Abstract
The proper maintenance of telomeres is essential for genome stability. Mammalian telomere maintenance is governed by a number of telomere binding proteins, including the newly identified CTC1–STN1–TEN1 (CST) complex. However, the in vivo functions of mammalian CST remain unclear. To address this question, we conditionally deleted CTC1 from mice. We report here that CTC1 null mice experience rapid onset of global cellular proliferative defects and die prematurely from complete bone marrow failure due to the activation of an ATR-dependent G2/M checkpoint. Acute deletion of CTC1 does not result in telomere deprotection, suggesting that mammalian CST is not involved in capping telomeres. Rather, CTC1 facilitates telomere replication by promoting efficient restart of stalled replication forks. CTC1 deletion results in increased loss of leading C-strand telomeres, catastrophic telomere loss and accumulation of excessive ss telomere DNA. Our data demonstrate an essential role for CTC1 in promoting efficient replication and length maintenance of telomeres.
Keywords: DNA damage, DNA replication, mouse model, stem cell, telomere
Introduction
Mammalian telomeres consist of TTAGGG repeats, ending in single-stranded (ss) G-rich overhangs that play important roles in the protection and replication of chromosome ends. Telomeres are protected by the telomere binding proteins TRF2–RAP1 and TPP1–POT1. Telomere length independent removal of these proteins results in the activation of a p53-dependent DNA damage response (DDR) at telomeres, engaging either non-homologous end joining (NHEJ) or homologous recombination (HR) repair pathways, respectively (Wu et al, 2006; Denchi and de Lange, 2007; Guo et al, 2007; Rai et al, 2010). Chromosomal rearrangements as a result of incorrectly repaired DNA breaks could lead to increased cancer formation (McKinnon and Caldecott, 2007).
Semi-conservative DNA replication is used to replicate most telomere DNA, while telomerase is required to elongate the lagging G-strand. In budding yeast, the Cdc13, Stn1 and Ten1 (CST) protein complex plays important roles in telomere length regulation (Nugent et al, 1996; Grandin et al, 1997, 2001; Puglisi et al, 2008). CST binds to the ss G-overhang, modulates telomerase activity and interacts with DNA polymerase α (Polα) to couple leading- and lagging-strand DNA synthesis (Qi and Zakian, 2000; Chandra et al, 2001; Bianchi et al, 2004; Grossi et al, 2004). The recent identification that the STN1 and TEN1 interacting protein CTC1 (conserved telomere maintenance component 1) binds ss DNA and stimulates Polα-primase activity suggests that mammalian CST also functions in mediating lagging-strand telomere replication (Goulian and Heard, 1990; Casteel et al, 2009; Miyake et al, 2009; Surovtseva et al, 2009; Dai et al, 2010). However, how mammalian CST promotes telomere replication is not known.
In addition to its putative function in telomere replication, mammalian CST is also implicated in telomere end protection. shRNA-mediated deletion of STN1 and CTC1 results in telomere deprotection and telomere erosion (Miyake et al, 2009; Surovtseva et al, 2009). These results, together with recent observations that STN1 associates with TPP1 (Wan et al, 2009), suggest that mammalian CST and shelterin might have overlapping functions in telomere end protection.
In this study, we report that formation of the CST complex is abrogated in the absence of CTC1. CTC1 null mice are viable but die prematurely from complete bone marrow (BM) failure due to the activation of a G2/M checkpoint. While end-to-end chromosome fusions are prominent in CTC1 null splenocytes and late-passage CTC1−/− mouse embryo fibroblasts (MEFs), they are absent during early passages. These results indicate that unlike deletion of shelterin components, deletion of CTC1 does not result in acute telomere deprotection. Rather, the DDR and chromosome fusions observed in the absence of CTC1 are a consequence of rapid, catastrophic telomere shortening, coupled with the accumulation of ss G-overhangs. We demonstrate that CTC1 functions to promote efficient replication of telomeric DNA. Our results therefore provide mechanistic insights into the functions of CTC1 in telomere replication and telomere length maintenance.
Results
Complete BM failure and premature death in CTC1 null mice
To ascertain the in vivo functions of CTC1, we generated conditional CTC1 knockout mice. The CTC1 gene has 24 exons and encodes a protein of 1121 amino acids (aa), with exon 2 containing the translation initiation start site. We engineered the CTC1 locus and flanked exon 6 with loxP sites in the targeting vector (Figure 1A; Supplementary Figure S1A and B). Deletion of exon 6 is predicted to generate a frameshift mutation, resulting in premature termination of the CTC1 open reading frame. Two independently targeted ES cell lines were generated and used to produce CTC1F/+ and CTC1F/F mice and MEFs (Supplementary Figure S1C). Expression of Cre-recombinase in CTC1F/F MEFs resulted in efficient deletion of exon 6 and the generation of a transcript encoding a severely truncated protein of only 234 aa, lacking all of the four predicted OB-folds required to interact with STN1 and TEN1 (Supplementary Figure S1B and D) (Theobald and Wuttke, 2004; Gao et al, 2007; Sun et al, 2009, 2011). Indeed, endogenous STN1 protein level was greatly reduced, and its localization to telomeres was undetectable in CTC1−/− MEFs (Supplementary Figure S2A and B). These results suggest that the CST complex is likely unstable in the absence of CTC1.
Figure 1.
Conditional deletion of CTC1. (A) Schematic showing CTC1 genomic locus, targeting construct and CTC1 null allele. Black boxes, coding exons, white box, non-coding exon; red box, exon 6; arrowheads, loxP sites; green rectangle, PGK-neo gene; EV, EcoRV; blue rectangles, left and right arm probes for genomic Southern. (B) Kaplan–Meier survival curve of WT and CTC1 null mice. Number of mice analysed is indicated. (C) Weight of spleens from WT and CTC1 null mice. P=0.0013. Two-tailed t-test was used to calculate statistical significance. (D) Histology of femurs derived from 25-day-old WT and CTC1 null mice. Scale bar, 100 μm. (E) Top: Expression of Sca-1 and c-Kit in multilineage negative BM cells from WT and CTC1−/− mice. Bottom: BrdU/7-AAD FACS profiles of BM LSK cells (Lin−Sca-1+ c-kit+) derived from aged WT and CTC1−/− mice. Results were expressed as percentage of total nucleated BM cells for each fraction. Number of mice analysed is indicated.
