Abstract
FtsA is an early component of the Z-ring, the structure that divides most bacteria, formed by tubulin-like FtsZ. FtsA belongs to the actin family of proteins, showing an unusual subdomain architecture. Here we reconstitute the tethering of FtsZ to the membrane via FtsA’s C-terminal amphipathic helix in vitro using Thermotoga maritima proteins. A crystal structure of the FtsA:FtsZ interaction reveals 16 amino acids of the FtsZ tail bound to subdomain 2B of FtsA. The same structure and a second crystal form of FtsA reveal that FtsA forms actin-like protofilaments with a repeat of 48 Å. The identical repeat is observed when FtsA is polymerized using a lipid monolayer surface and FtsAs from three organisms form polymers in cells when overexpressed, as observed by electron cryotomography. Mutants that disrupt polymerization also show an elongated cell division phenotype in a temperature-sensitive FtsA background, demonstrating the importance of filament formation for FtsA’s function in the Z-ring.
Keywords: actin, bacterial cytoskeleton, cell division, cytomotive, FtsZ
Introduction
A key step in bacterial cell division is the formation of the Z-ring composed of polymers of the tubulin-like protein FtsZ (for recent reviews: Dajkovic and Lutkenhaus, 2006; Adams and Errington, 2009). FtsZ finds the middle of the cell through two independent mechanisms in E. coli, nucleoid exclusion (Woldringh et al, 1991; Wu and Errington, 2004; Bernhardt and ) and the oscillating MinCDE system (Raskin and ; Lutkenhaus, 2007). The Z-ring constricts and through interaction with other components engages in remodelling cell wall and membranes in order to yield two daughter cells.
During this process FtsZ is known to interact with FtsA and then recruits other components of the divisome, the cell division apparatus. The divisome is currently known to contain more than 20 components and can be divided into inner and outer divisome (Vicente and Rico, 2006). This distinction reflects the fact that it needs to remodel the inner cell membrane, but also the rigid cell wall, and a direct link with enzymes for cell wall remodelling is made. The mechanism of constriction is currently unclear, but the potential or theoretical sources of energy in the system (apart from binding energies) are nucleotides associated with FtsA and FtsZ (inner divisome), as well as cell wall synthesis from precursors in the periplasm (outer divisome).
FtsA belongs to the actin/HSP70 family of proteins (Bork et al, 1992), although its fold was shown to considerably deviate from the canonical actin fold by a subdomain deletion and addition (van den Ent and ). It has been shown in vivo that a deletion of 12 amino acids from the highly conserved C-terminal tail of E. coli FtsZ failed to complement chromosomal ftsZ mutants and failed to interact with FtsA (Ma and Margolin, 1999). Interestingly, the same FtsZ interaction site is used in E. coli by another protein, ZipA (Ma and Margolin, 1999; Mosyak et al, 2000; Haney et al, 2001), that also shows some functional overlap with FtsA, despite being a very different protein in sequence and the exact functions of these proteins, FtsA and ZipA and their relationship with FtsZ have remained enigmatic.
Genetic screening has identified residues within the 2B subdomain of FtsA required for binding to FtsZ, indicating that this could be the interaction site on FtsA with FtsZ (Pichoff and Lutkenhaus, 2007). Moreover, analysis of FtsA sequences revealed a conserved C-terminal motif (Löwe and ), which is predicted to form an amphipathic helix and has been shown to localize FtsA (and with it FtsZ) to the membrane (Pichoff and Lutkenhaus, 2005).
As FtsA belongs to the actin/HSP70 family of proteins, with many members being able to form filaments, it is a long-standing question of whether FtsA is able to polymerize (Löwe et al, 2004). Three pieces of evidence currently exist that it could: expression of a mutant form of FtsA in E. coli leads to massive cylindrical structures formation in the cytoplasm and changes in cell shape (Gayda et al, 1992). GFP–FtsA C-terminal truncations lead to filamentous structures in cells as seen by fluorescence microscopy (Pichoff and Lutkenhaus, 2007). In another case, FtsA from Streptococcus pneumoniae has been demonstrated to form helical filaments in vitro, but no information is available on how that would be possible given that the canonical actin fold is incomplete in FtsA and on what FtsA protofilaments would look like (Lara et al, 2005).
Here, we reconstitute the FtsZ/FtsA/membrane system in vitro using Thermotoga maritima proteins and provide a crystal structure of the FtsA/FtsZ complex. This led us to the discovery that FtsA might be able to form actin-like filaments. We then investigated the polymerization of the FtsA actin-like protein and for the first time demonstrate that an actin-like protein of different subdomain architecture can form a canonical actin-like protofilament. Polymerization of FtsA is required for its proper biological function, as shown through genetically engineered Bacillus subtilis strains.
Results
The last 8–16 residues of T. maritima FtsZ are necessary and sufficient to interact with T. maritima FtsA in vitro
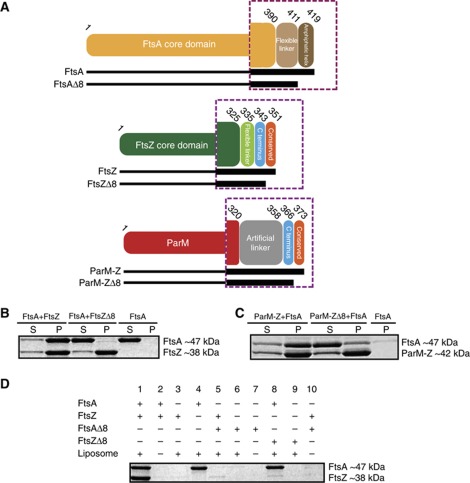
In order to investigate the interaction between the cell division proteins FtsZ and FtsA, we reconstituted the system in vitro using purified T. maritima FtsA (TmFtsA) and FtsZ (TmFtsZ) proteins as well as liposomes made from E. coli total lipid extract. TmFtsA and TmFtsZ were purified as either full-length (TmFtsa 1–419, TmFtsZ 1–351) or C-terminally truncated proteins (TmFtsAΔ8 1–411, TmFtsZΔ8 1–343, Figure 1A). The C-terminal tail of FtsZ is known to interact with FtsA (Ma and Margolin, 1999), whereas the C-terminal tail of FtsA is thought to directly interact with the membrane via an amphipathic helix (Pichoff and Lutkenhaus, 2005). No extra residues were introduced at either end and great care was taken not to lose the tails through proteolysis during purification, as we found to happen easily when monitored by high-resolution mass spectrometry. The proteins were then tested in pelleting assays in the presence of GTP (Figure 1B). In the presence of GTP, TmFtsZ forms polymers that easily sediment, and if these polymers interact with TmFtsA, both proteins will be found in the pellet. This assay revealed that the last eight amino acids of TmFtsZ are necessary for the interaction with TmFtsA, as TmFtsZΔ8 failed to recruit TmFtsA to the pellet fraction. To discover whether or not the highly conserved C-terminal TmFtsZ fragment is sufficient for this interaction, we constructed a hybrid protein consisting of the actin-like protein ParM followed by an artificial linker and the last 16 residues of TmFtsZ. ParM polymerizes in an ATP-dependent manner (van den Ent et al, 2002) and does not interact with FtsA alone (not shown). A pelleting assay in the presence of ATP (Figure 1C) demonstrated that the hybrid protein ParM-Z efficiently interacted with TmFtsA when polymerized; however, when the last eight residues of this hybrid protein are removed (ParM-ZΔ8), the interaction is significantly reduced and the trace amount of FtsA in the pellet fraction is probably a result of some unspecific interactions. We conclude that the last 8–6 residues of TmFtsZ are both necessary and sufficient for interaction with TmFtsA.
