Abstract
The kinetochore is responsible for accurate chromosome segregation. However, the mechanism by which kinetochores assemble and are maintained remains unclear. Here we report that de novo CENP-A assembly and kinetochore formation on human centromeric alphoid DNA arrays is regulated by a histone H3K9 acetyl/methyl balance. Tethering of histone acetyltransferases (HATs) to alphoid DNA arrays breaks a cell type-specific barrier for de novo stable CENP-A assembly and induces assembly of other kinetochore proteins at the ectopic alphoid site. Similar results are obtained following tethering of CENP-A deposition factors hMis18α or HJURP. HAT tethering bypasses the need for hMis18α, but HJURP is still required for de novo kinetochore assembly. In contrast, H3K9 methylation following tethering of H3K9 tri-methylase (Suv39h1) to the array prevents de novo CENP-A assembly and kinetochore formation. CENP-A arrays assembled de novo by this mechanism can form human artificial chromosomes (HACs) that are propagated indefinitely in human cells.
Keywords: CENP-A, centromeres, chromosomes, epigenetic regulation, heterochromatin
Introduction
The kinetochore is responsible for accurate chromosome segregation. During mitosis, kinetochores assemble on specialized centromere chromatin (Cleveland et al, 2003; Allshire and Karpen, 2008) composed of nucleosomes containing the essential histone H3 variant CENP-A (Earnshaw and Rothfield, 1985). Recent studies have identified several factors, including the Mis18 complex and HJURP (Hayashi et al, 2004; Camahort et al, 2007; Fujita et al, 2007; Mizuguchi et al, 2007; Stoler et al, 2007; Pidoux et al, 2009; Williams et al, 2009), involved in the deposition of newly synthesized CENP-A at pre-existing CENP-A chromatin regions (Okada et al, 2006; Fujita et al, 2007; Jansen et al, 2007; Dunleavy et al, 2009; Foltz et al, 2009). However, the mechanism by which centromere chromatin assembles and is stabilized at specific genomic loci remains unclear.
Centromeric DNA sequences are competent to form de novo functional kinetochores in yeasts, mouse and some human cell lines (Clarke and Carbon, 1980; Hahnenberger et al, 1989; Harrington et al, 1997; Ikeno et al, 1998; Moralli et al, 2006; Okada et al, 2007). Human centromeric alpha-satellite (alphoid) DNAs can induce high efficiency de novo CENP-A and functional kinetochore assembly and subsequent human artificial chromosome (HAC) formation when introduced into HT1080 human fibrosarcoma cells. HAC kinetochore formation is highly dependent on regular arrays of alphoid DNA sequences with CENP-B binding capacity (Ohzeki et al, 2002; Okamoto et al, 2007), although de novo kinetochore assembly is not a simple DNA-protein reaction.
Chromatin modifications are thought to regulate functional kinetochore assembly and maintenance by an epigenetic mechanism. Recent studies of normal centromeres also suggest a possible involvement of canonical histone H3-containing nucleosomes in kinetochore function. In humans, CENP-A nucleosomes are localized to only a portion of the megabase-sized alphoid DNA arrays, where they are organized as multiple clusters interspersed with histone H3 nucleosomes (Blower et al, 2002; Sullivan and Karpen, 2004; Ribeiro et al, 2010). Canonical H3 nucleosomes co-purify with CENP-A in oligonucleosomes (Ando et al, 2002), and some classes of CENPs (e.g. CENP-T, -W) are suggested to bind only to H3 nucleosomes (Hori et al, 2008). Thus, epigenetic CENP-A-mediated kinetochore assembly could also be affected by the surrounding H3 chromatin state. Thus, functional kinetochore formation and maintenance may be influenced by additional factors that determine the modification status of centromeric chromatin.
The fundamental question addressed by this study is how different chromatin fates are generated on alphoid DNA in human cells and what kind of chromatin directs functional centromere/kinetochore assembly. We found that competency for stable CENP-A assembly and de novo kinetochore assembly are correlated with the acetylation status of H3K9 on alphoid DNA in several different cell types. We therefore decided to manipulate H3K9 modifications during de novo kinetochore assembly using a synthetic alphoid DNA array carrying multiple tet operator (tetO) sequences that allow the tethering of chromatin modifiers into the array as tet repressor (tetR) fusions (Nakano et al, 2008; Cardinale et al, 2009; Bergmann et al, 2011).
Tethering of tetR-EYFP-p300 or tetR-EYFP-PCAF, two histone acetyltransferase (HAT) domains that promote acetylation of H3K9, results in assembly of newly synthesized CENP-A on exogenous alphoid DNA arrays. Remarkably, HAT induction of de novo CENP-A chromatin assembly requires HJURP but bypasses the need for hMis18α, and spontaneously nucleates assembly of an outer kinetochore on the artificial DNA arrays. Indeed, in a technological breakthrough, these HAT-induced de novo CENP-A arrays can even lead to the formation of stable HACs that are maintained indefinitely in human cell lines that have previously proven refractory to HAC formation. Together, our data reveal that CENP-A assembly appears to be controlled by a histone H3K9ac/me3 balance that acts upstream of HJURP.
Results
Cell-type-dependent chromatin assembly on transfected human alphoid DNA
De novo kinetochore assembly is efficient in HT1080 cells. However, neither stable de novo kinetochore formation nor CENP-A assembly on exogenous alphoid DNA occurs in many other commonly used human cell lines, including HeLa (Figure 1A and Supplementary Figure S1).
Figure 1.