We crossed CTC1F/F mice with the zona pellucida 3 (ZP3)-Cre deleter mouse to generate CTC1−/− animals. While CTC1 null mice appeared grossly normal right after birth, they were consistently smaller than their wild-type (WT) littermates, developed sparse fur coverings and had a median lifespan of only 24 days (Figure 1B; Supplementary Figure S2C). Endogenous STN1 protein levels were markedly reduced or undetectable in all CTC1−/− tissues examined (Supplementary Figure S2D). Gross inspections revealed that compared with WT controls, CTC1 null mice possess significantly smaller thymi and spleens, although other organs appeared to be of normal size and weight in relationship to body weight (Figure 1C; Supplementary Figure S3). Histological examination of femurs derived from CTC1−/− mice just before they died revealed complete BM failure, with a total absence of trilineage haematopoiesis and replacement of the BM by stromal adipose tissues that is likely the cause for premature death (Figure 1D). This BM failure phenotype is reminiscent of the haematopoietic defects observed in Pot1b−/−; mTR+/− mice, although in comparison BM defects in CTC1−/− mice occurred much more rapidly (Hockemeyer et al, 2008; He et al, 2009). Since BM failure in Pot1b−/−; mTR+/− mice is due to compromised haematopoietic stem cell (HSC) function (Wang et al, 2011), we examined the HSC status of CTC1 null mice. FACS analyses of BM derived from 4/5 CTC1 null mice revealed severe depletion of both LK (Lin−, Sca-1−, c-kit+) and LSK (Lin−, Sca-1+, c-kit+) cells, populations enriched in HSCs and multipotent progenitors (0.022% in CTC1−/− BM versus 2.54% for WT controls). These results suggest that HSCs were depleted in a majority of CTC1 null mice, likely resulting in complete BM failure (Figure 1E). Interestingly, in vivo BrdU labelling of a younger CTC1 null mouse revealed apparently normal numbers of LK cells, but with a significantly higher percentage of abnormal LSK cells in the G2/M-phase of the cell cycle compared with WT controls (42.3% for CTC1−/− BM versus 2.51% for WT controls) (Figure 1E). We surmised that in the absence of CTC1, proliferating HSCs experience a G2/M cell-cycle arrest. Because the majority of CTC1 null mice already display this phenotype at birth, we isolated fetal livers from 15.5-day-old WT and CTC1−/− embryos. Compared with WT controls, a three-fold reduction in the total number of LSK cells was observed in 100% of CTC1−/− fetal livers, with a two-fold increase in the number of cells arrested in G2/M (Supplementary Figure S4A). Taken together, these data indicate that CTC1 deletion resulted in a profound G2/M arrest in HSCs, leading to BM failure and premature death.
Proliferative defects in CTC1 null cells
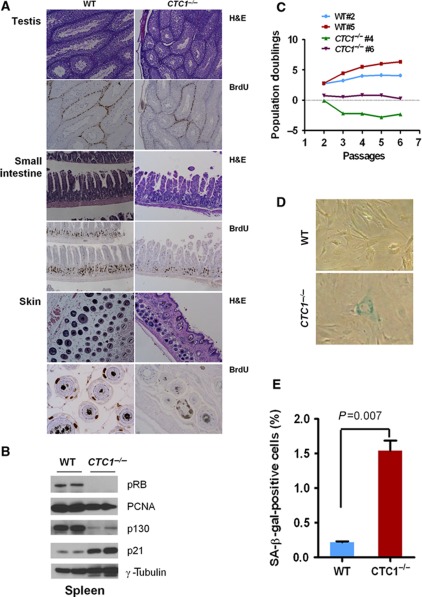
To further examine the impact of CTC1 deletion on cellular proliferation, we performed in vivo BrdU incorporation experiments in WT and CTC1 null mice. Deletion of CTC1 resulted in an overall decrease in cellular proliferative capacity, with reduced BrdU incorporation in highly proliferative tissues including the skin, small intestine and testis (Figure 2A; Supplementary Figure 4B). Analysis of CTC1−/− splenocytes revealed a cell-cycle profile indicative of a growth arrest phenotype, including the diminished expression of the proliferative markers phosphorylated pRB, p130 and PCNA and increased expression of p21, which promotes cell-cycle arrest in the setting of DNA damage (Figure 2B). Profound proliferative arrest was also observed in CTC1 null MEFs, manifested as rapid growth arrest and a 20-fold increase in senescence-like phenotype by passage 2 (Figure 2C–E).
Figure 2.
Deletion of CTC1 results in attenuated proliferation. (A) Tissues isolated from in vivo BrdU-labelled mice were harvested and serial sections processed for H&E and immunostaining with an anti-BrdU antibody. (B) Immunoblots of pRB, p130, PCNA and p21 in splenocytes of the indicated genotypes. γ-tubulin served as loading control. (C) Proliferative defects in primary CTC1 null MEFs. Two independent lines per genotype are shown. (D) SA-β-galactosidase staining and (E) quantification of WT and CTC1 null MEFs at passage 2. Mean values were derived from at least three experiments. Two-tailed t-test was used to calculate statistical significance. Error bars: standard error of the mean (s.e.m.).
Chromosome fusions and progressive telomere loss in the absence of CTC1
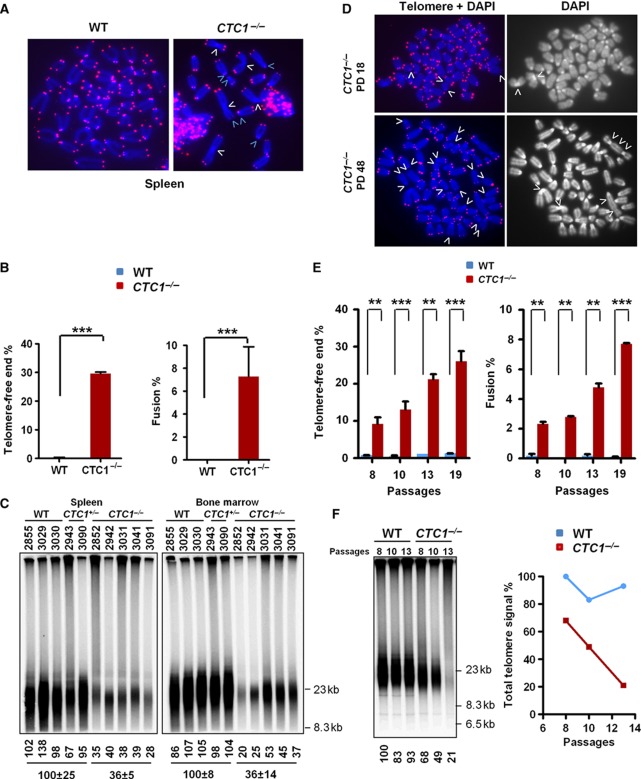
The increased p21 expression in CTC1−/− splenocytes suggested that the observed proliferative arrest could occur as a consequence of increased DNA damage. Since dysfunctional telomeres are recognized as damaged DNA, we examined the status of telomeres in CTC1 null cells. A profound defect in telomere end protective function was observed in CTC1 null cells. Compared with WT controls, 30% of chromosome ends in CTC1 null splenocytes lacked telomeric signals, and end-to-end chromosome fusions were observed in 7% of all chromosomes examined (Figure 3A and B). None of these fusions possessed any telomere signals at fusion sites, suggesting that they arose as a consequence of critical telomere attrition. In support of this notion, TRF Southern analysis revealed extensive telomere loss in 5/5 of spleens and BMs isolated from CTC1 null mice (Figure 3C; Supplementary Figure S5). An ∼3-fold progressive increase in the number of fused chromosomes and telomere-free chromosome ends were observed in CTC1 null MEFs from passages 10 to 19, along with a dramatic decline in total telomere length by passage 13, to 21% of total telomere signal observed in WT controls (Figure 3D–F). These results indicate that rapid and catastrophic telomere attrition occurred in the absence of CTC1, culminating in critical telomere shortening, chromosome fusions and compromised cellular functions.