Figure 1.
In vitro reconstitution of the FtsZ/FtsA/membrane complex using T. maritima proteins. (A) Schematic representation of T. maritima proteins used. Dotted violet squares represent enlarged views of the C-terminal regions, which are crucial for the experimental design. (B) FtsA-FtsZ co-pelleting assay. S: supernatant, P: pellet. The C-terminal eight residues of FtsZ are necessary to interact with FtsA. (C) The C-terminal 16 residues of FtsZ are sufficient to interact with FtsA as shown in a ParM-Z-FtsA co-pelleting assay. A hybrid protein consisting of R1 ParM and the last 16 residues of FtsZ binds to FtsA. (D) Full-length FtsA binds to the membrane via its C-terminal amphipathic helix and tethers FtsZ to the membrane as shown by a liposome co-sedimentation assay. FtsZ does not have affinity for lipids itself.
What residues are then required for FtsA to tether itself and FtsZ to the membrane? To address this question we reconstituted the FtsZ/FtsA/membrane complex by performing a co-sedimentation assay in the absence of any nucleotides (Figure 1D). This showed that TmFtsA recruits TmFtsZ to the membrane (lane 1) and TmFtsZ does not have an affinity for the lipid bilayer itself (lane 3). Also, this assay confirms that both truncated TmFtsA and TmFtsZ are not functional: TmFtsZΔ8 is not able to interact with TmFtsA (lane 8), whereas TmFtsAΔ8 does not bind to liposomes (lane 6), emphasizing the role of the C-terminal amphipathic helix, as had previously been deduced for the E. coli proteins in vivo (Pichoff and Lutkenhaus, 2005).
Crystal structure of the FtsA binding motif of TmFtsZ bound to TmFtsA at 1.8 Å resolution
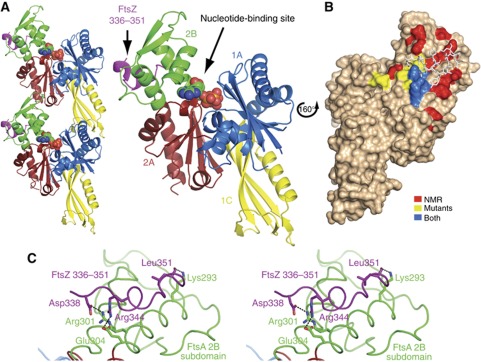
Crystallization trials using the full-length proteins failed, probably due to the flexible linker connecting FtsZ’s core and C-terminal subdomains, so we replaced FtsZ with the peptide corresponding to the last 16 residues of the protein. The complex crystallized with two TmFtsA molecules per asymmetric unit, each with the TmFtsZ peptide bound on the surface of subdomain 2B between helices H6 and H8 of TmFtsA (Table I, Figure 2A, left). The FtsZ peptide is far away from the nucleotide-binding site of FtsA (Figure 2A, right), which contains an endogenous ATP molecule at 0.65 occupancy. We identified three salt bridges between the peptide and residues belonging to the H8 helix of the 2B domain, TmFtsA(Arg301) to TmFtsZ(Asp338), TmFtsA(Glu304) to TmFtsZ(Arg344) and one involving TmFtsA(Lys293) and the carboxyl group of the very last Leu351 of the TmFtsZ peptide. The peptide is in predominantly helical conformation (Figure 2C), but is not just a single, straight helix. Binding of the FtsZ peptide to FtsA is in the micromolar range with a K d of 45–58 μM, as determined by two independent isothermal titration calorimetry experiments (ITC, Supplementary Figure 1).
Table 1. Data collection and refinement statistics.
| TmFtsA, UNIPROT Q9WZU0 + TmFtsA peptide 336-EGDIPAIYRYGLEGLL-351 +ATP | TmFtsA, UNIPROT Q9WZU0+ATPγS | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 56.72, 75.72, 100.27 | 48.00, 64.59, 73.00 |
| β (deg) | 93.64 | 102.93 |
| Resolution (Å) | 1.80 (1.90–1.80) | 1.80 (1.90–1.80) |
| R merge | 0.072 (0.578) | 0.073 (0.329) |
| I/σI | 10 (2.4) | 10.6 (3.6) |
| Completeness (%) | 99.8 (100) | 99.6 (99.9) |
| Redundancy | 3.6 (3.7) | 3.5 (3.6) |
| Refinement | ||
| Resolution (Å) | 1.8 | 1.8 |
| No. of reflections | 74 365 | 38 267 |
| R work/R free | 0.21/0.24 | 0.19/0.23 |
| Model | ||
| Protein | FtsA: A 7-27, 32-319, 328-390 B 7-27, 29-74, 77-322, 327-390FtsZ: C 338-351 D 337-351 | FtsA: 7-392 |
| ATPγS | — | 1 |
| ATP | 2 | — |
| Mg2+ | — | 1 |
| Waters | 459 | 201 |
| B-factors (Å2) | ||
| Protein | 34.70 | 32.65 |
| ATPγS | — | 31.62 |
| ATP | 35.04 | — |
| Mg2+ | — | 52.30 |
| Water | 41.46 | 43.26 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.024 |
| Bond angles (deg) | 1.078 | 2.179 |
| Ramachandran (%) | 91.2 | 91.4 |
| PDB ID | 4A2A | 4A2B |
Figure 2.
Crystal structure and solution NMR studies show that FtsZ binds to FtsA’s surface between helices H6 and H8 within subdomain 2B. (A) Crystallographic asymmetric unit containing two FtsA molecules, each with the C-terminal 16-residue FtsZ peptide bound (left). FtsA has been colour-coded according to the conserved actin family of proteins subdomain architecture. The peptide (purple) is bound on the surface of subdomain 2B (green). ATP is shown as spheres. (B) Solution NMR studies identifying amide resonances perturbed upon addition of the FtsZ peptide (red). These results support the crystal structure as well as previous in vivo mutations by Pichoff-Lutkenhaus (2007) (yellow). Residues highlighted by both in vivo and NMR studies are in blue. (C) A stereo representation of the FtsA–FtsZ interacting site. The peptide is depicted in purple, and 2B subdomain in green. Three salt bridges have been identified: FtsA(Arg301)–FtsZ(Asp338), FtsA(Glu304)–FtsZ(Arg344) and FtsA(Lys293)–FtsZ(Leu351) (C-terminal carboxyl group).
Identification of the TmFtsZ binding site on TmFtsA by solution NMR
Although the peptide fitted very well into the electron density map (Supplementary Figure 2), we then aimed at confirming the binding site by solution NMR. Triple labelled 15N13C2H TmFtsA (47 kDa) yielded high-quality TROSY spectra at 50 °C (Supplementary Figure 3). Comparison of the spectra recorded in the absence and presence of the same 16 residue FtsZ peptide revealed chemical shift perturbations (c.s.ps) as well as resonances that were exchange broadened beyond detection, as a result of FtsZ peptide binding (Supplementary Figure 4A). On the basis of a standard set of TROSY triple resonance experiments, over 80% of backbone resonances of TmFtsA’s primary sequence were assigned (see Materials and methods and Supplementary Figure 4B). This enabled us to identify all perturbed resonances (Supplementary Figure 4C) and map these onto the crystal structure (Figure 2B). Satisfyingly, both solution NMR and the X-ray structure agree on the binding site of the FtsZ C-terminal tail on the surface of subdomain 2B of FtsA, and both agree well with prior knowledge from mutants (Pichoff and Lutkenhaus, 2007).