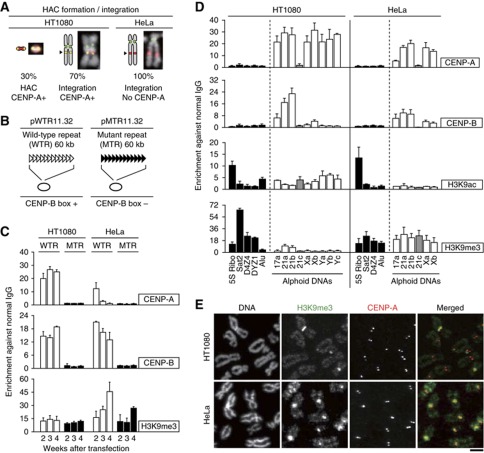
Cell type specific chromatin modifications on transfected and endogenous alphoid DNA. (A) Summary of the HAC formation assay. The pWTR11.32 plasmid, which contains 60 kb of α21-I 11mer repeat (shown in panel B), was transfected to HT1080 or HeLa cells. Single transformants were isolated and analyzed for chromosomal events by FISH and microscopy. Examples of HAC and integration are shown as merged images. Signals in pictures indicate DNA (gray), BAC plasmid DNA (red) and CENP-A (green). (B and C) Time-course ChIP analysis. The pWTR11.32 or pMTR11.32 plasmid (panel B) was transfected to HT1080 or HeLa cell. Transfectants were cultured under presence of selective drug (G418), and harvested at 2, 3 and 4 weeks after transfection. ChIP assay was carried out with normal IgG and indicated antibodies (panel C). Primer set for synthetic 11mer repeats was used for quantitative PCR. Error bars, s.d. (n=2). (D) Chromatin modifications on human repetitive DNAs. ChIP assay was carried out with normal IgG and indicated antibodies. Primer sets used for quantitative PCR are specific to 5S ribosomal DNA (5S Ribo), satellite 2 (Sat2), D4Z4 repetitive DNA (D4Z4), DYZ1 repetitive DNA (DYZ1), Alu elements (Alu), 17 alphoid (17a), 21-I alphoid (21a, 21b), 21-II alphoid (21c), X alphoid (Xa, Xb) and Y alphoid DNA (Ya, Yb, Yc) sequences. More information for these primers is shown in Supplementary Figure S3A and Supplementary Table S2. Columns indicate non-alphoid repetitive DNA controls (black), type I alphoid DNA (white) and type II (gray), respectively. Error bars, s.d. (n?3). (E) Examples of metaphase chromosome staining. Mitotic cell spreads were stained with DAPI (gray), anti-H3K9me3 (green) and anti-CENP-A antibody (red). Scale bar, 3 μm.
Surprisingly, HeLa cells, TIG7 human fetal primary, hTERT-BJ1 immortalized fibroblasts and U2OS osteosarcoma cells, all efficiently assemble CENP-A chromatin de novo, but CENP-A levels declined rapidly during subsequent cell culture (Figure 1B, C, Supplementary Figure S2 and S3C). The decrease in CENP-A levels on transfected alphoid DNA in HeLa cells was accompanied by a progressive increase in the heterochromatin-associated modification, H3K9me3 (Figure 1C).
Detailed ChIP analysis of the chromatin modification status at several endogenous centromeres revealed that alphoid DNA appears more euchromatic in HT1080 cells than in HeLa (Figure 1D). Using CENP-A and CENP-B as controls, H3K9ac, a euchromatic modification, was readily detected on HT1080 alphoid DNA, but was much lower at HeLa centromeres (Figure 1D). In addition, HT1080 cells had substantially lower levels of H3K9me3 on alphoid DNA than on other repetitive DNA sequences, including satellite 2, D4Z4 and DYZ1. In contrast H3K9me3 levels on alphoid DNA were significantly higher in HeLa, TIG7, hTERT-BJ1 and U2OS cells (Figure 1D and SupplementaryFigure S3). The ChIP data were confirmed by a stronger H3K9me3 staining intensity at mitotic centromeres in HeLa cells (Figure 1E).
Suv39h1 negatively regulates de novo CENP-A assembly on alphoid DNA at ectopic site
The histone methyltransferase Suv39h1 may be one critical factor responsible for this difference between HT1080 and HeLa alphoid DNA chromatin. HT1080 cells express only 50% of the relative level of Suv39h1 mRNA found in HeLa cells (Figure 2A). Suv39h1 over-expression increased both levels of the enzyme itself and H3K9me3 on centromeric alphoid DNAs in HT1080 cells (Figure 2B). These results fit with the observations that mouse cells doubly null for Suv39h1 and Suv39h2 (Suv39hdn) have low levels of centromeric H3K9me3 (Peters et al, 2001).
Figure 2.
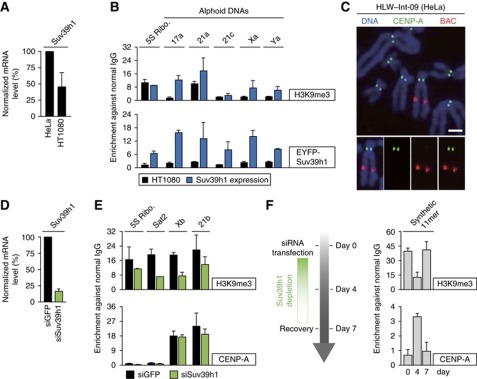
Suv39h1, histone H3K9 tri-methylase, negatively regulates ectopic CENP-A assembly. (A) Suv39h1 expression level. Total RNA was purified from each cell line, reversely transcribed and quantified by real-time PCR. Suv39h1 mRNA amounts were normalized by HPRT transcripts. Both Suv39h1 and HPRT genes are on X chromosome. (B) Exogenous Suv39h1 expression induced H3K9me3 modification on centromeric alphoid DNAs in HT1080 cells. EYFP-tagged Suv39h1 gene was transfected and cells were harvested more than four weeks after transfection. ChIP was carried out with normal IgG and a set of indicated antibodies. Primer sets shown at the top were used for quantitative PCR. Error bars, s.d. (n=2). (C) Examples of no ectopic CENP-A assembly in HeLa integration cell (HLW-Int-09). Mitotic cells were spread on cover glass, and stained with DAPI (blue), anti-CENP-A antibody (green), and BAC DNA probe (red). Scale bar, 5 μm.(D) Depletion of Suv39h1 with siRNA. siRNAs for the GFP gene (siGFP; control) or Suv39h1 (siSuv39h1) was transfected to HLW-Int-09 cell. Total RNA was purified and quantified by real-time PCR. Suv39h1 mRNA levels were normalized by HPRT RNA. Vertical axis indicates relative Suv39h1 mRNA level against a negative control (siGFP). Error bar, s.d. (n=3). (E, F) ChIP assay was carried out with HLW-Int-09 cells treated by siGFP or siSuv39h1. Normal IgG and a set of different antibodies were used for ChIP. Indicated primer sets were used for quantitative PCR (top). Error bars in panel E, s.d. (n=2). (F) HLW-Int-09 cells were harvested at three time points, 0, 4 and 7 days after siSuv39h1 transfection and used for ChIP analysis. Error bars, s.d. (n=3).
