicing were monitored in mRNA purified from kidney. (G) bad+/+ or bad−/− mice were injected as described in (B). XBP-1 mRNA splicing was monitored in the liver extracts after indicated time points. As control bad and actin mRNA levels were monitored. Figure source data can be found with the Supplementary data.
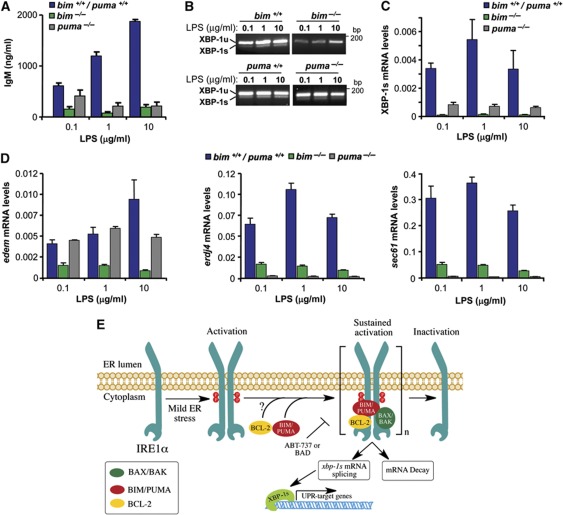
BH3-only proteins modulate immunoglobulin secretion and XBP-1 mRNA splicing in lipopolysaccharide-stimulated primary B cells
Specialized secretory cells such as B lymphocytes rely on continuous protein folding and quality control in their ER, and this process require XBP-1 and IRE1α (Reimold et al, 2001; Iwakoshi et al, 2003; Zhang et al, 2005), but not PERK activation (Gass et al, 2008). To assess the possible impact of BIM in the control of XBP-1 mRNA splicing in a physiologically model of ER stress, we first monitored the rate of IgM secretion in bim−/− primary B cells that were stimulated with different concentrations of lipopolysaccharide (LPS). As compared with WT controls, bim−/− B cells secreted much less IgM upon LPS stimulation (Figure 7A). A similar phenotype in IgM secretion was observed in LPS-stimulated puma−/− primary B cells (Figure 7A). This secretory defect was associated with a marked reduction in LPS-induced XBP-1 mRNA splicing in both bim−/− and puma−/− B cells as monitored by RT–PCR (Figure 7B). Since physiological levels of XBP-1 mRNA splicing are low, we confirmed these results with a quantitative assay using real-time PCR (Figure 7C). These effects on XBP-1 mRNA splicing directly correlated with a drastic attenuation in the upregulation of the XBP-1s-target genes edem, erdj4 and sec61 in LPS-stimulated B cells (Figure 7D). In contrast, bad−/− B cells secreted more IgM that WT control cells, associated with enhanced XBP-1 mRNA splicing (Supplementary Figure S7A and B). Taken together, these data indicate that BH3-only proteins regulate XBP-1 mRNA splicing in a physiological model of ER stress associated with a high demand of protein folding and secretion.
Figure 7.
BIM and PUMA expression regulates XBP-1 MRNA splicing and immunoglobulin secretion in primary B cells. (A) Primary B cells were purified from spleens of bim/puma+/+, bim−/− and puma−/− mice. Then, cells were stimulated with LPS (0.1, 1.0 and 10 μg/ml). After 2 days of culture, IgM concentrations were measured in the supernatants by ELISA assay. (B) In samples presented in (A), mRNA was extracted and XBP-1 mRNA splicing was monitored after LPS stimulation by RT–PCR. (C) As in (B), XBP-1s mRNA processed form was quantified by real-time PCR. (D) XBP-1s-target genes edem, erdj4 and sec61 were monitored by real-time PCR in samples described in (B). Bars represent the average and standard error of three parallel cultures. (E) Working model: BH3-only proteins regulate the sustained signalling of IRE1α. ER stress triggers IRE1α dimerization and phosphorylation, leading to engagement of its ribonuclease activity, which process XBP-1 mRNA and lead to the decay of certain mRNAs. The sustained signalling of IRE1α after prolonged ER stress requires the interaction with accessory proteins including activator BH3-only proteins (i.e., PUMA and BIM) and BCL-2. The interaction of BH3-only protein and IRE1α is mediated by the BH3 domain and regulates the phosphorylation and oligomerization status of the stress sensor. As a functional consequence, the maintenance of IRE1α signalling leads to the expression of an active pool of XBP-1s in the nucleus, leading to the engagement of UPR downstream transcriptional responses. Inactivator/sensitizer BH3-only proteins (i.e., BAD) or by treatments with the BH3-mimetic drug ABT-737 attenuates XBP-1 mRNA splicing. The exact mechanisms underlying the regulation of IRE1α by BCL-2 remains to be determined. This model suggests an additional regulatory checkpoint in IRE1α signalling and reveals a novel biological function of BH3-only proteins at the ER membrane where they determine the kinetic and amplitude of IRE1α signalling.