FtsA forms actin-like protofilaments
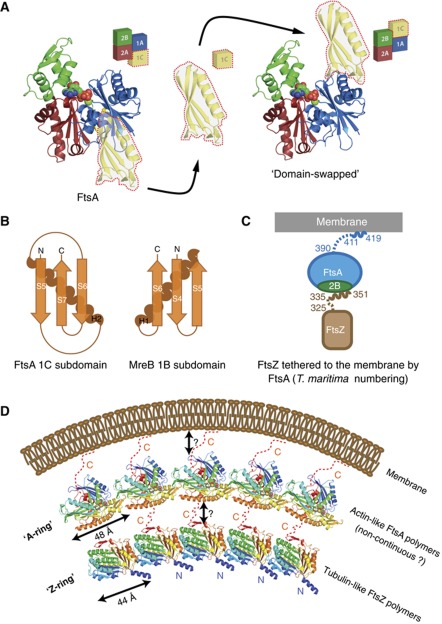
On the basis of its fold, FtsA was originally classified as a member of the actin/HSP70 family of proteins, alongside actin and prokaryotic actin-like MreB (Bork et al, 1992), although a large deviation from the canonical fold was reported, replacing subdomain 1B with 1C on the other side of the protein (van den Ent and ). This raised the important question of whether FtsA would be able to polymerize into filaments. The canonical actin fold is shown in Figure 3A using MreB (van den Ent et al, 2001): it consists of subdomains 1A, 1B, 2A and 2B (Figure 3A and B), and in the protofilament, the conserved subfilament that all polymerizing members of the actin family share, polymerization utilizes a contact between subdomains 1B and 2B on one subunit, and 1A and 2A on the other. FtsA has a distinct subdomain architecture: it contains subdomains 1A, 2A and 2B, but subdomain 1B is replaced with 1C, located on the other side of the protein, when interpreting it in terms of the protofilament architecture (Figures 2A and 3).
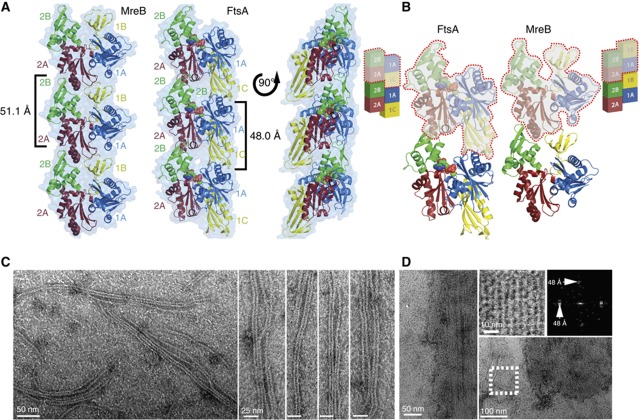
Figure 3.
Crystallographic and EM analysis reveal that FtsA, in spite of its unusual subdomain architecture, is able to form actin-like protofilaments. (A) FtsA, in the presence of ATPγS, crystallized as a continuous polymer (crystallographic unit cell edge a is aligned with the longitudinal axis of the protofilament). The packing remarkably resembles that of MreB, which has the canonical actin-like fold (subdomains 1A, 1B, 2A, 2B) and forms actin-like, straight protofilaments. The longitudinal spacings are 51.1 and 48.0 Å for MreB and FtsA, respectively. (B) A more detailed view of FtsA and MreB dimers. Monomeric subunits are rimmed in red. FtsA and MreB adopt a similar subdomain architecture; however, the 1B subdomain of MreB (yellow) is missing in FtsA and instead subdomain 1C in FtsA is located on the other side of the molecule. It therefore appears that FtsA is a subdomain variation of the actin fold that still enables the formation of canonical protofilaments. (C) FtsA polymerizes on a lipid monolayer forming long filaments, which often form doublets. (D) Occasionally, FtsA forms 2D sheets on the lipid monolayer. Fourier transformation of a sheet (white dotted square) reveals the same longitudinal spacing of about 48 Å, which matches the spacing present in the crystal structure.
Inspection of our new FtsA/FtsZ crystal structure (Figure 2A, left) reveals that the TmFtsA dimer resembles an actin-like protofilament. Furthermore, when co-crystallized with ATPγS, TmFtsA produced another crystal form containing continuous protofilaments that closely resemble the previous dimer and are again actin-like (Table I, Figure 3A and Supplementary Figure 5). No FtsZ peptide electron density could be found in this case. In the ATPγS crystals, the longitudinal axis of the protofilament is aligned with the crystallographic a axis, providing the repeat of 48.0 Å (Table I). The protofilament structure clearly resembles that of MreB (longitudinal spacing 51.1 Å; van den Ent et al, 2001) and all other polymerizing actin-like proteins (Figure 3A).
This leads to the question of how FtsA forms the same protofilament when it has a different subdomain architecture. FtsA and MreB comprise the 1A, 2A and 2B subdomains, which are similarly arranged in both proteins. However, the 1B subdomain of MreB, which in the polymer structure makes a contact with the 1A subdomain of the succeeding subunit (van den Ent et al, 2001), in FtsA is replaced with the 1C subdomain occurring on the opposite side of the molecule, which in the polymer structure contacts the 1A subdomain of the preceding subunit (Figure 3B). In other words: the subdomain can be loosely described as having been swapped from one side of the FtsA molecule to the other, but its relative position within the protofilament has been conserved when compared with canonical actin-like protofilaments (Figure 6A).
This alteration has implications for the polymer appearance. First, the ends of the polymers will look very different, with the 1C subdomain of FtsA explicitly projecting at one terminus and the 2B subdomain at the other. Second, as FtsA’s 1C subdomain contains nearly twice the number of residues compared with MreB’s 1B subdomain and has a dissimilar topology (Figure 6B), there are important differences between the two polymer structures. MreB protofilaments are flat when observed along the longitudinal axis, whereas FtsA’s 1C subdomain protrudes out, placing monomeric FtsA subunits at an angle with respect to the longitudinal axis and making the polymer structure thicker (Figure 3A right, Supplementary Figure 5). Also, in the structure of the MreB filament, the tip of the subdomain 2A consisting of helices H8 and H9 is inserted in the cleft between subdomains 1B and 2B. A similar arrangement in the FtsA protofilament is obviously not possible, so the 2A subdomain stays in contact only with the 2B subdomain and is positioned somewhat behind it when looking from the front of the molecule (Figure 3A). However, given that FtsA and MreB (also actin and ParM) are only ∼15% identical in sequence, their protofilaments are remarkably analogous. We conclude that it appears that FtsA is a variation of the actin fold that still enables the formation of canonical actin-like protofilaments.
TmFtsA protofilaments formed on lipid monolayers are identical to the filaments observed in crystals
To demonstrate that FtsA filaments exist outside crystals we used purified TmFtsA for in vitro electron microscopy. Conventional, negatively stained electron microscopy on carbon-coated grids failed, as the proteins (TmFtsA and TmFtsAΔ8) did not exhibit any polymeric structures but remained apparently monomeric. We then took advantage of FtsA’s affinity for membranes through polymerization on a lipid monolayer (Figure 3C and D). By using this technique we revealed several hundred nanometre long filaments, often present as doublets or higher-order structures. Interestingly, no exogenous nucleotide was required to obtain these, but it is important to note that the protein contains some bound ATP carried over from the purification, as is evident from the ATP in the crystal structure, which was not added. Occasionally, two-dimensional sheets were observed (Figure 3D). Fourier transformations of sheets (Figure 3D, top right) revealed a longitudinal spacing of around 48 Å, corresponding to the spacing found in the crystal. This implies that the TmFtsA polymers visualized by electron microscopy and seen in atomic detail in the crystal structures are very similar or identical.