Suv39h1 depletion by RNAi revealed a remarkable inverse correlation between CENP-A and H3K9me3 levels on an alphoid DNA array integrated ectopically on a chromosomal arm in HeLa cells (HLW-Int-09; Figure 2C and F).
These results suggest that Suv39h1 suppresses ectopic CENP-A incorporation, presumably by maintaining H3K9me3 levels on alphoid DNA. However, Suv39h1 depletion alone and the accompanying transient increase in CENP-A were not sufficient for functional kinetochore formation on ectopic alphoid DNA arrays (Okada et al, 2007). Additional regulatory factors must be required for functional kinetochore formation de novo on alphoid DNA.
HAT recruitment breaks the barrier for de novo kinetochore assembly
Several observations suggest that histone acetyltransferases may be required for functional CENP-A assembly and subsequent kinetochore formation de novo (Nakano et al, 2003; Okamoto et al, 2007). Furthermore, the acetyltransferases p300 and PCAF [p300/CBP associated factors (Yang et al, 1996)] both localize at functional, but not at inactive, centromeres (Supplementary Figure S4) (Craig et al, 2003; Choi et al, 2009).
To test the hypothesis that histone acetylation might antagonize H3K9me3 and promote functional CENP-A assembly, we expressed tetR-EYFP fused to the histone acetyl-transferase (HAT) domains of p300 or PCAF in HeLa cells (Figure 3A). Into the tetR-EYFP expressing cells, we then introduced a 50 kb synthetic DNA array based on the α21-I alphoid dimer sequence with a tetO site where the CENP-B box would be on one monomer (pWTO2R; Figure 3A and Supplementary Figure S5) (Ebersole et al, 2005; Kim et al, 2009). In this system, tetR fusion proteins bound to tetO sites within the synthetic alphoid DNA arrays can directly modify the chromatin environment at a single centromere or locus in human cells.
Figure 3.
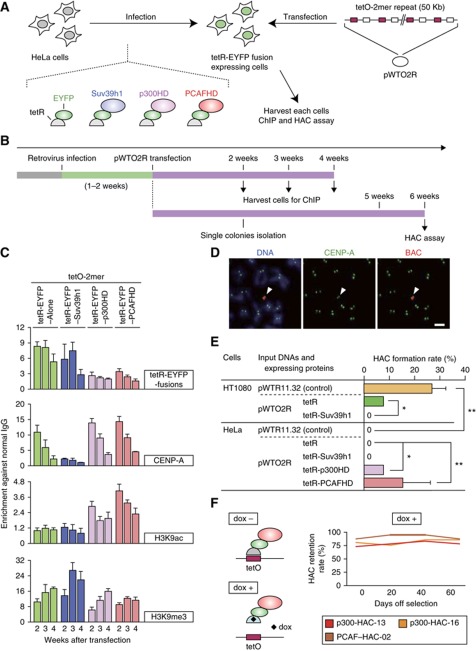
Recruiting of histone acetyl-transferases induced de novo kinetochore formation in HeLa cell. (A) The expression constructs and BAC plasmid used in this Figure. TetR-EYFP gene was fused with Suv39h1, p300 HAT domain (p300HD) or PCAF HAT domain (PCAFHD). HeLa cell lines expressing these tetR-EYFP fusions were generated by retrovirus infection, and these cells were transfected with α21-I alphoidtetO DNA containing plasmid (pWTO2R; see Supplementary Figure S5). (B) Schematic timetable for ChIP and HAC assay. (C) Time-course ChIP analysis. Cells transfected by plasmid pWTO2R were harvested at 2, 3 and 4 weeks after transfection. Normal IgG and a set of specific antibodies were used for ChIP. A set of primers for α21-I alphoidtetO 2mer (tetO-2mer) was used for quantitative PCR. Columns indicate the results obtained with cells expressing tetR-EYFP (green), tetR-EYFP-Suv39h1 (blue), tetR-EYFP-p300HD (pink) or tetR-EYPF-PCAFHD (red) fusions, respectively. Error bars, s.d. (n=3). (D) Examples of a HAC (p300-HAC-13) formed in HeLa cell. Metaphase cells were spread and stained with DAPI (blue), anti-CENP-A antibody (green) and BAC DNA probe (red). BAC DNA probe visualizes a vector region of the introduced pWTO2R construct. Scale bar, 3 μm. (E) Summary of HAC formation. Bars indicate a frequency of HAC formation in the cells expressing protein fusions. Error bars, s.d. (n?2). Chi-square test of the predominant pattern for HAC formation frequency indicated significant differences. Asterisks * or ** indicate P values, (P<0.05) or (P<0.005), respectively. (F) HAC stability without HAT tethering. HAC containing cells were cultured for 60 days under presence of doxycycline (no tetR binding condition; left panel) and absence of selective drug (permissive condition for HAC loss). The number of HAC retention rate in 30–50 spread metaphase cells was scored by FISH using input BAC DNA specific probes (right panel). HAC loss rate was calculated with HAC retention rates at day 0 (N0) or at day 60 (N60) using the following formula:N60=N0×(1–R)60 (Ikeno et al, 1998). All HAC cell lines showed high stability (HAC loss rate >0.001).
Tethering of either HAT domain fusion (tetR-EYFP-p300HD or tetR-EYFP-PCAFHD) to the synthetic alphoid DNA enhanced H3K9ac modification and CENP-A assembly, as demonstrated by time-course ChIP assays (Figure 3B, C and Supplementary Figure S6). In contrast, tethering of the tetR-EYFP-Suv39h1 fusion increased H3K9me3 levels and also decreased CENP-A assembly. This raised the question whether HAT domain recruitment could stimulate de novo kinetochore formation.
Remarkably, stable HACs bearing the synthetic α21-I alphoidtetO repeat were detected in HeLa cell lines expressing tetR-EYFP-p300HD or tetR-EYFP-PCAFHD (in 8 or 14% of cell lines, respectively; Figure 3B, D and and Supplementary Figure S7). Importantly, HAC formation was never detected when the synthetic α21-I alphoidtetO repeat was introduced into cells expressing tetR-EYFP or tetR-EYFP-Suv39h1 (Figure 3E). Similarly, alphoidtetO-based HAC formation was never observed in HT1080 cells expressing tetR-EYFP-Suv39h1 (Figure 3E and Supplementary Figure S8).