Figure 3.
Chromosomal fusions and telomere deletion in CTC1 null cells. (A) Telomere-FISH analysis on metaphase chromosome spreads of CTC1+/+ and CTC1−/− splenocytes using Tam-OO-(CCCTAA)4 telomere peptide nucleic acid (red) and 4,6-diamidino-2-phenylindole (DAPI, blue). A minimum of 180 metaphases were analysed per genotype. White arrows, chromosome fusion sites; green arrows, representative signal-free ends. (B) Quantification of chromosome aberrations in (A). Left, telomere signal-free chromosome ends; right, chromosome fusions. A minimum of 30 metaphases were examined. Mean values were derived from at least three experiments. Two-tailed t-test was used to calculate statistical significance. P values are indicated. Error bars: s.e.m. (C) HinfI/RsaI digested spleen and BM genomic DNA of the indicated genotypes were hybridized under denatured conditions with a 32P-labelled [CCCTAA]4-oligo to detect total telomere DNA. Telomere signal intensity (%) was quantified using EtBr staining intensity as the total DNA loading control. Error bars: s.e.m. (D) Telomere-FISH and DAPI (left) and DAPI only (right) analysis on metaphase chromosome spreads of immortalized CTC1+/+ and CTC1−/− MEFs at passages 10 and 19 using Tam-OO-(CCCTAA)4 telomere peptide nucleic acid (red) and DAPI (blue). White arrows, left panel, telomere signal-free ends, right panel, chromosome fusion sites. (E) Quantification of increased chromosome aberrations with passage in (D). Left, telomere signal-free chromosome ends; right, chromosome fusions. A minimum of 30 metaphases were examined. Mean values were derived from a minimum of 2 cell lines per genotype. Two-tailed t-test was used to calculate statistical significance. P values: **: <0.005, ***: <0.0005. Error bars: s.e.m. (F) left, HinfI/RsaI digested CTC1+/+ and CTC1−/− MEF genomic DNA of the indicated passages were hybridized under denatured conditions with a 32P-labelled [CCCTAA]4-oligo to detect total telomere DNA. Telomere signal intensity (%) was quantified by setting WT total telomere DNA as 100%. Right, graph showing rapid telomere loss with passage observed in CTC1−/− MEFs.
CTC1 deletion results in increased 3′ G-overhang
A consequence of CTC1 deletion is increased formation of the 3′ ss G-overhang. We therefore examined the status of the G-overhang in CTC1 null tissues and MEFs. Non-denaturing in-gel hybridization revealed that CTC1 null splenocytes and total BM exhibited a 23- to 34-fold increase in the signal intensity of the ss G-overhang (Figure 4A; Supplementary Figure S5). This dramatic increase in both overhang signal intensity and length heterogeneity correlated inversely with overall telomere length, with the greatest amount of the ss G-overhang signal observed in tissues with the shortest total telomere length. ss TTAGGG repeats were sensitive to Exonuclease I treatment, suggesting that they originated at the 3′ end (Figure 4B). In contrast to the increased ss TTAGGG repeats observed in POT1b−/−; mTR+/− mice (He et al, 2009), increased ss G-overhang was observed only in rapidly proliferating CTC1 null splenocytes and BM and not in quiescent liver cells (Supplementary Figure S6). We next asked how quickly this increase in ss G-overhang signal intensity occurred after CTC1 deletion. Tamoxifen treatment of CAG-CreER-CTC1F/F MEFs revealed that the increase in ss TTAGGG repeat intensity occurred rapidly, within 9 days after activation of Cre-recombinase, prior to the onset of any observable telomere shortening or evidence of significant telomere loss at chromosome ends (Figure 4C; Supplementary Figure S7A–D). This result suggests that acute removal of CTC1 induced ss G-strand formation that is not accompanied immediately by telomere dysfunction.
Figure 4.
Deletion of CTC1 results in increased ss G-overhang. (A) In-gel hybridization analysis of genomic DNA isolated from BM and spleens of CTC1+/+ and CTC1−/− mice under native (top panel) and after denaturation (bottom panel) with a 32P-labelled [CCCTAA]4-oligo probe to detect ss (top) and total (bottom) telomere DNA. Overhang signal intensity (%) was normalized to the total telomeric signals. (B) In-gel hybridization analysis of genomic DNA isolated from CTC1+/+ and CTC1−/− MEFs treated with (+) or without (−) Exo I under native (top) and after denaturation (bottom). Gels were hybridized with a 32P-labelled [CCCTAA]4-oligo probe. (C) In-gel hybridization analysis of genomic DNA isolated from CreER-CTC1F/F (two independent lines) and CTC1+/+ MEFs treated with 4-HT for the indicated times. Overhang signal intensity (%) was normalized to the total telomeric signals and compared with CTC1F/F MEFs without 4-HT and WT MEFs plus 4-HT. (D) 2D gel electrophoresis analysis (first dimension: molecular weight, second dimension: DNA conformation) of genomic DNA isolated from CTC1+/+ and CTC1−/− MEFs and splenocytes. Left panels, schematic of DNA conformations revealed under native (top) and denaturing (bottom) conditions. Right panels, 2D analysis of restriction enzyme digested genomic DNAs under native (top) and denaturing (bottom) conditions. White arrowheads, single-strand-G-DNA; thick white arrowhead, double-strand linear DNA; black arrowheads, t-circles. DNAs were first fractionated under native conditions, hybridized with a 32P-labelled [CCCTAA]4-oligo probe to detect ss DNA species, then denatured and probed for all DNA conformations.