FtsA proteins from T. maritima, E. coli and B. subtilis all form filaments in E. coli cells when overexpressed
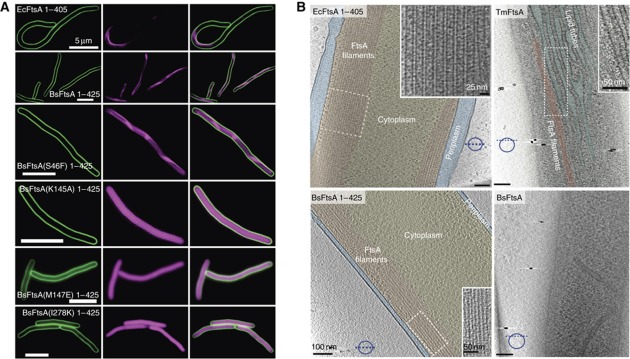
Our previous experiments showed that TmFtsA polymerizes to form actin-like protofilaments in vitro on lipid monolayers. We investigated a broader range of species by including E. coli FtsA (EcFtsA) and B. subtilis FtsA (BsFtsA) proteins in our in vivo analysis. First, we overexpressed mCherry-tagged FtsAs deprived of the C-terminal amphipathic helices in E. coli and visualized them with conventional EPI as well as high-resolution SIM (Structured Illumination Microscopy) fluorescence microscopy. The amphipathic helices were removed for fluorescence microscopy in order to see filaments in the cytoplasm rather than membrane-associated polymers, which would be difficult to observe. Cells displayed strong fluorescence and contained long filaments: mCherry–EcFtsA 1–405, when overexpressed in E. coli, forms distinct, elongated polymer structures that tend to bend the cells into a tight curvature (Figure 4A, top panels, Supplementary Figure 6). Note that an earlier study found cytoplasmic aggregated structures formed by a similar C-terminal EcFtsA truncation (Gayda et al, 1992). mCherry–BsFtsA 1–425 produced large polymers as well (Figure 4A, second top panels, Supplementary Figure 6); however, in this case the cells remained straight. Overexpression of mCherry–TmFtsA 1–411 formed inclusion bodies (data not shown) so it was not included in subsequent analysis by electron cryotomography.
Figure 4.
FtsA polymers can be visualized in living cells by fluorescence microscopy and electron cryotomography. (A) N-terminal mCherry fusions to EcFtsA and BsFtsA (C-terminal amphipathic helix removed) as well as to four BsFtsA mutants have been overexpressed in E. coli using a T7 expression system. Membranes were stained with FM1-43. The three top panels show distinct, polymeric structures and the three bottom panels show that some mutations introduced within the polymerization interface prevent polymerization, leading to diffuse localization. The locations of these mutations are shown in Supplementary Figure 7A. All images using SIM. For conventional EPI images and more examples please consult Supplementary Figure 6. (B) Untagged E. coli, B. subtilis and TmFtsA proteins were overexpressed in E. coli cells and imaged by electron cryotomography. EcFtsA without the amphipathic helix (top left) forms long, straight bundles in the middle of the cell (navy blue dotted line represents cross-section’s orientation). Full-length TmFtsA (top right) causes membrane distortion and forming of lipid tubes (turquoise) that are coated with protein polymers (red) in the cell membrane’s proximity (see Supplementary movie 1). The bottom panels show either full-length BsFtsA (right) or BsFtsA truncated for the amphipathic helix (left). As expected, they trigger membrane distortion and straight filaments, respectively. White dotted areas are shown enlarged in the insets.
To investigate the subcellular organization and appearance of FtsA filaments in more detail, we imaged EcFtsA 1–405, BsFtsA 1–425, BsFtsA and TmFtsA in intact cells by electron cryotomography. Both BsFtsA and TmFtsA polymers, containing the C-terminal amphipathic helices, bind to the inner surface of the cytoplasmic membrane, thereby distorting it (Figure 4B, right panels). This causes the formation of elongated lipid tubes coated with protofilaments roughly parallel to the long axis of these tubes (Supplementary movie 1). It is highly likely that the protofilaments are made of FtsA. In the absence of the amphipathic helix, EcFtsA 1-405 and BsFtsA 1-425 formed long, straight filaments visible in the cytoplasm that do not invaginate the membrane (Figure 4B, left panels). EcFtsA 1–405 filaments were exceptionally prominent and assembled into large bundles aligned along the direction of the long axis of the cell (Figure 4B, top left panel). Fourier transform of these gave a longitudinal repeat of 57 Å, which does not match the TmFtsA spacing of 48 Å that we determined above. Because of the low resolution of the tomograms it is not clear from the pictures that this repeat actually measures along the protofilament axis, as the E. coli polymers appear to be helical (Figure 4B, top left panel inset). In cells overexpressing BsFtsA 1–425, similar fibres and localization patterns to those cells overexpressing EcFtsA 1–405 were observed (Figure 4B, bottom left panel).
BsFtsA filaments are required for efficient cell division
Next we investigated whether FtsA polymerization is needed for its function in cell division. For this, we rationally designed potential polymerization-deficient mutants (Supplementary Figure 7A) based on manual inspection of the protofilament crystal structures described above (Table I, Figure 3A). The mutated proteins were then analysed by fluorescence microscopy after expressing them as mCherry fusion proteins in E. coli (Figure 4A, Supplementary Figure 6). The in vivo fluorescence assay seems to be the most reliable test for FtsA polymerization, as it shows that the protein is made, does not form inclusion bodies and discrimination between fibres and uniform fluorescence is unambiguous, certainly using SIM. Western blot analysis with anti-B. subtilis FtsA antibodies was used to confirm that comparable amounts of mutants are made (Supplementary Figure 7B). BsFtsA(S46F) 1–425 showed the same pattern as BsFtsA 1–425 (Figure 4A, third top panels, Supplementary Figure 6) demonstrating that it still polymerizes. In contrast, three BsFtsA 1–425 mutants K145A, M147E and I278K displayed evenly distributed fluorescence in cells and did not feature any distinct polymer-like structures (Figure 4A, three bottom panels). We conclude that BsFtsA(K145A), M147E and I278K are unable to form polymers, whereas BsFtsA(S46F) forms polymers similar or identical to the wild type. Moreover, BsFtsA(I47D), P144K and G279R formed inclusion bodies (data not shown). Also, a recent report from the Lutkenhaus’s group (Pichoff et al, 2012, Supplementary Figure 8) strengthens our argument by producing mutants deficient in FtsA self-interaction that map nicely to the FtsA interaction surfaces in the protofilament structure described here.