Although exogenous HAT activity is required for initial de novo kinetochore formation in HeLa cells, once established, the de novo kinetochores no longer require this exogenous activity to maintain their structure and function. We initially observed that HACs were stably maintained in cell clones that no longer expressed the tetR-EYFP-HAT fusion construct, presumably due to the silencing of retrovirus integration sites. We therefore, directly tested whether de novo kinetochores remained functional following forced dissociation of the HAT domain fusions by culturing cells for more than 60 days in the presence of doxycycline (Figure 3F). Microscopic and ChIP analyses showed that the HACs remained mitotically stable and capable of recruiting inner and outer kinetochore proteins CENP-A, -C, -T, hKNL1, Hec1, hDsn1 and hMis12 (Supplementary Figure S9A, B) in the absence of bound exogenous HAT fusion proteins (Figure 3F; Loss of tetR fusion binding to the HAC was confirmed by ChIP—Supplementary Figure S9C).
Thus, HAT domain recruitment to the synthetic α21-I alphoidtetO array renders HeLa cells competent for de novo kinetochore formation.
Centromere chromatin modifications regulate newly synthesized CENP-A assembly
Kinetochore maintenance requires the targeting of newly synthesized CENP-A to centromeres during mitotic exit/early G1 (Jansen et al, 2007). To test whether the same chromatin modifiers that potentiate de novo kinetochore assembly also affect newly synthesized CENP-A assembly at an established HAC kinetochore, we transiently co-transfected constructs expressing HA-tagged CENP-A (HA-CENP-A) plus various tetR-EYFP-fusion proteins into tetO-HAC containing HeLa cells (HeLa-HAC-R5; Figure 4A and B). We then asked if HA-CENP-A (a mark for newly assembled CENP-A) assembled on the HAC and endogenous centromeres at 24 h (i.e. one complete cell cycle in HeLa cells) after transfection.
Figure 4.
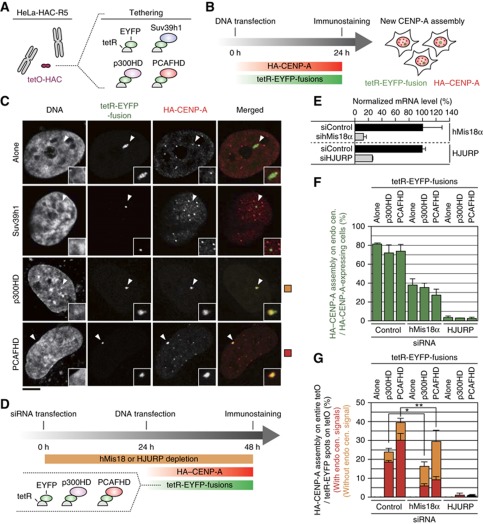
HAT tethering on tetO-HAC induced expansion of newly synthesized CENP-A assembly through HJURP. (A) A HAC cell line (HeLa-HAC-R5) was transfected with a set of tetR-EYFP-fusion expressing vectors. (B) Timetable for the experiment. HA-tagged CENP-A expression vector (pCDNA5-HA-CENP-A) was co-transfected with tetR-EYFP-fusion expressing vector. (C) Representative images of newly synthesized CENP-A assembly. Cells were stained with DAPI, anti-GFP (recognize EYFP; green) and anti-HA (red). Arrowheads indicate tetO-HAC position. Scale bar, 5 μm. (D) Schematic timetable for gene depletion and new CENP-A assembly assay. HeLa-HAC-R5 cells were firstly transfected with siRNA. After 24 h incubation, HA-CENP-A and a set of tetR-EYFP-fusion expression vectors were co-transfected. Cells were stained with DAPI, anti-GFP and anti-HA. (E) hMis18α or HJURP depletion using siRNA. siRNAs for hMis18α (sihMis18α) and for HJURP (siHJURP) as well as for a negative control (siControl: siNegative, ambion) were used for transfection. Total RNA was purified two days after transfection and quantified by real-time PCR. hMis18α or HJURP mRNA levels were normalized by HPRT transcripts. Horizontal axis indicates relative hMis18α or HJURP mRNA level against a negative control (siControl). Error bar, s.d. (n=3). (F) HA-CENP-A assembly frequency on endogenous centromere was counted in each sample (n?100). Error bar, s.d. (n=3). (G) A frequency of expanded HA-CENP-A assembly induced by HAT tethering (example is shown in panel C bottom) was counted in each sample (n?100). Error bar, s.d. (n=3). Column colors indicate subpopulations of cells, which had CENP-A assembly at endogenous centromere (red) and had no assembly (orange). *P-values of t-test are 0.001 (red column) and 0.017 (orange column). **P-values of t-test are 0.006 (red column) and 0.033 (orange column).
Tethering of tetR-EYFP alone did not affect the assembly of newly synthesized HA-CENP-A onto either the HAC or endogenous centromeres (Figure 4C and Supplementary Figure S10A). In contrast, tethered tetR-EYFP-Suv39h1 specifically reduced HA-CENP-A assembly on the HAC centromere (Figure 4C and Supplementary Figure S10B). This was coupled with destabilization of the HAC, detected as lagging chromosomes and micronuclei (Supplementary Figure S10C-G).
Unexpectedly, tethering of p300HD or PCAFHD induced HA-CENP-A hyper-assembly not only at the HAC centromere, but covering the entire alphoidtetO signal on the HAC in a significant proportion of cells (34 and 40% in Figure 4C and Supplementary Figure S10B).
We next tested whether the known canonical CENP-A deposition factors hMis18α and HJURP are involved in this HAT-induced CENP-A assembly. We first depleted hMis18α or HJURP by siRNA knockdown and then tethered tetR-fused HAT proteins to the synthetic alphoid array (Figure 4D, E). hMis18α depletion reduced HA-CENP-A assembly at both endogenous centromeres and the HAC centromere (Figure 4F). However, stable HA-CENP-A assembly continued on alphoidtetO DNA with tethered HAT fusions following hMis18α depletion, which blocks CENP-A assembly on endogenous centromeres (Figure 4G, orange bars). The cell population, which had newly assembled HA-CENP-A on alphoidtetO DNA but no HA-CENP-A signals on endogenous centromeres, was relatively increased after hMis18α depletion (Figure 4G, orange bars. P<0.05). These results indicate that tethering of HAT fusions can partially rescue HA-CENP-A assembly in the absence of hMis18α.