The marked ss TTAGGG repeat heterogeneity observed in CTC1 null cells prompted us to use native and denaturing 2D gel electrophoresis to determine whether other DNA conformations (linear or circular forms) are generated at telomeres following CTC1 deletion. 2D gels performed on DNAs from WT MEFs and splenocytes under native conditions revealed an arc of ss G-rich telomeric DNA, corresponding to the 3′ ss G-overhang (Figure 4D) (Wu et al, 2006; Nabetani and Ishikawa, 2009; Oganesian and Karlseder, 2011). Under denaturing conditions, both ss and double-stranded (ds) telomeric DNAs were observed (Figure 4D). In CTC1 null MEFs and splenocytes, the same DNA conformations were observed, with an increased amount of ss G-DNA present even when total ds telomeric signals were barely detectable (Figure 4D). Surprisingly, we detected the presence of prominent extrachromosomal telomeric repeat (ERCT) DNA in the form of circular ds telomere (t)-circles in CTC1 null MEFs (Figure 4D). T-circles are a hallmark of cells that maintain their telomeres via the HR based alternative lengthening of telomeres (ALT) mechanism of telomere maintenance (Cesare et al, 2009; Nabetani and Ishikawa, 2009; Oganesian and Karlseder, 2011), suggesting that HR at telomeres is elevated in the absence of CTC1.
DDR in cells lacking CTC1
Protection of the ss telomeric overhang requires the coordinated activities of RPA and POT1 (Flynn et al, 2011). Since POT1 is typically limiting in mammalian cells (Kibe et al, 2010), we hypothesized that in the absence of CTC1, the POT1–TPP1 complex would be insufficient to fully protect the excessive ss telomeric DNA from interacting with RPA, resulting in subsequent activation of DNA damage checkpoint proteins ATR and its downstream kinase Chk1. In support of this notion, shRNA-mediated depletion of CTC1 results in the initiation of a DDR, manifested as increased phosphorylation of ATR and Chk1 (Figure 5A; Supplementary Figure S7E and F). In CTC1−/− MEFs, activation of both ATR, ATM, Chk1 and Chk2 were observed, accompanied by rapid growth arrest and a senescence-like phenotype (Figure 2C–E and Figure 5A–C). Overexpression of CTC1, but not STN1 alone, in CTC1 null MEFs completely repressed checkpoint protein activation and rescued the proliferative defect, indicating that the DDR was due to CTC1 deletion (Figure 5C and D). Deletion of CTC1 did not affect the localization of shelterin complex at telomeres (Figure 5E; Supplementary Figure S8). Interestingly, telomere dysfunction induced DNA damage foci (TIFs) were not observed in CTC1 null MEFs. Instead, γ-H2AX-positive foci were visible in the nucleus without obvious association with telomeric signals (Figure 5F). CTC1 null MEFs were able to elicit robust TIF formation when endogenous POT1a and POT1b were removed from telomeres by overexpression of the dominant negative allele TPP1ΔRD (Figure 5F). These results suggest that either the DDR observed was not due to telomere defects, or that they occurred on a subset of telomeres too short to be visualized by the TIF assay. Evidence of elevated global DNA damage (increased number of broken chromosomes, chromatids and fragments) was not detected in metaphase spreads from CTC1 null MEFs (Figure 3A and D), suggesting that the observed DDR originated from chromosome ends devoid of telomere sequences. To test this hypothesis, we examined the co-localization of γ-H2AX and telomere-FISH signals on mitotic chromosomes (Cesare et al, 2009; Thanasoula et al, 2010). WT control MEFs infected with shRNA against TRF2 or expressing TPP1ΔRD displayed prominent γ-H2AX co-staining with telomeres, indicating that uncapped telomeres devoid of TRF2 or POT1–TPP1 activate a DDR (Figure 5G and H and data not shown). In contrast, in CTC1 null MEFs γ-H2AX staining was observed only at chromosome ends missing telomere signals (Figure 5G and H). These results suggest that unlike cells missing shelterin components, the DDR in CTC1 null MEFs occurs only at chromosome ends devoid of telomeres.
Figure 5.
CTC1 deletion activates a DDR. (A) Immunoblot of pATR, p53 and p21 in WT MEFs treated with the indicated shRNAs. γ-Tubulin served as loading control. (B) Immunoblot of pATM, pChk1 and pChk2 in two independent lines of MEFs of the indicated genotypes. γ-Tubulin served as loading control. (C) Immunoblot of pATM, pChk1, pChk2, Flag–CTC1, HA–STN1 and endogenous STN1 in CTC1 null MEFs reconstituted with indicated cDNAs. γ-tubulin served as loading control. (D) Proliferation of SV40 immortalized CTC1 null MEFs reconstituted with the indicated cDNAs. (E) Telomere-PNA FISH demonstrating the localization of shelterin components to telomeres in CTC1−/− MEFs. Cells were stained with the indicated antibodies (green), telomere-PNA FISH (red) and DAPI (blue). Localization of the indicated shelterin proteins on telomeres (telo) in CTC1 null MEFs. (F) γ-H2AX-positive TIFs in WT (top) and CTC1−/− (bottom) MEFs, expressing either vector or TPP1ΔRD cDNA. Cells were fixed 72 h after retroviral infection, stained with anti-γ-H2AX antibody (green), Tam-OO-(CCCTAA)4 telomere peptide nucleic acid (red) and DAPI (blue). (G) WT MEFs expressing TPP1ΔRD or CTC1−/− MEFs were arrested in mitosis with colcemid, metaphase spreads prepared and stained with anti-γ-H2AX antibodies (green), Tam-OO-(CCCTAA)4 telomere peptide nucleic acid (red) and DAPI (blue). Arrows point to sites of γ-H2AX localization. Insets show enlarged images. (H) CTC1−/− MEFs (two independent lines), WT MEFs treated with shRNA against TRF2 or expressing TPP1ΔRD were assayed for TIF formation. Quantification of the number of TIFs with or without visible telomere signals co-localizing with γ-H2AX staining are indicated.