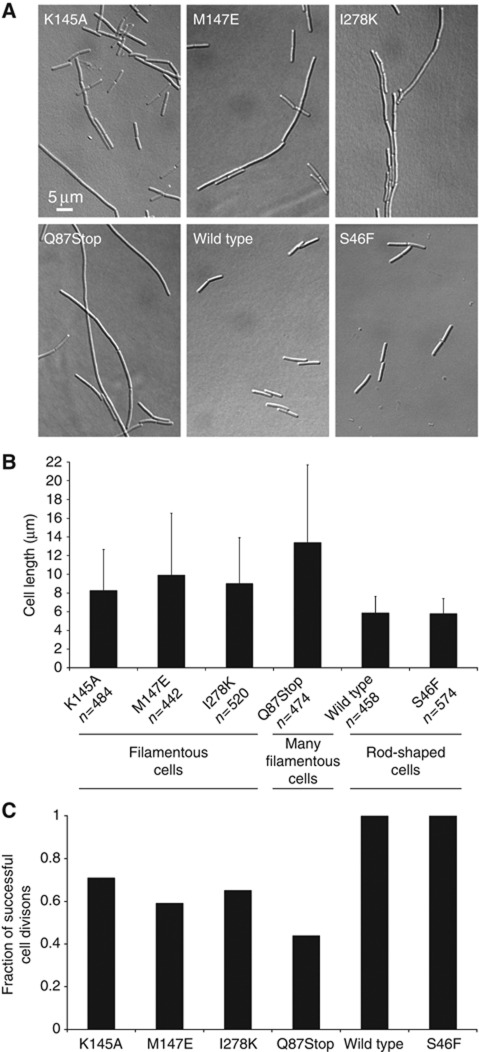
Having identified BsFtsA polymerization-deficient mutants, we wanted to study whether or not FtsA polymers are of biological significance in vivo. A complementation experiment was performed in a BsFtsA temperature-sensitive (ts) background at 42°C (see Materials and methods, Beall and Lutkenhaus, 1992 and Karmazyn-Campelli et al, 1992). Under elevated temperature, the ts-FtsA protein is unable to support cell division and the cells acquire a filamentous appearance that is straightforward to distinguish and quantify. We then complemented the endogenous ts-FtsA with a series of mutant versions of FtsA, inserted into the aprE locus, under the control of the IPTG-inducible Pspac promoter. We tested six different variants: wild-type FtsA, FtsA(Q87Stop; truncated protein product), FtsA(K145A), FtsA(M147E), FtsA(I278K) and FtsA(S46F). We used this experimental setup as it has been shown (Beall and Lutkenhaus, 1992) that wild-type ftsA expressed from the Pspac promoter complemented the filamentation phenotype triggered by the ts-FtsA protein, and we were wondering if our polymerization-deficient mutants would behave similarly. As is shown in Figure 5, the wild-type and S46F cells were of normal length, whereas the Q87Stop and the three polymerization-deficient mutants K145A, M147E and I278K grew as elongated, filamentous cells (Figure 5A). These were long cells of varying length and it stays in accordance with previous studies (Jensen et al, 2005) on an ftsA-null B. subtilis strain where the average cell length was 22.6±11.9 μm. Quantification also reflects that fact (Figure 5B) with significant standard deviations associated with the FtsA polymerization-deficient mutants and the Q87Stop mutant. The average cell length of the FtsA mutants is larger than that of the wild type; however, the phenotype is not as severe as in case of Q87Stop. Also, assuming that the length per nucleoid is constant and the wild-type FtsA cells undergo correct cell divisions, we calculated a fraction of successful cell divisions (Figure 5C). In case of Q87Stop about 0.43 cytokineses are fine, and for the FtsA polymerization-deficient mutants it varies between 0.59 (M147E) and 0.71 (K145A). This suggests that cell division in the absence of FtsA filaments (monomeric FtsA is still present) is much less efficient than usual. This is not totally unexpected and it seems as if the Z-ring binding to the membrane by monomeric FtsA is not as effective as by FtsA protofilaments. It implies a role for FtsA polymerization in bacterial cell division.
Figure 5.
Replacing thermo-sensitive FtsA with either polymerization-deficient mutants or a non-functional version has dramatic effects on cell division in B. subtilis. (A) DIC images of genetically engineered B. subtilis strains showing that non-polymerizing FtsAs (K145A, M147E, I278K) do not fully complement the thermo-sensitive spoIIN279(ts) allele. As a control, non-functional FtsA (Q87Stop) shows a very severe cell division defect. S46F, which was found to still polymerize (Figure 4A), shows no effect on cell division. (B) A chart showing the average cell lengths of strains bearing tested FtsA variants. The wild type was 5.89±1.74 μm and S46F exhibits similar values. Cells of the Q87Stop variant were 13.40±8.30 μm, and the three polymerization-deficient mutants were as follows: K145A 8.29±4.36 μm, M147E 9.93±6.6 μm and I278K 9.04±4.86 μm. (C) Fractions of successful cell divisions were calculated referring to the wild-type FtsA variant.
Discussion
We have reconstituted in vitro the T. maritima membrane-bound complex of FtsA and FtsZ, and regions indispensable for complex formation have been identified. This work confirms and refines previous in vivo findings and leads to the FtsZ-tethering model depicted in Figure 6C.
Figure 6.
FtsA is a subdomain variation of the actin fold that still enables the formation of similar protofilaments. (A) The 1C subdomain is present on the other side of a monomeric subunit in respect to 1B subdomain of MreB. (see also Figure 3A and B). (B) Both the 1C subdomain of FtsA and the 1B subdomain of MreB consist of an α-alpha helix and a three-stranded β-sheet, but the topology is different. For 1C it is S5-H2-S6-S7 and for 1B it is S4-S5-H1-S6. (C) Architecture of the FtsA/FtsZ membrane-bound complex, as analysed here for T. maritima. (D) The Z-ring made of polymerized tubulin-like FtsZ may be attached to the membrane by an ‘A-ring’, in turn made of polymerized actin-like FtsA or short stretches of FtsA polymers, based on the relative number of FtsZ and FtsA molecules in cells.
Importantly, the structural work on the FtsA/FtsZ complex led us to the discovery that FtsA is able to assemble into filaments that resemble actin-like protofilaments and we show that these filaments are formed by FtsAs from three different model organisms. The protofilaments are formed despite FtsA having unusual subdomain architecture. Finally, we demonstrate that polymerization-deficient FtsA mutants show a modest phenotype in cell division in a thermo-sensitive-FtsA complementation assay in B. subtilis.
It has previously been shown that the last 15–20 residues of FtsZ are highly conserved and form a region for interactions with other proteins, such as ZipA, MinC and FtsA (Shen and Lutkenhaus, 2009), and an E. coli FtsA R286W mutant protein could be co-pelleted with FtsZ (Beuria et al, 2009). We have now shown that the interaction between unmodified and wild-type FtsZ and FtsA can be observed in vitro and we could identify and characterize the interactions in detail. We show the tail of FtsZ binding to subdomain 2B of FtsA, as was implied by earlier mutant work (Pichoff and Lutkenhaus, 2007). We could not find any effect of FtsZ tail binding on FtsA’s nucleotide binding or hydrolysis activity, nor on FtsA polymerization, and it currently seems to be a purely architectural interaction leading to the membrane tethering of the Z-ring. Our model in Figure 6C does suggest that the Z-ring of bacterial cell division is some distance from the membrane, and this has been reported earlier by electron tomography data (Li et al, 2007).
The structure of E. coli FtsZ peptide in complex with ZipA has previously been solved (Mosyak et al, 2000). Comparing the two structures unveils some similarities, although they are largely confined to a small α-helical region and the termini point in different directions (Supplementary Figure 9). Certainly, the binding pockets on ZipA and FtsA for the same FtsZ tail seem to be quite diverse (as can be judged from looking at structures from different organisms). Therefore, it seems likely that the FtsZ tail adopts different conformations, at least partly, to bind to ZipA and FtsA, not simultaneously (Supplementary Figure 9). This is conceivable in the light of overlapping functions of ZipA and FtsA (Pichoff and Lutkenhaus, 2002).