Importantly, HJURP depletion dramatically reduced HA-CENP-A assembly both on endogenous host centromeres and on the HAC. Furthermore, neither was rescued by tethering of HAT-fusion proteins to the HAC alphoidtetO array (Figure 4F, G). Thus, HJURP is required for HAT-mediated CENP-A assembly.
Given that HAT tethering can potentiate de novo kinetochore formation on a HAC and induce HA-CENP-A hyper-assembly covering non-centromeric regions of the HAC (Figures 3 and 4), we next tested whether HAT-tethering can induce de novo CENP-A assembly on a chromosomal arm. We did this using a stable cell line (HeLa-Int-03), which carries an ectopic alphoidtetO chromosomal integration on which we have failed to detect any essential kinetochore-specific proteins other than CENP-B (which binds to the CENP-B box) (Supplementary Figure S11).
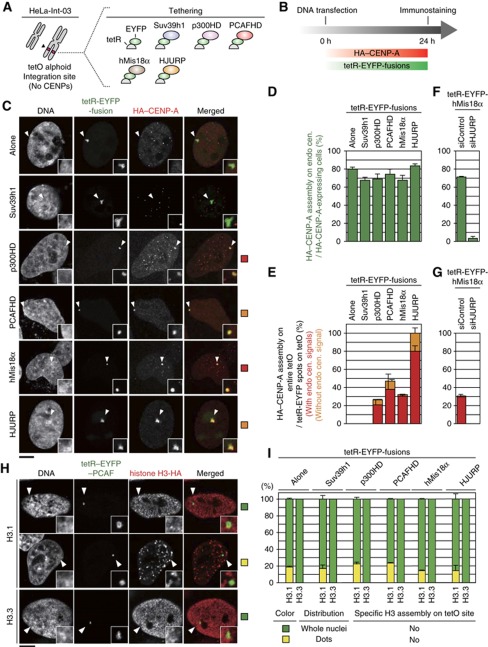
Tethering of tetR-EYFP-p300HD or tetR-EYFP-PCAFHD induced HA-CENP-A hyper-assembly on the ectopic array in 27 and 47% of cells, respectively (Figure 5A and E). A similar effect was observed after tethering the CENP-A assembly factors, tetR-EYFP-hMis18α or tetR-EYFP-HJURP (HA-CENP-A hyper-assembly in 32 and 100% of cells, respectively—Figure 5C and E). CENP-A assembly at the ectopic site induced by tetR-EYFP-hMis18α tethering was diminished by HJURP depletion (Figure 5F), consistent with Barnhart et al (2011). In controls, tethering of tetR-EYFP alone or tetR-EYFP-Suv39h1 did not induce HA-CENP-A assembly at the ectopic site (Figure 5C–E). Moreover, no specific enhancement of the assembly of newly expressed HA-tagged histone H3.1 nor H3.3 was observed on the ectopic alphoidtetO array by the HAT tetherings in addition to the usual assembly patterns of those histone H3 (Figure 5H and Supplementary Figure S12).
Figure 5.
HAT and CENP-A deposition related factor could induce de novo ectopic CENP-A assembly. (A) Schematic diagram. HeLa-Int-03 cell line had ectopic integration site of alphoidtetO DNA on host chromosome (left). This ectopic site had no CENP-A assembly. In addition to the previous four constructs, two new tetR-EYFP-fusions were used for the experiment (right). (B) Schematic timetable for new CENP-A assembly assay. HeLa-Int-03 cells were co-transfected with HA-CENP-A and a set of tetR-EYFP-fusion expression vectors. (C) Representative images of newly synthesized CENP-A assembly on ectopically integrated alphoidtetO DNA. Cells were stained with DAPI, anti-GFP (green) and anti-HA (red). Arrowheads indicate alphoidtetO DNA integration sites. Scale bar, 5 μm. (D) A frequency of HA-CENP-A assembly on endogenous centromere per total HA-CENP-A expressing cells was counted in each sample (n?100). Error bar, s.d. (n=3). (E) Frequency of de novo HA-CENP-A assembly on ectopic alphoidtetO DNA integration site. HA-CENP-A signals on tetR-EYFP spot per total tetR-EYFP spots were counted in each sample (n?100). Error bar, s.d. (n=3). (F and G) hMis18α tethering assay under HJURP depletion. The frequencies shown in panel D and E were counted (n?100). Error bar, s.d. (n=3). (H) Representative images of newly synthesized histone H3.1 and H3.3 localization. HA-tagged histone H3 was expressed with the same procedure to panel B. Cells were stained with DAPI, anti-GFP (green) and anti-HA (red). Scale bar, 5 μm. (I) Distribution of histone H3s. Histone H3.1 showed two localization pattern, whole nuclei (green column) and dots (yellow column). No specific enrichment was observed at tetO alphoid DNA other than the usual assembly pattern of these histone H3. Cells containing tetR-EYFP spots were counted in each sample (n?100). Error bar, s.d. (n=3).
Next, we determined when during the cell cycle HAT tethering induces CENP-A assembly. We tethered tetR-EYFP fusion proteins for 2 h by controlling the presence and absence of doxycyline, and detected new CENP-A assembly using a SNAP-CENP-A pulse labeling technique (Supplementary Figure S13A, B). Within 2 h of tethering the appropriate tetR-EYFP fusions, labeled SNAP-CENP-A assemblies were detected not only on endogenous centromeres but also on an ectopic alphoidtetO integration site (Supplementary Figure S13C–E). We determined the cell cycle phase of cells that had assembled SNAP-CENP-A at the ectopic site by using Cyclin B staining (Fujita et al, 2007; Silva et al, 2012). SNAP-CENP-A assemblies induced on ectopic sites by tethering p300HD, PCAFHD and hMis18α were observed only in Cyclin B negative (G1) cells (Supplementary Figure S13C–E). This is the cell cycle phase that is normally permissive for CENP-A assembly at endogenous centromeres (Silva et al, 2012). HJURP tethering induced ectopic CENP-A assembly both in the Cyclin B positive and negative cells (Supplementary Figure S13E). Taken together, HAT activity is sufficient to trigger the specific assembly of newly synthesized CENP-A on alphoid DNA in G1 phase.
HAT tethering induces de novo functional kinetochore assembly at the ectopic site
We next investigated whether ectopic CENP-A assembly driven by chromatin acetylation or tethered hMis18α or HJURP can induce assembly of the outer kinetochore in HeLa cells (Figure 6A).