CTC1 is required for efficient replication and restart of stalled replication forks at telomeres
The repetitive nature of telomere DNA sequences poses a special challenge to the DNA replication machinery (Gilson and Geli, 2007; Sampathi and Chai, 2011). Telomere binding proteins are thought to be required to promote efficient telomere replication and prevent replication fork stalling at telomeres (Martinez et al, 2009; Sfeir et al, 2009; Badie et al, 2010). Given that the excessive ss G-overhang DNA induced by CTC1 deficiency could arise as a defect in the synthesis of C-strand telomeres, we suspected that deletion of CTC1 hindered efficient replication of telomeres. To test whether CTC1 promotes efficient replication of telomere DNA, we examined WT and CTC1 null MEFs for the presence of fragile telomeres, a hallmark of telomere replication defects (Sfeir et al, 2009). A significant increase in the number of fragile telomeres was observed in the absence of CTC1, with the number of fragile telomeres observed in CTC1−/− MEFs equivalent to those observed in aphidicolin-treated WT controls (Figure 6A and B). CTC1−/− MEFs treated with aphidicolin grew extremely poorly and generated few metaphases containing fragile telomeres. These results suggest that CTC1 is required for proper telomere replication. To further test this hypothesis, we measured the efficiency of BrdU incorporation into telomeres during the S-phase, using BrdU labelling and CsCl separation of replicated leading/lagging telomeres (Dai et al, 2010). Cells were synchronized at the G1/S boundary and then released into S-phase in the presence of BrdU, and cells were collected after replication completed (Supplementary Figure S9A). Replicated and unreplicated DNA was separated on CsCl density gradients, and telomeres detected by slot-blot with a telomere probe (Supplementary Figure S9B). Acute deletion of CTC1 in CAG-CreER; CTC1F/F MEFs was accomplished by treatment with 4-hydroxy tamoxifen (4-HT) during the synchronization process for 3 days. CTC1 deletion resulted in decreased replication of both leading- and lagging-strand telomeres, accompanied by an increase in unreplicated telomeres (Figure 6C; Supplementary Figure S9C). To examine whether the long-term loss of CTC1 preferentially affect C-strand synthesis at the lagging telomeres, we utilized chromosome-orientation FISH (CO-FISH) to monitor the status of leading- and lagging-strand telomeres in CTC1 null MEFs. Compared with WT and early passage CTC1 null MEFs, a significant decrease in the intensity of leading C-strand telomere signals was observed in late-passage CTC1 null MEFs (Figure 6D and E). Taken together, these results suggest that replication of telomere DNA was less efficient in the absence of CTC1, with a preferential decrease in the amount of leading C-strand telomeres observed at late passages.
Figure 6.
CTC1 is required for efficient replication and efficient restart of stalled replication fork at telomeres. (A) Left, examples of the fragile telomere phenotype observed in CTC1 null MEFs and WT MEFs treated with 0.2 μM aphidicolin. Arrows point to fragile telomeres. (B) Quantification of fragile telomeres observed in (A). (C) Summary of telomere replication in CTC1F/F and CAG-CreER; CTC1F/F MEFs treated with or without 4-hrdroxytamoxifen (4-HT) to delete CTC1. Two-tailed t-test was used to calculate statistical significance. n≥4, Error bars: s.e.m. (D) Examples of CO-FISH images of partial metaphase spreads from WT (PD 49), CTC1−/− (PD14) and CTC1−/− (PD58) MEFs. Red signals: G-strand telomeres, green signals: C-strand telomeres. Red arrowheads point to loss of lagging-strand telomeres and green arrowheads point to loss of leading-strand telomeres. (E) Quantification of signal intensity of leading- and lagging-strand telomeres in (D). Two independent cell lines were analysed, and a minimum of 35 metaphases and 1500 chromosomes were scored per genotype and passage. Mean values were derived from two cell lines. The two-tailed t-test was used to calculate statistical significance. Error bars: s.e.m. (F) Distribution of telomere signals on CsCl gradient after releasing from HU treatment. BrdU pulse-labelled telomere DNA following HU treatment was separated on CsCl gradient. Fractions were collected from the bottom of the gradient and slot-blot was used to quantitate the amount of telomeres in each fraction. Telomere signals from telomeres with density ≥1.755 g/ml are amplified in the insert. (G) Percentage of BrdU-containing replicated telomeres. One-tailed t-test was used to calculate statistical significance. n=3. Error bars: s.e.m.
Since telomeres resemble difficult to replicate fragile sites, additional factors might be required to restart stalled telomere replication. To examine whether CTC1 plays a role for efficient restart of stalled forks at telomeres, cells were synchronized and released into S-phase. At mid-S-phase, cells were treated with hydroxyurea (HU) for 1 h to arrest replication forks. After the removal of HU, BrdU was added to media for 1 h to pulse label daughter strands replicated from restarted forks (Supplementary Figure S9D). Cells were then collected immediately after BrdU labelling and the heavier BrdU-containing telomere DNA was then separated from non-BrdU-incorporated telomeres using the CsCl density separation technique (Figure 6F). The percentage of BrdU-labelled telomeres among the total telomere molecules represents the efficiency of the restart of stalled fork specifically at the telomeric region. Since the time for pulse labelling was short, the majority of telomeres was unlabelled and the population of BrdU-incorporated telomeres was low (Figure 6F). Comparison of BrdU-labelled telomeres revealed that CTC1deletion led to a statistically significant reduction in telomeric BrdU incorporation after the removal of HU (Figure 6G). Taken together, our data suggest that CTC1 may play a role for the efficient restart of stalled replication forks at telomeres.
Discussion
In this study, we generated a conditional CTC1 knockout mouse to demonstrate an essential role for CTC1 in regulating telomere replication. Deletion of CTC1 in mice resulted in G2/M cell-cycle arrest in haematopoietic stem cells, culminating in complete BM failure and premature death. This growth arrest phenotype is reminiscent of the Mec1-mediated G2/M cell-cycle arrest observed when the CST complex is disrupted in budding yeast (Garvik et al, 1995; Grandin et al, 1997, 2001; Jia et al, 2004). Mechanistically, our data suggest that CTC1 facilitates telomere replication by promoting efficient restart of stalled replication forks. CTC1 deletion results in increased loss of leading C-strand telomeres, catastrophic telomere loss and accumulation of excessive ss telomere DNA that activates an ATR-dependent DDR. Collectively, these findings suggest that mammalian CTC1 is required to prevent rapid telomere loss by promoting its efficient and complete replication.
The CST complex appears to possess evolutionarily conserved functions in DNA replication (Giraud-Panis et al, 2010; Nakaoka et al, 2011). In yeast, Cdc13 and Stn1 interact with Polα and couple telomeric C- and G-strand synthesis, preventing the formation of excessively long G-overhangs. CST could either mediate C-strand fill in following G-strand extension by telomerase and/or prevent excessive C-strand resection by nucleases (Qi and Zakian, 2000; Grossi et al, 2004; Petreaca et al, 2006; Puglisi et al, 2008). In mouse cells, the shelterin component POT1b functions to prevent C-strand resection by unknown nucleases (Hockemeyer et al, 2008; He et al, 2009). Since the G-overhang appears to be of normal length in quiescent CTC1−/− liver cells, CTC1 is unlikely to play a significant role in mediating C-strand resection in all cell types. We favour a role for CTC1 in facilitate priming and synthesis of telomere DNA chains on the lagging-strand template, and/or promoting efficient re-initiation of lagging telomere DNA synthesis by DNA Polα at stalled replication forks. These functions might be especially important if a replication stall uncouples DNA Polα and replication helicase activities the fork (Yao and O’Donnell, 2009). In the absence of CTC1, fork stalling at telomeres would give rise to ss G-strand DNA that are insensitive to Exonuclease I digestion. Replication fork stalling appears to be a particularly vexing problem at telomeres, since the aberrant G-rich secondary structures are postulated to cause steric hindrance, making them difficult substrates to replicate. Therefore, besides the CST complex, several other telomere binding proteins and associated factors have evolved to facilitate replication fork progression through telomeres (Crabbe et al, 2004; Miller et al, 2006; Martinez et al, 2009; Sfeir et al, 2009; Badie et al, 2010; Ye et al, 2010). While we postulate that the CST complex is required for efficient replication of telomeres, we cannot rule out the possibility that they also participate in general DNA replication and the restart of stalled forks at non-telomere regions.