Our work shows that FtsA is more actin-like than was previously assumed. FtsA forms protofilaments on lipid monolayers and in cells when overexpressed. FtsA forms actin-like protofilaments, although they diverge significantly from the ones formed by actin, MreB and ParM. These differences are most likely caused by the aberration in structure, fold and size of subdomain 1C that replaces 1B on the other side of the molecule (van den Ent and ). The topology of FtsA’s 1C subdomain is S5-H2-S6-S7, whereas MreB’s 1B subdomain is S4-S5-H1-S6 (Figure 6B). The origin of the 1C subdomain during evolution is unknown, although it seems unlikely that it is related to 1B. This suggests that 1C is not simply a domain swap (moving the subdomain into another loop region of the actin fold) but a more complicated domain replacement with something different, in a various location that still enables similar filament formation. A DALI search for the most similar fold to subdomain 1C returns RNA polymerase II chain G (PDB id: 3H0G) with a Z-score of 6.3 and 10% sequence identity over 61 aligned residues (Supplementary Figure 10). This is a significant hit, but to us does not provide any clues.
Tendency of FtsA to dimerize, oligomerize or polymerize has been studied using genetics methods (Yim et al, 2000; Rico et al, 2004; Shiomi and Margolin, 2007, 2008); however, we now provide structural data on how the filament formation is possible in light of the deviant FtsA domain architecture. Previous reports are based mainly on either yeast or bacterial two-hybrid assays that do not distinguish between dimerization and polymerization. Importantly, two recent reports (Krupka et al, 2012; Pichoff et al, 2012) support our model of the FtsA protofilament. Krupka et al (2012) show that the 1C subdomain is present within the polymerization interface. Pichoff et al (2012) list mutants, deficient in FtsA self-interaction, that map nicely to the FtsA–FtsA interfaces generated by our protofilament structure. Interestingly, the FtsA mutants impaired in self-interaction, identified in the study by Pichoff et al (2012), can bypass the requirement for ZipA, which is another Z-ring membrane anchor in E. coli and still support cell division. This is most likely possible because recruitment of downstream proteins by FtsA is unaffected and the protein is still functional in the absence of ZipA. In contrast, FtsA in B. subtilis is presumably the only membrane-tethering agent for FtsZ and as shown in this work, polymerization-deficient mutants trigger elongated cells.
FtsA and FtsZ are the only nucleotide-binding components of the inner divisome machinery, with the potential to produce energy by nucleotide turnover. FtsZ’s GTP binding and hydrolysis is now relatively well-understood (Oliva et al, 2004) and nucleotide is efficiently hydrolysed only upon filament formation. In contrast, less is known about FtsA’s ATPase activity. Clearly, TmFtsA can bind to ATP and ATPγS as demonstrated by the crystal structures (presented here and van den Ent and ), but it still has not been shown conclusively that the protein, from any organism, exhibits ATPase activity (with one exception, which might represent contamination: Feucht et al, 2001). In the course of this work, we performed ATPase assays with purified TmFtsA in the presence of miscellaneous ligands, including divalent metal cations, the FtsZ C-terminal peptide and liposomes, and they did not yield any detectable activity of the protein (data not shown). Also, in our in vitro experiment on lipid monolayers, polymers formed in the absence of exogenous nucleotide, although some ATP from purification is bound to the protein. Similarly, previous work on S. pneumoniae FtsA showed ATP-dependent polymerization (Lara et al, 2005); however, no membrane was present. It is worth noting that our assay was performed with Thermotoga protein and this might be a special case because of the extreme temperature adaptation of this protein. Full length FtsA proteins containing the C-terminal amphipathic helix are very difficult to be produced in a pure and well-behaved form, particularly when they originate from mesophilic organisms. This situation is somewhat similar to MreB, where it was very recently shown that an N-terminal amphipathic helix hinders biochemical experiments on most proteins, except Thermotoga (Salje et al, 2011). However, removal of the amphipathic helix will alter the protein’s properties and polymerization may then not be observed in vitro. In vivo, the system may be balanced so that membrane interaction and charge compensation are required for efficient polymerization, and it seems that ATP hydrolysis is not required for polymerization on the lipid surface (see Figure 3C and D). However, in this case membrane binding might be the energy-reducing factor acting in lieu of ATPase activity, which might still be essential in solution or cells. To address this question more work using putative ATPase mutants in vivo will be needed.
Why might FtsA polymerization be required in cells? It is thought that cell division in most bacteria proceeds through constriction of the Z-ring. The ring might either produce mechanical force or direct the position of cell-wall synthesizing machinery in the periplasm, where force may then be generated. It has been shown in C. crescentus by tomography that the Z-ring may consist of short (∼100 nm) filaments situated randomly near the division sites rather than one, long filament encircling the cell (Li et al, 2007) and this has been recently supported by PALM microscopy in E. coli (Fu et al, 2010). Also, it has been estimated in B. subtilis that there are around 1000 and 5000 molecules of FtsA and FtsZ, respectively, per cell (Feucht et al, 2001) and it is unclear whether this is enough to cover the entire cell circumference.
Our work shows that the Z-ring might be tethered to the membrane by an ‘A-ring’ made of FtsA polymer(s) (Figure 6D). Because there is most likely less FtsA in a cell than FtsZ, the A-ring is probably non-continuous. This would lead to a putative constriction model, whereby FtsZ and FtsA polymers are parallel to the inner surface of the cytoplasmic membrane, and the mismatch of spacings between the two proteins (FtsA ∼48 Å and FtsZ ∼44 Å) could trigger FtsZ/FtsA filament curvature and in turn membrane constriction (Li et al, 2007). Alternatively, FtsZ filaments could be attached to the membrane by short stretches of FtsA polymers only every several FtsZ subunits. Depolymerization of such rarely membrane-attached FtsZ protofilaments would also lead to constriction. As a third alternative, FtsA could polymerize perpendicular to the FtsZ filament axis, restricting the number of FtsZ filaments in a bundle. To distinguish between these models, more electron cryotomography will be required to determine how FtsA and FtsZ protofilaments are mutually oriented in cells, how they are arranged with respect to the membrane and how the flexible linkers in both proteins are organized.
Finally, how did the 1C/1B subdomain variation happen? The FtsA 1B/1C subdomains’ fold topology is reversed and they vary in size. Analysis of actin-like protein sequences and structures, including actin, MreB (van den Ent et al, 2001) and ParM (van den Ent et al, 2002), reveals that the 1B subdomain is the most variable and FtsA is the most evolutionary distant from other actin-like proteins. All these proteins play key roles in distinct cellular processes relying on their actin-like filamentous nature. Opposite orientations of the 1C and 1B subdomains could be explained by the fact that the former is preceded by S1-S2-S3-H1-S4 of the FtsA 1A subdomain, whereas the latter is preceded by only S1-S2-S3 and is followed by H2-S7 of the MreB 1A subdomain, thus it seems they are ‘inserted’ in slightly different regions of 1A’s primary sequence. It would be compelling to find out whether the last common ancestor of actin and FtsA was more ‘actin-like’ or ‘FtsA-like’ or it rather existed as a three-subdomain protein (1A, 2A, 2B) and later acquired either the 1C or 1B subdomain.
Materials and methods
Plasmids and strains
Plasmids and strains used in this work are listed in Supplementary Tables 1 and 2, respectively.