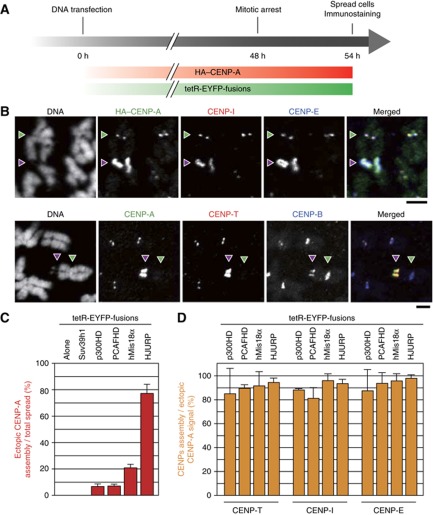
Figure 6.
Ectopic kinetochore proteins assembly induced by CENP-A recruiting factors. (A) Schematic diagram of metaphase cell preparation. HeLa-Int-03 was co-transfected with HA-CENP-A and a set of tetR-EYFP fusion protein expressing vectors. Six tetR-EYFP-fusions are shown in Figure 5A. After 48 h incubation, cells were arrested in metaphase and spread on cover glass for immuno-staining. (B) Examples of high order centromere proteins assembly at ectopic alphoidtetO DNA integration site. Spread mitotically arrested cells were stained with DAPI, anti-HA (green), anti-CENP-I (red) and anti-CENP-E (blue) (top). Another staining was also carried out with DAPI, anti-CENP-A (green), anti-CENP-T (red) and anti-CENP-B (blue) (bottom). Arrowheads indicate endogenous centromere (green) and ectopic alphoidtetO DNA integration site (red). The indicated images were obtained with tetR-EYFP-HJURP tethering. The results obtained with other fusions are shown in Supplementary Figure S14. Scale bars, 3 μm. (C) A frequency of ectopic HA-CENP-A assembly per total spread cells was counted (n?100). Error bar, s.d. (n=3). (D) A frequency of ectopic kinetochore proteins (CENP-T, CENP-I and CENP-E) assembly per total ectopic CENP-A signal positive cells (n=8∼100). Error bar, s.d. (n=3).
CENP-A assembled on ectopic alphoidtetO arrays was maintained in metaphase cells, where the ectopic HA-CENP-A was always detected as an extended region weakly stained with DAPI (Figure 6B, C). HA-CENP-A-coated arrays were observed in 21 or 77% of metaphase cells expressing tetR-EYFP-hMis18α or tetR-EYFP-HJURP, respectively (compared to 32 and 100% in interphase cells). HAT-induced CENP-A assembly was apparently less stable on mitotic chromosomes (7% of mitotic cells, compared with 27∼47% of interphase cells—Figures 5E and C).
Remarkably, the essential inner or outer kinetochore markers CENP-T, -I and -E (Hori et al, 2008; Santaguida and Musacchio, 2009) assembled on the ectopic array following CENP-A assembly (red arrowheads, Figure 6B, D and Supplementary Figure S14). These proteins accumulated at greater levels than at the centromeres of host chromosomes (Figure 6B, green arrowheads). Such an induced hyper-assembly of kinetochore proteins at the ectopic sites appears to result in bundling of an excess amount of microtubules and results in aberrant spindle formation. As a result these cells appear to be arrested in mitosis (Supplementary Figure S15C). In contrast, kinetochore assembly was not observed on CENP-A assembled nonspecifically on whole chromosomal arm regions (Supplementary Figure S16), consistent with Van Hooser et al (2001) and Gascoigne et al (2011).
A HAC assay with α21-I alphoidtetO BAC accompanied by transient transfection of these tetR-EYFP fusion-expressing plasmids confirmed that induced kinetochores like in those shown in Figure 6 can acquire stable maintenance and full function as stable artificial chromosomes (Supplementary Figure S17A). In HeLa cells, HAC formation was efficiently supported by co-transfection with tetR-EYFP-PCAFHD, tetR-EYFP-hMis18α and tetR-EYFP-HJURP but not with tetR-EYFP alone (Supplementary Figure S17C). Furthermore, tetR-EYFP-PCAFHD also supported HAC formation in U2OS cells where again no HAC formation was seen with tetR-EYFP alone (Supplementary Figure S3, S17B and C). Interestingly, HAC formation was not dramatically increased in HT1080 cells if the α21-I alphoidtetO BAC was introduced together with a construct expressing tetR-EYFP-PCAFHD.
These results demonstrate that tethering of HAT activity or Mis18α can induce HJURP-dependent de novo CENP-A chromatin assembly and subsequent assembly of a functional kinetochore on the alphoid DNA array.
Centromere H3K9 acetylation normally occurs in a short time window following metaphase
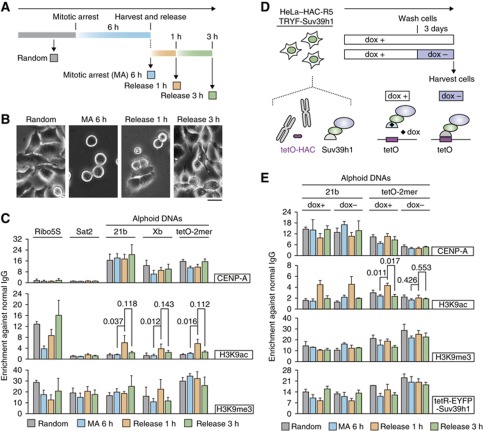
Although forced HAT tethering can induce CENP-A and kinetochore protein assembly on alphoidtetO DNA, the level of H3K9 acetylation on endogenous alphoid DNA is normally very low—almost undetectable in unsynchronized HeLa cells (Figure 1D). This raises the question of whether CENP-A assembly induced by acetylation of H3K9 is biologically relevant. If centromere acetylation does normally occur, it may be during only a brief cell cycle window—possibly coinciding with the localization of hMis18α and HJURP to centromeres. HJURP centromere localization is maximal at two hours after release from a metaphase arrest, and rapidly decreases thereafter (Dunleavy et al, 2009).