Stalled replication forks at telomeres might also initiate aberrant recombination events that could account in part for the catastrophic loss of total telomeric DNA observed in CTC1 null cells (Lustig, 2003; Miller et al, 2006). The loss of bulk telomeric DNA observed in the BM of CTC1 null mice occurred much more rapidly than the progressive loss of telomere sequences observed in POT1b−/−; mTR+/− mice, another mouse model in which rapid telomere attrition results in haematopoietic defects (He et al, 2009; Wang et al, 2011). In contrast to the POT1b−/−; mTR+/− mouse, in which telomere shortening and increased G-overhang was observed in all organs examined, telomere defects in CTC1 null animals were limited only to rapidly proliferative tissues (haematopoietic system, skin, small intestine) and were not observed in the quiescent liver. These results further support a defect in telomere replication as a mechanism for the phenotypes observed in CTC1 null mice. Since HR is the primary pathway utilized to restart stalled replication forks, we postulate that increased aberrant HR at telomere replication forks in the absence of CTC1 results in catastrophic telomere shortening. In support of this notion, T-circles, a product of cells that utilize HR for telomere maintenance (Wang et al, 2004; Nabetani and Ishikawa, 2009; Oganesian and Karlseder, 2011), are prominent in CTC1 null MEFs, suggesting that CTC1 is required to repress aberrant HR at telomere replication forks. It is intriguing to note that Arabidopsis CTC1 mutants also display elevated recombination and extensive telomere loss prior to end-to-end chromosome fusions (Surovtseva et al, 2009).
The mammalian CST complex is thought to play a role in telomere end protection (Miyake et al, 2009; Surovtseva et al, 2009). However, our studies suggest that telomere protection mediated by the CST complex is distinct from that mediated by the shelterin complex. Removal of mammalian TRF2–RAP1 or TPP1–POT1 complexes results in immediate telomere dysfunction, manifested as recruitment of ATM and ATR to chromosome ends, respectively, robust TIF formation, and initiation of NHEJ and HR-mediated repair reactions, while overall telomere length remains intact (Wu et al, 2006; Denchi and de Lange, 2007; Guo et al, 2007; Deng et al, 2008; Dimitrova et al, 2008; Rai et al, 2010). By contrast, increased TIF formation was not observed in CTC1 null MEFs, even at late passages. We postulate that in the absence of CTC1, the increased ss G-overhang generated over time could not be completely protected by TPP1–POT1, leading to the recruitment of RPA and subsequent activation of ATR/Chk1 (Flynn et al, 2011, 2012). Increased telomere attrition ultimately results in the displacement of the shelterin complex from chromosome ends, leading to the recruitment of γ-H2AX and the activation of ATM/Chk2. Our results suggest that in contrast to the shelterin complex, which is involved in both telomere end protection and replication, the CST complex promotes telomere replication and does not play a direct role in repressing a DDR at mammalian telomeres. It would be interesting to understand whether shelterin cooperates with CST to regulate mammalian telomere replication.
Several inherited human BM failure syndromes are due to defects in telomere maintenance. For example, Dyskeratosis congenita (DC) is characterized by very short telomeres, BM failure, cutaneous phenotypes and increased cancer incidence (reviewed in Young, 2010). Recently, whole-exome sequencing revealed that missense mutations in CTC1 results in the human disorder Coats plus (Anderson et al, 2012; Polvi et al, 2012; reviewed in Savage, 2012). Coats plus is an autosomal recessive disorder characterized by retinal telangiectasia, intracranial calcifications, osteopenia and gastrointestinal bleeding (Tolmie et al, 1988; Crow et al, 2004). Interestingly, white blood cells from Coats plus patients with compound heterozygous mutations have very short telomeres, suggesting that telomere maintenance function is compromised in these patients. Indeed, some Coats plus patients develop a normocytic anaemia that reflects a degree of BM failure (Anderson et al, 2012). These exciting clinical results suggest that similar to the phenotypes observed in our CTC1 null mice, human CTC1 missense mutations also confer haematopoietic defects. Understanding how these point mutations impact upon CTC1 function should reveal additional insights into the function of the CST complex at telomeres. We postulate that patients with a family history of unexplained BM failure and without the characteristic DC mutations affecting telomere length maintenance might have mutations in CTC1, and perhaps in other members of the CST complex as well. CTC1 mutations should therefore be examined in patients with inherited BM failure lacking mutations in genes encoding shelterin components and proteins involved in telomerase function.
Materials and methods
Generation of CTC1 conditional knockout mice
A long-range PCR strategy was used to obtain three correct genomic fragments for generating a conditional targeting vector. A 3.3-kb KpnI–XhoI fragment (5′ arm), a 5.1-kb SalI–NotI fragment (3′ arm), and a 1.2-kb HindIII–SalI fragment containing exon 5 and the second loxP at 3′ end were inserted into pKOII vector (Wu et al, 2006). The targeting vector was linearized with NotI and electroporated into AB2.1 ES cells. ES cells were screened by long-range PCR and two selected ES cell candidates (1A11, 3E3) with correct recombination were further confirmed by Southern analysis and injected into C57BL/6 blastocysts. Chimeric mice were mated with WT C57BL/6 to generate CTC1F/+ offspring. CTC1F/+ mice were intercrossed to obtain CTC1F/F homozygous mice. Female CTC1F/F mice were mated with ZP3-Cre or chicken actin (CAG)-Cre male mice to give rise to CTC1F/+;ZP3-Cre or CTC1F/+;CAG-CreER mice. Female CTC1F/+;ZP3-Cre mice were crossed with WT mice to obtain CTC1+/− heterozygous mice. The homozygous CTC1−/− mice were generated by intercrossing CTC1+/− mice. All mice were maintained according to Yale University IACUC-approved protocols.