Discussion
Although IRE1α constitutes the phylogenetically most conserved branch of the UPR, little is known about the regulation of its activity. Under ER stress conditions, XBP-1s controls the induction of a vast spectrum of UPR-related genes involved in almost every aspect of the secretory pathway (Lee et al, 2003b; Shaffer et al, 2004; Acosta-Alvear et al, 2007). Recent evidence from several laboratories indicates that IRE1α activation is specifically regulated by a set of positive and negative regulators many among which have previously been linked to apoptosis (reviewed in Hetz et al, 2011; Woehlbier and Hetz, 2011). In the control of apoptosis, BH3-only proteins act as direct or indirect activators of BAX and BAK at mitochondria, hence stimulating the permeabilization of the outer mitochondrial membrane (reviewed in Youle and Strasser, 2008). Here, we report a novel function of BH3-only proteins where they modulate specific UPR-related events. Careful kinetic analyses of cells exposed to ER stress, led to the conclusion that BIM/PUMA specifically affects the sustained activation of IRE1α. At the molecular level, we mapped the effect of BIM and PUMA on the phosphorylation status of IRE1α and the dissociation of clusters upon prolonged ER stress, thus modulating IRE1α RNAse activity. Interestingly, a recent report revealed the existence of an allosteric site on IRE1α that affects its ribonuclease activity (Wiseman et al, 2010). It remains to be determined if BCL-2 family members regulate IRE1α activity through this site.
Our results suggest an intriguing scenario where BIM/PUMA expression may have distinct consequences under ER stress depending on their subcellular localization and the intensity of the stimuli. Under mild ER stress, BIM and PUMA would act as stress sentinels and stimulate the activation of early adaptive responses to ER stress by the maintenance of IRE1α signaling. In fact, the expression of BIM or PUMA modulate the secretion of IgM by primary B cells, a biological process associated with the occurrence of physiological and non-apoptotic levels of ER stress, which is strictly dependent on the IRE1α/XBP-1 signalling branch of the UPR (Reimold et al, 2001; Iwakoshi et al, 2003; Zhang et al, 2005; Gass et al, 2008). In addition, the effects of BIM and PUMA on XBP-1 splicing occurred before the induction of cell death (Supplementary Figure S2E) and overall the effects of BCL-2 family members on apoptosis clearly dissociated from their impact on the UPR (Supplementary Figure S7C). Conversely, under chronic or irreversible ER stress, BIM/PUMA would enforce mitochondrial-mediated apoptosis and hence eliminate irreversible damaged cells.