Indeed, ChIP analysis revealed that H3K9 acetylation levels increased temporarily on endogenous and HAC centromere alphoid DNAs at one hour after release from a metaphase arrest, but fell again by three hours after the release (Figure 7A–C). The temporary increase in H3K9ac can be blocked by tethering tetR-EYFP-Suv39h1 and CENP-A levels also fell following tethering of tetR-EYFP-Suv39h1 (Figure 7E and Supplementary Figure S18; compare doxycycline+/doxycycline-).
Figure 7.
Centromere acetylation occurs within a short time window following metaphase. (A) Schematic diagram for cell sample preparation. Cells were arrested in metaphase for six hours, and then harvested and released to G1 phase. Mitotically arresting (pale blue), one hour post release (orange), three hours post release (green) and random culture cells (gray) were harvested. (B) Examples of phase contrast microscope images for cells at each time points. Scale bar, 20 μm. (C) Centromere acetylating activity in HeLa cell. ChIP assay was carried out with normal IgG and a set of specific antibodies using samples indicated in panel A. A set of primers was used for quantitative PCR (top). Error bars, s.d. (n=3). P-values obtained with t-test are indicated. (D) Schematic diagrams for Suv39h1 tethering. A HeLa-HAC-05 cell line expressing tetR-EYFP-Suv39h1 was established in the presence of doxycycline (dox). Three days before sample preparation, tetR-EYFP-Suv39h1 tethering to HAC was induced by dox washout. Then a set of four cell samples shown in panel A was harvested and used for ChIP. (E) Suv39h1 tethering represses the increase of centromeric H3K9ac level. HeLa-HAC-R5 cells expressing Suv39h1 were cultured with presence or absence of doxycycline for three days, and then harvested similar to that shown in panel A. ChIP assay was carried out with a set of specific antibodies. A set of primer was used for quantitative PCR (top). Error bars, s.d. (n=3). P-values obtained with t-test are indicated.
These results confirm the presence of intrinsic acetylation activity on centromere chromatin, and show that this activity is apparently restricted to a short time window from anaphase through early G1. Taken together these results indicate that CENP-A assembly is regulated by the H3K9 acetyl/methyl balance. If H3K9 is acetylated, the chromatin can bind chaperones such as HJURP and assemble CENP-A chromatin. If it is trimethylated, CENP-A assembly is inhibited.
Discussion
Centromeric chromatin acetylation induces de novo heritable kinetochore assembly
Tethering of histone acetyltransferases (HATs) induces de novo assembly of CENP-A and functional kinetochore on ectopic alphoidtetO DNA, and can culminate in de novo formation of stable human artificial chromosomes (HACs). HAT-induced de novo CENP-A assembly appears to mimic the natural process. It requires the activity of specific CENP-A deposition factor HJURP. The HAT normally responsible for de novo CENP-A assembly and its key substrates in addition to H3K9 remain to be identified(Supplementary Figure S19). Nonetheless, this observation that tethered HAT activity in canonical H3 chromatin can induce de novo CENP-A and outer kinetochore assembly by adjusting the modification status of H3K9 represents a major step towards understanding the epigenetic regulation of kinetochore assembly.
Recent exciting studies demonstrated that tethering of CENP-C and CENP-T (Gascoigne et al, 2011) or HJURP (Barnhart et al, 2011) to an ectopic LacO array induced the assembly of a functional outer kinetochore. However, whether those kinetochore-like structures were stably inherited was not tested. Here, we show that kinetochores formed de novo by targeted induction of CENP-A assembly direct accurate segregation of the resulting HACs for many generations without any requirement for continued tethering of the exogenous HAT. Thus, our data suggest that proper assembly of CENP-A chromatin is sufficient for long-term epigenetic maintenance of centromere activity.
H3K9 ac/me3 are positive and negative regulators of CENP-A assembly, respectively
The notion that CENP-A assembly may normally be linked to chromatin acetylation (Nakano et al, 2003; Fujita et al, 2007; Okamoto et al, 2007) is strongly supported by our detection of a pulse of histone H3 acetylated on lys 9 (H3K9ac) during a brief window following release from a mitotic arrest. This timing corresponds remarkably well with the observed localization of hMis18α and HJURP at kinetochores (Fujita et al, 2007; Foltz et al, 2009; Dunleavy et al, 2009) and is the cell cycle window in which CENP-A assembly normally occurs (Jansen et al, 2007; Silva et al, 2012).
Although Suv39h1 over-expression increased levels of H3K9me3 on centromeric alphoid DNA, the functions of endogenous centromeres on host chromosomes were not impaired. However, tethering of Suv39h1 to the alphoidtetO kinetochore blocked the pulse of centromeric H3K9 acetylation normally seen during mitotic exit and interfered with the assembly of newly synthesized CENP-A on the established HAC centromere. Thus, although kinetochores do contain limited H3K9me3-containing chromatin regions (Ribeiro et al, 2010), the CENP-A chromatin core in the active kinetochore must be protected from Suv39h1-induced H3K9 tri-methylation during mitotic exit.
Regulation of the balance between H3K9ac (promoting CENP-A assembly) and H3K9me3 (inhibiting it) may be critical not only for de novo kinetochore assembly in our artificial system, but also for genome stability. The extremely large secondary kinetochores formed by induction of CENP-A assembly at ectopic sites apparently caused abnormal bundling of spindle microtubules and resulted in a mitotic arrest. This suggests that kinetochore geometry must be regulated appropriately on endogenous alphoid DNA—possibly to avoid formation of merotelic attachments. Adjusting the balance between H3K9 acetylation and methylation might provide a modulation mechanism to minimize inappropriate CENP-A assembly and the formation of ectopic centromeres on native chromosomes.
The role of centromeric heterochromatin may vary in different organisms. In fission yeast, heterochromatin is important not only for sister chromatid cohesion, but also for de novo CENP-A assembly (Hayashi et al, 2004; Grewal and Jia, 2007; Ishii et al, 2008; Kagansky et al, 2009). Understanding this contrast between fission yeast and human CENP-A assembly clearly requires additional study.
Breaking the HAC barrier
Since the first HAC formation assay, it has been unclear why de novo kinetochore formation could occur in HT1080 cells but not in other popular cell lines, such as HeLa. Indeed, in some quarters, this was taken to suggest that HAC formation in HT1080 might in some way be an aberrant process. Here, we suggest a very simple H3K9 acetyl/methyl balance model to explain this host cell specificity for HAC formation. Assembly of a core of CENP-A sufficient to establish an epigenetically stable active centromere appears to require H3K9ac, and if the balance is tipped in favor of H3K9me3, then the CENP-A that assembles initially is gradually lost and stable kinetochores do not form.