Protein purification
TmFtsA (from plasmid pSZ8) and FtsZ (pSZ7) proteins as well as the ParM-FtsZ (pSZ19) hybrid protein for in vitro assays were purified using the IMPACT system (NEB). The proteins were expressed at 37 °C in C41(DE3) cells for 4 h. Cells were lysed in buffer A (50 mM Tris, 10 mM EDTA, 2 mM 1,10-phenantroline, 1 mM PMSF, pH 8.5) by passing through a Constant Systems cell disruptor at 25 kpsi. The soluble fraction was loaded on a chitin column (NEB), which was then thoroughly washed with buffer B (A+50 mM NaCl) and finally with 2 column volumes of buffer C (B+50 mM DTT) and left overnight at 4 °C to cleave off the intein tag. The untagged proteins were eluted with buffer B, and pooled fractions were passed over an ion-exchange Q column (HiTrap, GE Healthcare). The column was washed with buffer D (20 mM Tris, 10 mM EDTA, 2 mM 1,10-phenantroline, 1 mM PMSF, pH 7.5) and the proteins were eluted by applying a gradient of buffer E (D+1 M NaCl). Peak fractions were pooled, concentrated and subjected to size-exclusion chromatography on Sephacryl S200 (GE Healthcare) in buffer F (50 mM Tris, 1 mM NaN3, 1 mM EDTA, pH 7.5). Peak fractions were concentrated to 10 mg/ml and stored at −80 °C. This protocol yielded full-length TmFtsA, TmFtsZ and ParM-Z. For the proteins lacking the C-terminal amino acids the same protocol was performed, but 1,10-phenantroline and PMSF were omitted, which resulted in efficient and specific cleavage of the last eight amino acids by an unknown E. coli protease. The protein’s identity and correct molecular masses were confirmed by N-terminal sequencing and electrospray mass spectroscopy displaying 38 307 Da for TmFtsZ, 37 404 Da for TmFtsZΔ8, 47 015 Da for TmFtsA, 45 968 Da for TmFtsAΔ8, 42 011 for ParM-Z and 41 109 form ParM-ZΔ8.
Co-pelleting assay
Purified proteins were mixed at a final concentration of 15 μM each. The samples were pre-spun at 57 k r.p.m. in a Beckman TLA100 rotor for 30 min, supernatants transferred to new tubes and GTP (in case of FtsZ) or ATP (in case of ParM-Z hybrid) to a final concentration of 2.5 mM was added. Next, the tubes were centrifuged at 57 k at 20 °C for 30 min. The supernatants were removed for analysis and the pellets washed with buffer containing the same components as the reaction buffer aside from proteins. Pellets were solubilized with SDS gel loading buffer (same volume as for the supernatants) and samples were analysed by SDS–PAGE.
Co-sedimentation assay
Vesicles were prepared using E. coli total lipid extract (Avanti Polar Lipids) in buffer F using sonication and extrusion, using a pore size of 1 μm. Vesicles were mixed with pre-spun proteins at a final concentration of 15 μM and centrifuged at 40 k r.p.m. at 20 °C for 15 min in a Beckman TLA100 rotor. The supernatants were removed for analysis, the pellets washed with buffer F and solubilized with SDS gel loading buffer (same volume as for the supernatants) and samples were analysed by SDS–PAGE.
Crystallization and structure determination
Full-length TmFtsA was co-crystallized with either the C-terminal TmFtsZ peptide (336 H2N-EGDIPAIYRYGLEGLL-COOH 351) or the peptide and ATPγS. The protein was at 10 mg/ml, the peptide at 10× molar excess and the nucleotide at 2 mM. FtsA and the peptide yielded crystals in the following conditions: 0.1 M bisTris, pH 5.5, 12% (w/v) PEG 10 k, 0.15 M ammonium acetate. Crystals were moved to the same conditions supplemented with 18% (v/v) PEG 200 and flash-frozen in liquid nitrogen. Crystals of FtsA in complex with ATPγS grew from 0.1 M Tris, pH 6.7, and 42% ethanol. Crystals were transferred into the same conditions supplemented with 25% (v/v) PEG 200 before flash freezing. Data were collected at 100 K at ESRF in Grenoble, France (beamlines ID29 and ID23.1). Both crystal forms diffracted to 1.8 Å. The data were processed with MOSFLM and SCALA (CCP4). Phases were calculated by molecular replacement with TmFtsA Protein Data Bank (PDB) entry 1E4F as a model using PHASER (McCoy et al, 2007). The structures were refined with REFMAC (Murshudov et al, 1997) or PHENIX (Adams et al, 2010). Residues (91.2%) for FtsA+peptide+ATP and residues (91.4%) for FtsA+ATPγS were in the most favored regions of the Ramachandran plot as determined by PROCHECK (CCP4 1994). Crystallographic data are summarized in Table I. Structures were deposited in the PDB with codes 4A2A and 4A2B.
Isothermal titration calorimetry
The experiment was performed at 35 °C on an ITC200 instrument. The sample cell was filled with 55 μM TmFtsA in buffer F and titrated with 500 μM FtsZ peptide in buffer F while stirring at 1000 r.p.m. Twenty aliquots of the peptide (2 μl) were injected at 120 s intervals. Data analysis was performed using the Origin-ITC analysis package in ‘one set of sites’ mode. The experiment was repeated using 70 μM TmFtsA and 730 μM FtsZ peptide.
Solution NMR
To assign the backbone resonances of the 47 kDa FtsA (pSZ1) protein from T. maritima, we overexpressed 15N13C2H triple-labelled FtsA protein growing bacteria on MOPS minimal media using 15NH4Cl as the sole nitrogen source, 13C2H glucose as the sole carbon source and 2H2O as the solvent. The protein was purified using standard nickel-NTA chromatography followed by size-exclusion chromatography on Sephacryl S200 in buffer F. 3D TROSY experiments (HNCO, HN(CA)CO, HNCA, HN(CO)CA, HN(CO)CACB and HNCACB) were recorded at 50 °C on a Bruker Avance II+ 700 MHz spectrometer at a sample concentration of 85 μM in buffer F. All data were processed in Topspin 2.1 (Bruker, Karlsruhe), analysed in SPARKY (Goddard-Kneller) and backbone connectivities were obtained with MARS (Jung and Zweckstetter, 2004), MAPPER (Guntert et al, 2000) or in-house scripts. To confirm and extend obtained assignments, 15N selectively labelled (15N Leu and 15N Ile) FtsA samples yielded amide resonances of Leu (Ile) residues, respectively. Weighted c.s.p. maps were based on TROSY HSQC spectra acquired in the absence or presence of the FtsZ C-terminal peptide (2.5 molar excess). Weighted c.s.ps larger than 0.04 p.p.m. or disappearing resonances were assumed to be involved in peptide binding and mapped onto the crystal structure. The assignment data were deposited in the Biological Magnetic Resonance Data Bank with accession code 18339.
Monolayer assay
2D lipid monolayers were prepared from E. coli total lipid extract (Avanti Polar Lipids). Teflon block wells were filled with 50 μl of buffer F, next, a drop of lipid solution in chloroform was applied on top and incubated for an hour to let the chloroform evaporate, leaving a monolayer of lipid on the surface of the buffer. EM carbon-coated grids (carbon side down) were placed on the top of each well and the protein applied into the side injection well, underneath the surface. After 1 h, the grids were stained with 2% uranyl acetate and visualized using a Philips electron microscope operating at 80 kV.