Several non-apoptotic activities for BCL-2 family members have been described over the last few years (reviewed in Hetz and Glimcher, 2008). For example, BAD and NOXA controls glucose metabolism (Danial et al, 2003, 2008; Lowman et al, 2010), whereas BID expression modulates DNA repair responses (Kamer et al, 2005; Zinkel et al, 2005). BCL-2 and BCL-XL can inhibit autophagy, a phenomenon that is antagonized by BH3-only proteins (Pattingre et al, 2005; Maiuri et al, 2007). BCL-2 and BCL-XL also modulate pro-inflammatory processes through direct interactions with NALP1 (Bruey et al, 2007), and BAX and BAK control mitochondrial morphogenesis (Karbowski et al, 2006). Finally, several members of the BCL-2 family regulate ER calcium fluxes (Rodriguez et al, 2011). The data presented here indicate yet another non-apoptotic and physiologically relevant function of BH3-only proteins, namely the control of the UPR. As a common denominator of their non-apoptotic functions, BCL-2 family members may act as specialized stress sentinels on a novel regulatory network that modulate adaptive responses and trigger cell death only when cell damage is deemed irreversible (Hetz et al, 2011). Our data support a model where a complex signalling platform is assembled at the level of IRE1α to determine its activation status in terms of signalling intensity and kinetics of activation/inactivation. As a stress rheostat, the UPRosome would involve multiple BCL-2 family proteins that, beyond their apoptotic effects on mitochondrial integrity, act at the level of ER membranes to determine the amplitude and kinetics of the UPR and hence the cell’s ability to adapt to ER injuries.
Materials and methods
Reagents
Tm, Thap, brefeldin A, actinomycin and zVAD-fmk were purchased from Calbiochem EMB Bioscience Inc. ABT-737 was obtained from Selleck Chemicals. Phos-tag™ was purchased from Wako Pure Chemical Industries. Cell culture media, fetal calf serum and antibiotics were obtained from Life Technologies (Maryland, USA). All other reagents used here were from Sigma or the highest grade available.
Cell culture and DNA constructs
All BIM/PUMA MEFs used here were recently described (Kim et al, 2009), and were maintained in Dulbecco’s modified Eagles medium supplemented with 5% fetal bovine serum, non-essential amino acids and grown at 5% CO2. IRE1α-deficient cells were described before (Calfon et al, 2002). Retroviral bicistronic expression vectors for HA-tagged BH3-only proteins and GFP, or expression vectors for combinations between BIM with anti-apoptotic proteins (hBCL-2-IRES-hBIM-EL) were described before (Kim et al, 2006). The production of amphotropic retroviruses using the HEK293GPG packing cell line was performed as described (Kim et al, 2006). Retroviral plasmids were transfected using Efectene (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocols. Cell viability was monitored by propidium staining and fluorescence-activated cell sorting (FACS) analysis as described previously (Lisbona et al, 2009). BAX and BAK DKO cells reconstituted with cytochrome b5-BAK or ER-targeted BIM and PUMA retroviral vectors were previously described (Klee et al, 2009). IRE1α–HA expressing retroviruses were previously described in the pMSCV-Hygro vector (Hetz et al, 2006), where IRE1α contains two tandem HA sequences at the C-terminal domain and a precision enzyme site before the HA tag. A HIS tag was also included after the signal peptide at the N-terminal region.
Calcium signalling was measured as previously described using Fura-2 (Rojas-Rivera et al, 2012). Results are expressed as the ratio between 340 and 400 nm (R340/400) signals (Grynkiewicz et al, 1985).
RNA isolation, RT–PCR and real-time PCR
Real-time PCR primers were previously described (Iwakoshi et al, 2003; Lee et al, 2003a; Hollien et al, 2009). XBP-1s mRNA was monitored by semi-quantitative time PCR using the following primers: 5′-AAGAACACGCTTGGGAATGG-3′ and 5′-CTGCACCTGCTGCGGAC-3′. For PstI splicing based assay was performed using the following primers: 5′-GGATCTCTAAAACTAGAGGCTTGGTG-3′ and 5′-AAACAGAGTAGCAGCGCAGACTGC-3′. These primers amplified a 600-bp cDNA, product that was digested by PstI to reveal a restriction site that is lost after IRE1α mediate the splicing of mXBP-1 (Calfon et al, 2002). For conventional splicing, densitometric analysis was performing using Image J software where the each time point analysed considering the addition of the spliced and non-spliced XBP-1 form as 100% to then calculate the percentage of splicing.
IPs and in vitro splicing assay
HEK cells were co-transfected with different DNA constructs and after 48 h protein extracts were prepared in CHAPS buffer (1% CHAPS, 100 mM KCl, 50 mM Tris (pH 7.5), 50 mM NaF, 1 mM Na3VO4, 250 mM PMSF and protease inhibitors). IPs were performed as described (Lisbona et al, 2009).