Generation of CTC1−/− and CTC1F/F, CAG-CreER MEF
CTC1−/− MEF were isolated from E13.5 embryos by intercrossing CTC1+/− mice. CTC1F/F; CAG-CreER or CTC1F/F MEFs were obtained from E13.5 embryos by intercrossing CTC1F/+;CAG-Cre with CTC1F/+ mice. Primary MEFs at passages 1 or 2 were immortalized with SV40 large-T antigen retrovirus. To acutely delete CTC1, CTC1F/F; CAG-CreER MEF was treated with 1 μM 4-HT for 2 days and maintained in the presence of 10 nM 4-HT for 1 week. All MEFs were cultured in DMEM with 10% serum.
Isolation of BM, splenocytes and fetal liver cells
Mice were injected with BrdU (i.p.) at the dosage of 150 mg/kg 2 h before euthanizing. BM cells were flushed from hind limb bones through a 21-gauge needle into HBSS+ (Hanks balanced salt solution (Invitrogen, Carlsbad, CA)), 2% FBS (Invitrogen) and 10 mM HEPES. Fetal liver from E15.5 embryo was dissociated by passing 21-gauge needle. Spleen was smashed between two pieces of glass slide. The cells were passed through 40 μM cell strainer to make sure of single-cell suspensions. Red blood cells were removed by the lysis buffer (3% acetic acid in methylene blue (Stem Cell Technologies, Vancouver, BC, Canada). Fetal liver cells were incubated in GMEM media with 10% serum in the presence of 10 μM BrdU for 2 h at 37°C. To obtain metaphase chromosome, splenocytes were suspended in DMEM with 10% serum at 37°C for 3 h and treated with Colcemid for 1 h.
Flow cytometry
In all, 1 × 107 total BM or fetal liver cells were centrifuged and resuspended in 200 μl HBSS+. Cells were stained for 15 min with a cocktail of antibodies including nine lineage markers: Ter-119, CD3, CD4, CD8, B220, CD19, IL-7Rα, GR-1 and Mac-1 (eBioscience, San Diego, CA) conjugated to APC-Cy-7 (47-4317 eBioscience). In addition, the cocktail also contained anti-Sca-1-PE (12-5981 eBioscience) and anti-c-Kit-APC (17-1171 eBioscience). After staining, cells were washed, resuspended in HBSS+ and analysed by flow cytometry (Calibur or LSRII, BD). LSK populations were selected based on low or negative expression of the mature lineage markers and dual-positive expression for Sca-1 and c-Kit. The analysis of BrdU incorporation was performed using the FITC BrdU flow kit (BD Pharmingen, Cat#552598). FlowJo software was used for all data analysis.
Generation of mouse STN1 antibody, expression plasmids and commercial antibodies used
Full-length mouse STN1 cDNA was obtained from RT–PCR and subcloned into pRSET-A (Invitrogen) vector. His-tagged STN1 fusion protein was expressed in BL21DE3 bacteria and purified by Nickel-charged agarose beads according to Invitrogen’s protocols. Rabbit antiserum were raised by injection of purified fusion protein (Ptlab) and affinity purified by cellulose membrane containing fusion protein. Full-length mouse CTC1 and STN1 expression vectors were generated by inserting full-length cDNAs into pQCXIP retrovirus vectors. Anti-mouse TRF1 and TRF2 were gifts from Jan Karlseder (Salk). Commercial antibodies used were Anti-Phospho-Chk1 (Ser345) (#2348), Anti-Phospho-ATR (Ser428) (#2853), Anti-Phospho-ATM (Ser1981) (#4526) from Cell Signaling; Anti-pRB (#554136) and Anti-Chk2 (#611570) from BD Pharmingen; Anti-p21 (sc-6246), Anti-p130 (sc-317) and Anti-PCNA (sc-7907) from Santa Cruz; Anti-Flag, HA, γ-tubulin from Sigma; Anti-γ-H2AX (#05-636) from Upstate; Anti-RAP1(#A300-306) from Bethyl.
Knockdown of STN1 or CTC1 with shRNA
Lentivirus-based shRNAs against mouse STN1 and CTC1 were purchased from Sigma (sequences available on request). The knockdown efficiency of different shRNA was tested by either RT–PCR or immunoblotting. Total RNA was extracted with Trizole reagent (Invitrogen) and reverse transcription was performed with Super transcriptase III Kit (Invitrogen). PCR was performed with SybrGreen PCR Kit (Biolab) and reactions were run on an ABI PCR machine.
Histology and immunohistochemistry
Tissues were fixed in 10% formalin, paraffin embedded, sectioned and stained for haematoxylin and eosin. BrdU staining was done by Yale Pathology Histology Service Lab.
PNA-FISH, CO-FISH, TIF and immunofluorescence-FISH assays
Cells were treated with 0.5 μg/ml of Colcemid for 4 h before harvest. Trypsinized cells were treated with 0.06 M KCl, fixed with methanol:acetic acid (3:1) and spread on glass slides. Metaphase spreads were hybridized with 5′-Tam-OO-(CCCTAA)4-3′ probe. For CO-FISH, cells were treated with BrdU for 14 h before addition of Colcemid. Metaphase spreads were sequentially hybridized with 5′-FITC-OO-(TTAGGG)4-3′ and 5′-Tam-OO-(CCCTAA)4 probes. For the TIF assay, cells were seeded in eight-well chambers and immunostained with primary antibodies and FITC-secondary antibody, then hybridized with 5′-Tam-OO-(CCCTAA)4-3′ probe. IF-FISH on metaphase spreads were performed as described (Thanasoula et al, 2010).
TRF Southern and 2D-gel
In all, 5–10 μg total genomic DNA were separated by pulse-field gel electrophoresis (Bio-Rad). The gels were dried at 50°C and prehybridized at 58°C in Church mix (0.5 M NaH2PO4, pH 7.2, 7% SDS) and hybridized with γ-32P-(CCCTAAA)4 oligonucleotide probes at 58°C overnight. Gels were washed with 4 × SSC, 0.1% SDS buffer at 55°C and exposed to Phosphorimager screens. After in-gel hybridization for the G-overhang under native conditions, gels were denatured with 0.5 N NaOH, 1.5 M NaCl solution and neutralized with 3 M NaCl, 0.5 M Tris–Cl, pH 7.0, then reprobed with (TTAGGG)4 oligonucleotide probes to detect total telomere DNA. To determine the relative G-overhang signals, the signal intensity for each lane was scanned with Typhoon (GE) and quantified by ImageQuant (GE) before and after denaturation. The G-overhang signal was normalized to the total telomeric DNA and compared between samples. For 2D-gel TRF Southern assay, 20 μg of total genomic DNA was resolved in the first dimension in a 0.4% agarose gel at 1.5 V/cm, EB stained and cut out. The gel slice was casted into a second 1% agarose gel at a 90° orientation from the first gel and resolved at 8 V/cm. The steps for hybridization were same as the described above.