Electron cryotomography
C41(DE3) cells containing the plasmid for FtsA over-expression (pSZ6, pSZ41, pSZ47, pMZ114) were grown in M9 minimal medium supplemented with 0.2% glycerol at 30 °C. FtsA over-expression was induced by adding 0.2% agarose to the cultures at an OD600 of 0.2. After 3 h, the cells were collected, washed in PBS once, mixed with 10 nm protein-A-coated gold beads (Sigma-Aldrich, USA), and plunge-frozen onto Quantifoil R3.5/1 holey carbon grids using an FEI Vitrobot (FEI Company, USA). Cells were imaged using an FEI Polara Electron Microscope operating at 300 kV, equipped with a Gatan imaging filter set at the zero-loss peak with a slit-width of 20 eV. A Gatan Ultrascan 4000 CCD camera binned to 2 k × 2 k was used for imaging with SerialEM software (Mastronarde, 2005). Cells were imaged at a magnification of 41 k, corresponding to a pixel size of 5.8 Å at the specimen. The specimen was tilted from approximately −50° to 50° with a 1° increment during tilt series acquisition, and the defocus was set at 10 μm. The total dose for each tilt series is between 150 to 200 e/Å2. Tomographic reconstruction was calculated using the IMOD tomography reconstruction package (Kremer et al, 1996).
Fluorescence microscopy
E. coli C41(DE3) cells were transformed with vectors encoding N-terminal mCherry fusions to the E. coli (pSZ49) or BsFtsA (pSZ50a-e) proteins, deprived of the C-terminal amphipathic helices, and the expression was induced with 1 mM IPTG at OD600 0.2 for 1 h at 37 °C. Cells were stained with FM1-43 stain (Invitrogen), mounted on an agarose pad and visualized using a Nikon N-SIM microscope equipped with a Nikon APO TIRF ×100/1.49 lens in both EPI and 2D-SIM modes with the 488- and 543-nm solid state lasers. The final pixel size was 64 nm in the EPI mode and 32 nm in the SIM mode.
Strain construction and in vivo complementation experiment
To obtain genetically modified B. subtilis strains, first, we constructed integration vectors (pSZ61a-f) consisting of a spectinomycin-resistance cassette and an FtsA point mutant (K145A, M147E, I278K, Q87Stop, S46F) in between the aprE flanking sequences (using pAPNC213 (Morimoto et al, 2002)). These constructs were used to transform the B. subtilis S62 strain (BGSC no.: 1S62, possessing the spoIIN279(ts) ftsA mutation (Beall and Lutkenhaus, 1992; Karmazyn-Campelli et al, 1992)). Transformants were selected on spectinomycin (100 μg/ml) containing LB plates and the correct location and orientation of the insert was checked by PCR and sequencing. The strains were grown at 42 °C (to inactivate the endogenous thermo-sensitive FtsA protein) in LB medium in the presence of 1 mM IPTG (to induce the synthesis of the FtsA mutants). At OD600 0.4–0.5, cells were harvested, washed with PBS and visualized using a Zeiss LSM5 microscope.
Cell measurements and statistical analysis
About 500 cells for each FtsA variant were manually inspected and measured. Then average cell lengths and standard deviations over the given number of cells were calculated for each FtsA variant and plotted as bars and error bars, respectively, shown in Figure 5B. Fractions of successful cell divisions were obtained as a quotient of the number of observed cells for each variant by the number of possible cells. The number of possible cells was calculated (assuming that the length per nucleoid is constant and the wild-type FtsA cells undergo correct cell divisions) by dividing the sum of all cell lengths for each variant by the average wild-type FtsA cell length.
Supplementary Material
Acknowledgments
We thank Patricia Dominguez-Cuevas (Newcastle University, UK) for the pAPNC213 plasmid, and Richard Daniel and Henrik Strahl (Newcastle University, UK) for help with SIM fluorescence microscopy. Anti-FtsA antibodies were a gift from Jeff Errington’s lab (Newcastle University, UK). Roger Williams and Mark van Breugel (MRC-LMB, Cambridge, UK) are acknowledged for help with X-ray data collection. PS was supported by a Boehringer Ingelheim Fonds PhD Fellowship. Support by EU FP7 grant DIVINOCELL (FP7-223431) is acknowledged. We also thank beamline staff at ESRF ID29 and ID23.1 for excellent support. This work was supported by the Medical Research Council (grant number U105184326).
Author contributions: PS performed all experiments, except NMR analysis, which was done in collaboration with SMVF, and electron cryotomography was carried out by QW. Experimental design and analysis were performed by PS and JL, as was manuscript preparation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J (1992) Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol 174: 2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuria TK, Mullapudi S, Mileykovskaya E, Sadasivam M, Dowhan W, Margolin W (2009) Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem 284: 14079–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89: 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dajkovic A, Lutkenhaus J (2006) Z ring as executor of bacterial cell division. J Mol Microbiol Biotechnol 11: 140–151 [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J (2001) Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol 40: 115–125 [DOI] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J (2010) In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PloS One 5: e12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayda RC, Henk MC, Leong D (1992) C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol 174: 5362–5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG SPARKY 3: San Francisco, USA: University of California, [Google Scholar]
- Guntert P, Salzmann M, Braun D, Wüthrich K (2000) Sequence-specific NMR assignment of proteins by global fragment mapping with the program MAPPER. J Biomol NMR 18: 129–137 [DOI] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P (2001) Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem 276: 11980–11987 [DOI] [PubMed] [Google Scholar]
- Jensen SO, Thompson LS, Harry EJ (2005) Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-Ring assembly. J Bacteriol 187: 6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Zweckstetter M (2004) Mars—robust automatic backbone assignment of proteins. J Biomol NMR 30: 11–23 [DOI] [PubMed] [Google Scholar]
- Karmazyn-Campelli C, Fluss L, Leighton T, Stragier P (1992) The spoIIN279(ts) mutation affects the FtsA protein of Bacillus subtilis. Biochimie 74: 689–694 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Krupka M, Rivas G, Rico AI, Vicente M (2012) Key role of two FtsA protein terminal domains in its bidirectional polymerization. J Biol Chem 287: 7755–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, Vicente M, Mingorance J, Massidda O (2005) Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol 55: 699–711 [DOI] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ (2007) The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26: 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76: 539–562 [DOI] [PubMed] [Google Scholar]
- Löwe J, van den Ent F (2001) Conserved sequence motif at the C-terminus of the bacterial cell-division protein FtsA. Biochimie 83: 117–120 [DOI] [PubMed] [Google Scholar]
- Löwe J, van den Ent F, Amos LA (2004) Molecules of the bacterial cytoskeleton. Annu Rev Biophys Biomol Struct 33: 177–198 [DOI] [PubMed] [Google Scholar]
- Ma X, Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181: 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N (2002) Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148: 3539–3552 [DOI] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J 19: 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Löwe J (2004) Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2002) Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55: 1722–1734 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2007) Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol 64: 1129–1138 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J (2012) FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83: 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M (2004) Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol Microbiol 53: 1359–1371 [DOI] [PubMed] [Google Scholar]
- Salje J, van den Ent F, de Boer P, Löwe J (2011) Direct membrane binding by bacterial actin MreB. Mol Cell 43: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J (2009) The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol 72: 410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W (2007) Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol 66: 1396–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W (2008) Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol Microbiol 67: 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Löwe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Löwe J (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J 19: 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Löwe J (2002) F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 21: 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Rico AI (2006) The order of the ring: assembly of Escherichia coli cell division components. Mol Microbiol 61: 5–8 [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142: 309–320 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
- Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, Ruysschaert JM, Vicente M (2000) Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J Bacteriol 182: 6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.