LTQ analysis of HA–IRE1α IP
IRE1α-deficient MEF cells stably transduced with retroviral expression vectors for IRE1α–HA or empty vector were incubated for 6 h in the presence or absence of 100 ng/ml of Tm. Cell cultures were scaled-up and started with four 100 mm plates of semi-confluent cells. Then, cell lysates were prepared for IPs as indicated for experiments with HEK cells. As control, to eliminate non-specific background binding, the experiments were performed in parallel in IRE1α KO cells. Protein complexes were eluted from anti-HA antibody-agarose (Roche) by heating the samples at 95–100°C for 5 min in the presence of 0.1% Rapigest (Waters Corp.) and 100 mM ammonium bicarbonate. Eluted proteins were reduced, alkylated and digested overnight at 37°C with sequencing grade trypsin (1:20, w/w, trypsin/protein; Promega). Tryptic digests were dried, resuspended in 5% acetonitrile and 0.1% formic acid and injected into an analytical reverse-phase column (0.150 × 150 mm2, 5 μm beads; Magic C18AQ, Michrom Bioresources, Inc.) and separated at a flow rate of 1 ml/min over 60 min. Mass spectra were acquired in a linear ion trap mass spectrometer (LTQ, Thermo Electron Corp.) with data-dependent acquisition (Vaisar et al, 2007). Peptide and protein identification were searched using the database and parameters described in Becker et al (2010).
Western blot analysis and phostag gels
Western blot analysis was performed using standard conditions (Hetz et al, 2006). The following antibodies and dilutions were used: anti-XBP-1, 1:1000 (Iwakoshi et al, 2003), anti-SP-1 1:1000, anti-HSP90 1:5000, anti-ATF4 1:2000, anti-CHOP 1:2000 (Santa Cruz, CA), anti-HA 1:1000 (Roche, Basel, Switzerland), anti-BCL-2 1:2000, anti-BCL-XL 1:2000 (Transduction Laboratories, KY), anti-BIM 1:5000 (Calbiochem), anti-BNip3 1:2000 (Abcam), anti-AU1 (Covance), anti-PUMA 1:1000 (Sigma), anti-BiP, anti-ERp57, antiERp72, anti-calnexin, anti-calreticulin 1:3000 (Stressgene, USA), and anti-IRE1α 1:1000, anti-Phosphorylated and total eIF2α 1:1000 (Cell Signaling Technology). Phostag assay was performed using 50 μg of total protein was loaded in 6% SDS–PAGE minigels containing 25 μM of Phostag in the presence of 25 μM of MnCl2 (Kinoshita et al, 2006; Yang et al, 2010). IRE1α oligomers were analysed using non-denaturing gels as previously described (Liu et al, 2002; Hetz et al, 2006).
bim knockout mouse and Tm injection in vivo
bim+/− mice were generated by Dr Stanley Korsmeyer (Ranger et al, 2003). Animals were given a single 50 ng/g body weight intraperitoneal injection of a 0.05 mg/ml suspension of Tm in 150 mM dextrose as we previously described (Hetz et al, 2006; Lisbona et al, 2009). After different time points, mice were killed by CO2 narcosis. Livers were removed, and protein extracts were prepared for immunoblot or cDNA for RT–PCR analyses. All animal experiments were performed according to procedures approved by the Institutional Review Board’s Animal Care and Use Committee of the Faculty of Medicine of the University of Chile (approved protocol CBA # 0208 FMUCH). Analysis of immunoglobulin secretion of primary B-cell cultures was performed as described previously (Lisbona et al, 2009).