We propose that tethering of HAT activity to the input alphoid DNA array breaks the kinetic barrier provided by the very brief window of acetylation that occurs during mitotic exit. This appears to allow sufficient time for CENP-A to assemble into ‘core’ regions of sufficient size to be stably maintained (Alonso et al, 2007; Okamoto et al, 2007). Thus the synthetic tetO-alphoid/tetR-fusion tethering system now allows us to induce de novo kinetochore assembly on both newly introduced synthetic alphoid DNA arrays as well as pre-existing arrays integrated into chromosome arms. This ability to induce the formation of stable minichromosomes in HeLa, U2OS and other popular cell lines offers a powerful approach to analyzing epigenetic centromere/kinetochore formation and maintenance.
Materials and methods
Cell culture and transfection
HT1080 (tetraploid) and HeLa cells were grown in Glutamax I (Invitrogen) supplemented with 10% FBS at 37 °C in 5% CO2 atmosphere. For transfections, Lipofectamin 2000 (Invitrogen), Lipofectamine (Invitrogen) or FuGENE HD (Roche) was used for siRNA, BAC plasmid DNAs (pWTR11.32, pMTR11.32, pW/M11.64 and pWTO2R) or usual plasmid vectors, respectively. Retrovirus infection method used for tetR-fusions expression was previously described (Nakano et al, 2008). For depletion experiments, siRNAs for Suv39h1 were obtained from Dharmacon as a pool (D-009604-01, D-009604-02, D-009604-04 and D-009604-06), and siRNA sequence for hMis18α or HJURP depletion was referred Fujita et al (2007) or Dunleavy et al (2009), respectively. siRNA sequence for p300 or PCAF was CAGAGCAGUCCUGGAUUAGTT and GGUGGUAUCUGUUUCCGUATT, respectively. Control siRNAs were obtained from Dharmacon (siGFP) and Ambion (siNegative). Cell lines used in this study are shown in Supplementary Table S4.
ChIP
Cells were trypsinized and harvested in a centrifuge tube. Cells were washed with PBS and then fixed with 0.61% (for tetR-EYFP or EFYP fusions) or 0.3% (for histones) formaldehyde at 25 °C for 10 min. ChIP procedure was previously described (Ohzeki et al, 2002). Antibodies used for ChIP are shown in Supplementary Table S1. Immuno-precipitated DNAs were de-fixed at 65 °C for more than 4 h and purified by phenol/chloroform extraction following proteinase K treatment. Purified DNA was quantified by the competitive PCR (Supplementary Figure S2) or real-time PCR (BIORAD). For real-time PCR detection, SYBR Green I containing reagent was used (BIORAD). PCR primer sequences used for ChIP assay are shown in Supplementary Table S2.
Preparation of mitotic cells and chromosome spreads
For mitotic arrest, cells were treated with 350 nM of the highly reversible microtubule destabilizing drug, TN-16 (WAKO; Kitagawa et al, 1995; Perpelescu et al, 2009), for 2 to 6 h in the growth medium. Mitotic cells were harvested by pipetting, washed with PBS, incubated in a hypotonic buffer (20 mM Tris pH7.4, 1 mM EGTA and 40 mM KCl) and then spread on cover glass (Matsunami) by Cytospin3 (Shandon). Following immuno-staining and/or FISH were carried out according to the previously described method (Ikeno et al, 1998; Ohzeki et al, 2002). For ChIP with mitotic or post mitotic cells (Figure 7), mitotically arrested cells were harvested with the method described above, firstly. Then a portion of the mitotic cells was fixed for ChIP, and the remaining were washed with PBS two times and plated in petri dish. After 1 h incubation, unattached mitotic cells were washed out with PBS by pipetting. Attached cells were harvested for ChIP analysis at each time point. ChIP procedure is described in above section.
Supplementary Material
Acknowledgments
We would like to thank N Nozaki, K Yoda, M Yanagida, I Cheeseman and T Yen for antibodies, T Jenuwein for giving us MEFs derived from wild type or from Suv39h1Suv39h2 double null (Suv39h dn) embryos, T Ebersole for fruitful discussions and K Sumi for technical assistance. This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA, and the Kazusa DNA Research Institute Foundation. WCE. is a Principal Research Fellow of the Wellcome Trust [grant number 073915], and the Wellcome Trust Centre for Cell Biology is supported by grant number 092076.
Author contributions: JO and HM designed methods and experiments. JO carried out HAC formation and ChIP experiments. JO and JHB carried out cytological experiments. JO, NK, VN and MN constructed plasmids and alphoid 2mer repeat. HK provided modified histone antibodies. JO, NK, VL, WCE and HM worked on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, Yoda K, Radlwimmer B, Ladurner AG, Warburton PE (2007) Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol 8: R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002) CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22: 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL (2007) Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC (2009) Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell 20: 4194–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Choe H, Min J, Choi JY, Kim J, Lee H (2009) BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J 28: 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH (2003) Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet 12: 3109–3121 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Ebersole T, Okamoto Y, Noskov VN, Kouprina N, Kim JH, Leem SH, Barrett JC, Masumoto H, Larionov V (2005) Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res 33: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K (2008) Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Hahnenberger KM, Baum MP, Polizzi CM, Carbon J, Clarke L (1989) Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA 86: 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16: 431–439 [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC (2009) Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Ebersole T, Kouprina N, Noskov VN, Ohzeki J, Masumoto H, Mravinac B, Sullivan BA, Pavlicek A, Dovat S, Pack SD, Kwon YW, Flanagan PT, Loukinov D, Lobanenkov V, Larionov V (2009) Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Res 19: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Masumoto H, Ikeda M, Okazaki T (1995) Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol Cell Biol 15: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C (2007) Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Moralli D, Simpson KM, Wade-Martins R, Monaco ZL (2006) A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep 7: 911–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H (2008) Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14: 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Okamoto Y, Ohzeki J, Masumoto H (2003) Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci 116: 4021–4034 [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, Desai A, Fukagawa T (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300 [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H (2007) A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J 26: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K (2009) Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol 185: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC (2009) Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC (2010) A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA 107: 10484–10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28: 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63 [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE (2007) Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA 104: 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 1 14: 3529–3542 [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani YA (1996) p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382: 319–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.