Synchronization of cell-cycle and CsCl gradient grade separation of leading and lagging daughter telomeres from MEFs
For synchronizing MEFs, cells were serum starved in DMEM-0.1% FBS for 48 h and then released into fresh DMEM-10% FBS containing 1 μg/ml aphidicolin for 24 h. Aphidicolin was then removed by washing cells with pre-warmed PBS (three times) before released into fresh DMEM-10% FBS. MEFs synchronized in G1/S boundary were released into DMEM-10% FBS supplemented with 100 μM BrdU for 9 h. Cells were collected, genomic DNA was isolated with DNAeasy DNA isolation kit (Qiagen), digested with AluI/RsaI/CfoI/MspI/HaeIII/HinfI at 37°C overnight, mixed with CsCl solution, centrifuged in vertical rotor VTi65 (Beckman Coulter) for 16–22 h (45 000 r.p.m., 21°C). Fractions were collected, denatured in 1 M NaOH for 10 min, neutralized with 6 × SSC, and used for slot-blot. Blot was hybridized to telomere probe at 42°C overnight. All procedures were performed in dark.
Measurement of the efficiency of restart of stalled replication fork
Cells were synchronized at G1/S boundary and then released to S-phase for 3 h. HU (4 mM) was added to stall replication fork for 1 h. Cells were then washed with pre-warmed media for three times and released into DMEM-10% FBS containing 100 μM BrdU for 1 h. Procedures following this step were performed in dark. Cells were collected and genomic DNA was isolated, digested with AluI/RsaI/CfoI/MspI/HaeIII/HinfI at 37°C for overnight, mixed with CsCl solution, and centrifuged at 39 000 r.p.m. 21°C for 16–22 h. Fractions were collected, denatured in 1 M NaOH, neutralized with 6 × SSC, and used for slot-blot. Blot was hybridized to telomere probe at 42°C overnight. Telomeres with the ≥1.755 g/ml were considered replicated DNA, since the complete BrdU-incorporated lagging telomeres were located at density ∼1.760 g/ml (Supplementary Figure S7C and D).
Supplementary Material
Acknowledgments
We are grateful to Dr Jan Karlseder for providing anti-mouse TRF1 and TRF2 antibodies. This work was supported by the NCI (RO1 CA129037) to SC and NIH R15CA132090, R15GM099008, and American Cancer Society grant IRG-77-003-32 to WC.
Author contributions: PG, WC and SC conceived the project and designed the experiments. PG, JM, YW, CH and WC performed the experiments and generated data for the figures. PG, WC and SC analysed and interpreted the data, composed the figures and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O’Sullivan J, Leibundgut EO, Muter J, Abdel-Salem GM, Babul-Hirji R, Baxter P, Berger A, Bonafé L, Brunstom-Hernandez JE, Buckard JA, Chitayat D et al. (2012) Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 4: 338–342 [DOI] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, Blasco MA, Jonkers J, Tarsounas M (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D (2004) Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB (2009) A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR (2009) Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat StructMol Biol 16: 1244–1251 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Crow YJ, McMenamin J, Haenggeli CA, Hadley DM, Tirupathi S, Treacy EP, Zuberi SM, Browne BH, Tolmie JL, Stephenson JB (2004) Coats’ plus: a progressive familial syndrome of bilateral Coats’ disease, characteristic cerebral calcification, leukoencephalopathy, slow pre- and post-natal linear growth and defects of bone marrow and integument. Neuropediatrics 35: 10–19 [DOI] [PubMed] [Google Scholar]
- Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W (2010) Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J 29: 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Deng Y, Chan SS, Chang S (2008) Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer 8: 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E (2010) CST meets shelterin to keep telomeres in check. Mol cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ (1990) The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem 265: 13231–13239 [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2001) Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J 20: 6127–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S (2007) Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J 26: 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, Chang S (2009) Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 29: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T (2008) Engineered telomere degradation models dyskeratosis congenita. Genes Dev 22: 1773–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Weinert T, Lydall D (2004) Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 166: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T (2010) Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol 30: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman Flynn R, Chang S, Zou L (2012) RPA and POT1: Friends or foes at telomeres? Cell Cycle 11: 652–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet 4: 916–923 [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW (2007) DNA strand break repair and human genetic disease. Ann Rev Genomics Hum Genet 8: 37–55 [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F (2009) Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol 29: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F (2011) Xenopus laevis Ctc1-Stn1-Ten1 (xCST) complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Oganesian L, Karlseder J (2011) Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell 42: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 8: 748–755 [DOI] [PubMed] [Google Scholar]
- Polvi A, Linnankivi T, Kivelä T, Herva R, Keating JP, Mäkitie O, Pareyson D, Vainionpää L, Lahtinen J, Hovatta I, Pihko H, Lehesjoki AE (2012) Mutations in CTC1, Encoding the CTS Telomere Maintenance Complex Component 1, Cause Cerebroretinal Microangiopathy with Calcifications and Cysts. Am J Hum Genet 90: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S (2010) The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J 29: 2598–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathi S, Chai W (2011) Telomere replication: poised but puzzling. J Cell Mol Med 15: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA (2012) Connecting complex disorders through biology. Nat Genet 44: 238–240 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yang Y, Wan K, Mao N, Yu TY, Lin YC, DeZwaan DC, Freeman BC, Lin JJ, Lue NF, Lei M (2011) Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res 21: 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev 23: 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanasoula M, Escandell JM, Martinez P, Badie S, Munoz P, Blasco MA, Tarsounas M (2010) p53 prevents entry into mitosis with uncapped telomeres. Cur Biol 20: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS (2004) Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure 12: 1877–1879 [DOI] [PubMed] [Google Scholar]
- Tolmie JL, Browne BH, McGettrick PM, Stephenson JB (1988) A familial syndrome with coats’ reaction retinal angiomas, hair and nail defects and intracranial calcification. Eye (Lond) 2: 297–303 [DOI] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D (2009) OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen MF, Chang S (2011) Essential roles for Pot1b in HSC self-renewal and survival. Blood 118: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Yao NY, O’Donnell M (2009) Replisome structure and conformational dynamics underlie fork progression past obstacles. Cur Opin Cell Biol 21: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, Verhoeyen E, Britton S, Calsou P, Salles B, Bizard A, Nadal M, Salvati E, Sabatier L, Wu Y, Biroccio A et al. (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142: 230–242 [DOI] [PubMed] [Google Scholar]
- Young NS (2010) Telomere biology and telomere diseases: implications for practice and research. Hematology Am Soc Hematol Educ Program 2010: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.