Yeast two hybrid
The interaction between the IRE1αΔN protein and BIM-EL or BIM-ELL150E was performed using the two-hybrid assay (James et al, 1996). Interaction of the proteins was performed with the MatchMaker two-hybrid systems according to the manufacturer’s protocols and the yeast protocol handbook (Clontech). The plasmid pGADT7 (bait) encoding the cytoplasmic domain of IRE1α (IRE1αΔN) was analysed for interaction with the plasmid pGBKT7 (prey) encoding either BIM-EL or BIM-ELL150E. AH109 yeast cells were co-transformed with both prey and bait plasmids. The positive interactions were assayed by growth on Leu/Trp/His-deficient media after 3–7 days at 30°C. Media lacking aminoacids were also supplemented with X-α-Galactoside (40 μg/ml). We used the interaction between pGBKT7-p53 and pGADT7-T as positive control (Clontech), and a pGADT7 empty vector co-transformed with pGBKT7-IRE1αΔN as negative control. Eight microliters of each suspension and three subsequent 10-fold serial dilutions were individually spotted onto or synthetic defined (SD) (−Leu/−Trp), SD (−Leu/−Trp/−His) and SD (−Leu/−Trp/−His/X-α-Gal) plates for selection. Cells were incubated at 30°C for 2 days.
Statistical analysis
Results were statistically compared using the Kruskal–Wallis ANOVA for unpaired groups followed by multiple comparison post-tests (Newman–Keuls multiple comparison test). Where pertinent, Student’s t-test was performed for unpaired or paired groups. A P-value of <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank David Ron for providing IRE1α−/− cells. We thank Dr Ann-Hwee Lee for providing and generating IRE1α KO-reconstituted cells. We also thank the Mass spectrometry resource (Department of Medicine, University of Washington) and the Mass spectrometry core of Diabetes and Endocrinology Research Center (University of Washington). We thank Dr Andres Stutzin (CEMC, University of Chile) for giving feedback and access to all equipment’s for calcium measurements. This work was supported by the FONDECYT no. 1100176, FONDAP grant no. 15010006, Millennium Institute no. P09-015-F, Muscular Dystrophy Association, Michael J Fox Foundation, Alzheimer’s Association, ALS Therapy Allianze and North American Spine Society (to CH); FONDECYT no. 3100033 (to DRG); Ligue nationale contre le cancer, and Agence Nationale pour la Recherche (to GK), National Institute of Health no. R01CA125562 and American Caner Society no. RSG-10-030-01-CCG (to EC); P&F award from Nutrition and Obesity Research Center (to TV); Grant SAF2008-00350 from Ministerio de Ciencia e Innovación, and Grant 200720I026 from CSIC (to FPM), Fondecyt 1095089 and ICM P05-001-F (to CG). Grant SAF2008-00350 from Ministerio de Ciencia e Innovacion (Spanish Government). ML is the holder of a JAE-Doc postdoctoral fellowship (Spain) co-funded by the European Social Fund. DRR, HU and FL are funded by a CONICYT PhD fellowship.
Author contributions: DAR and CH wrote the manuscript, conceived or designed the experiments, performed the experiments and analysed the data; SZ, FL, DRR, HU, JCR, RA, DRH, TI and ML conceived and designed the experiments, performed the experiments, and analysed the data; EC, TV, CGB, AL, FXP and GK conceived or designed the experiments and analysed the data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD (2007) XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27: 53–66 [DOI] [PubMed] [Google Scholar]
- Bailly-Maitre B, Belgardt BF, Jordan SD, Coornaert B, von Freyend MJ, Kleinridders A, Mauer J, Cuddy M, Kress CL, Willmes D, Essig M, Hampel B, Protzer U, Reed JC, Bruning JC (2010) Hepatic Bax inhibitor-1 inhibits IRE1alpha and protects from obesity-associated insulin resistance and glucose intolerance. J Biol Chem 285: 6198–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, Farmer D, Reed JC (2006) Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci USA 103: 2809–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Gharib SA, Irwin AD, Wijsman E, Vaisar T, Oram JF, Heinecke JW (2010) A macrophage sterol-responsive network linked to atherogenesis. Cell Metab 11: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, Matsuzawa S, Terskikh AV, Faustin B, Reed JC (2007) Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell 129: 45–56 [DOI] [PubMed] [Google Scholar]
- Brunelle JK, Letai A (2009) Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci 122: Pt 4437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ (2003) BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424: 952–956 [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116: 205–219 [DOI] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, Deeney JT, Robertson K, Morash J, Kulkarni A, Neschen S, Kim S, Greenberg ME, Corkey BE, Shirihai OS, Shulman GI et al. (2008) Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med 14: 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW (2008) The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol 45: 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260: 3440–3450 [PubMed] [Google Scholar]
- Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E (2004) Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J Biol Chem 279: 49689–49693 [DOI] [PubMed] [Google Scholar]
- Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A (2010) HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol 8: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR (2009) IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138: 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13: 89–102 [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312: 572–576 [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher L (2008) The daily job of night killers: alternative roles of the BCL-2 family in organelle physiology. Trends Cell Biol 18: 38–44 [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH (2009) Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell 35: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1{alpha}. Physiol Rev 91: 1219–1243 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee A-H, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH (2003) Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4: 321–329 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A (2005) Proapoptotic BID is an ATM effector in the DNA-damage response. Cell 122: 593–603 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ (2006) Role of Bax and Bak in mitochondrial morphogenesis. Nature 443: 658–662 [DOI] [PubMed] [Google Scholar]
- Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 8: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2009) Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell 36: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5: 749–757 [DOI] [PubMed] [Google Scholar]
- Klee M, Pallauf K, Alcala S, Fleischer A, Pimentel-Muinos FX (2009) Mitochondrial apoptosis induced by BH3-only molecules in the exclusive presence of endoplasmic reticular Bak. EMBO J 28: 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A-H, Iwakoshi NN, Anderson KC, Glimcher LH (2003a) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA 100: 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH (2003b) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16: 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee B, Lee AS (2006) Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem 281: 7260–7270 [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318: 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P (2009) Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One 4: e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, Reed JC, Glimcher LH, Hetz C (2009) BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell 33: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Wong HN, Schauerte JA, Kaufman RJ (2002) The protein kinase/endoribonuclease IRE1alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J Biol Chem 277: 18346–18356 [DOI] [PubMed] [Google Scholar]
- Lowman XH, McDonnell MA, Kosloske A, Odumade OA, Jenness C, Karim CB, Jemmerson R, Kelekar A (2010) The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell 40: 823–833 [DOI] [PubMed] [Google Scholar]
- Luo D, He Y, Zhang H, Yu L, Chen H, Xu Z, Tang S, Urano F, Min W (2008) AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. J Biol Chem 283: 11905–11912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G (2007) Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26: 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM et al. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A (2007) ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding Q, Chen Y, Wu D, Xie D, Lin X, Gao X, Kaufman RJ, Li W, Liu Y (2010) A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal 3: ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, Kutok JL, Le Beau MM, Greenberg ME, Korsmeyer SJ (2003) Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci USA 100: 9324–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimertz C, Kogel D, Rami A, Chittenden T, Prehn JH (2003) Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J Cell Biol 162: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307 [DOI] [PubMed] [Google Scholar]
- Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2010) BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 330: 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D, Rojas-Rivera D, Hetz C (2011) Integrating stress signals at the endoplasmic reticulum: the BCL-2 protein family rheostat. Biochim Biophys Acta 1813: 564–574 [DOI] [PubMed] [Google Scholar]
- Rojas-Rivera D, Armisén R, Colombo A, Martínez G, Eguiguren AL, Díaz A, Kiviluoto S, Rodríguez D, Patron M, Rizzuto R, Bultynck G, Concha ML, Sierralta J, Stutzin A, Hetz C (2012) TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death Differ doi: 10.1038/cdd.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300: 135–139 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee A-H, Qian S-B, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21: 81–93 [DOI] [PubMed] [Google Scholar]
- Tabas I, Ron D (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF et al. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest 117: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D (2010) Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell 38: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlbier U, Hetz C (2011) Modulating stress responses by the UPRosome: a matter of life and death. TIBS 38: 329–337 [DOI] [PubMed] [Google Scholar]
- Yang L, Xue Z, He Y, Sun S, Chen H, Qi L (2010) A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PLoS One 5: e11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ (2005) The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ (2005) A role for proapoptotic BID in the DNA-damage response. Cell 122: 579–591 [DOI] [PubMed] [Google Scholar]
- Zong WX, Li C, Hatzivassiliou G, Lindsten T, Yu QC, Yuan J, Thompson CB (2003) Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol 162: 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.