Abstract
Highly motile Phytophthora sojae (P. sojae) zoospores of an oomycete plant pathogen and antioomycete candidate chemicals were encapsulated into microdroplets. Random fast self-motion of P. sojae zoospores was overcome by choosing an appropriate flow rate for a zoospore suspension. To influence stochastic loading of zoospores into a microfluidic channel, a zoospore suspension was directly preloaded into a microtubing with a largely reduced inner diameter. A relatively high single zoospore encapsulation rate of 60.5% was achieved on a most trivial T-junction droplet generator platform, without involving any specially designed channel geometry. We speculated that spatial reduction in the diameter direction of microtubing added a degree of zoospore ordering in the longitudinal direction of microtubing and thus influenced positively to change the inherent limitation of stochastic encapsulation of zoospores. Comparative phenotypic study of a plant oomycete pathogen at a single zoospore level had not been achieved earlier. Phenotypic changes of zoospores responding to various chemical concentration conditions were measured in multiple droplets in parallel, providing a reliable data set and thus an improved statistic at a low chemical consumption. Since each droplet compartment contained a single zoospore, we were able to track the germinating history of individual zoospores without being interfered by other germinating zoospores, achieving a high spatial resolution. By adapting some existing droplet immobilization and concentration gradient generation techniques, the droplet approach could potentially lead to a medium-to-high throughput, reliable screening assay for chemicals against many other highly motile zoospores of pathogens.
INTRODUCTION
Microfluidics has been attracting significant interest due to its vast potential to develop miniature tools for analyzing and controlling biochemical systems.1 Particularly, droplet-based microfluidics is capable of creating multiple identical compartments and parallel processing, providing an excellent platform technology for various chemical and biological applications.2, 3, 4 Attractive features of the droplet-based microfluidics include minute consumption of reagent, fast reaction, avoidance of dispersion of residence time, and high spatial resolution. Many chemical reactions performed within droplets have recently been reported to synthesize nanomaterials with desired structures and morphologies.5, 6 Encapsulation of single biological cells into droplets has also been demonstrated successfully for parallel single cell analysis and drug screening.7, 8, 9, 10, 11 It is noteworthy that these cells encapsulated using droplet approaches often have a low motility in media solutions.
Plant pathogens from the genus Phytophthora cause destructive diseases in a variety of crop plant species.12 For example, the oomycete pathogen P. sojae results in damping off and seedling death of soybean.13 The pathogenic P. sojae zoospores generally germinate on root surface and then penetrate the plant, causing serious economical losses to soybean crop.14 Chemical control such as by seed treatments with fungicidal drugs is a common practice.15 There have been several methods to test antioomycete or antifungal drug sensitivity and resistance in plant pharmacology.16, 17, 18, 19 Classical colorimetric method is based on measurement of fungal metabolic activity using color indicators, where the presence of a population of fungi can change the indicator color.16 Broth microdilution antifungal susceptibility test is another method to determine the lowest concentration of drugs, which yields significant inhibition of fungal growth.17 Molecular genetic analysis of antioomycete activity has been considered to be a more accurate drug screening method than many other classical methodologies.18 These technologies have contributed much to an acceleration of developing and screening for new antioomycete or antifungal chemicals for plant, but they often suffer from large sample requirement, lack of reliable statistics, relatively low throughput, and/or high cost.
We are interested in developing phenotypic screening assays for antioomycete candidate chemicals, by encapsulating highly motile pathogenic zoospores into microdroplets and by examining phenotypic changes of zoospores in response to different chemical conditions inside droplets. However, unlike biological cells, P. sojae zoospores are very agile in aqueous solutions and move fast with random trajectories. Efficient encapsulation of highly motile biological species was understudied previously. Also, droplet encapsulation of single bio-particles (e.g., cells) is a stochastic process that follows the Poisson distribution given by formula f (λ, k)= λk exp(−λ)/(k!) where f is probability of having k particles in a droplet given an average particle loading of λ particles per droplet.20 Several excellent techniques have been developed to improve single cell encapsulation rate by overcoming inherent limitations of stochastic encapsulation of cells within droplets.20, 21, 22 For example, by using a special high aspect-ratio channel, cells can self-organize into two evenly spaced streams whose longitudinal order is shifted by half the particle-particle spacing, resulting in a very high single cell encapsulation rate.20
In this paper, we report on encapsulating highly motile P. sojae zoospores into droplets for phenotypic screening of an antioomycete candidate chemical. The first part of the paper is to demonstrate a simple method to obtain a relatively high single zoospore encapsulation rate on a most trivial T-junction droplet generator platform. Specifically, a zoospore suspension was directly preloaded in a microtubing with a highly reduced inner diameter, for delivery of the suspension into a T-junction droplet generator. We hypothesized that spatial reduction in the diameter direction of the microtubing would allow zoospores to tunnel through the microtubing with a better order in the longitudinal direction than using a large inner diameter microtubing. In the second part of the paper, we demonstrated that zoospore germination and germling growth were feasible inside droplets under various chemical conditions. Phenotypic changes of zoospores were recorded and analyzed in parallel by interfacing the droplet device with a microscopic recording system and a zoospore growth tracking program. Comparative phenotypic study of a plant oomycete pathogen at a single zoospore level had not been achieved earlier. Besides a potential high zoospore spatial resolution, improved statistics could also be achieved through acquiring a medium-to-large-sized data set of zoospore germination and germling growth.
MATERIALS AND METHODS
Microfluidic device fabrication
The microfluidic device used in this study was fabricated using a conventional soft-lithography technique.23 Briefly, a silicon (Si) wafer was patterned with SU-8 photoresist (Microchem, MA, USA) to create a master mould for microfluidic channels. A pre-polymer mixture of polydimethylsiloxane or PDMS (Sylgard 184, Dow Corning, USA) and its curing agent with a weight ratio of 10:1 was poured onto the master mould and was thermally cured. Then, the hardened PDMS polymer was pealed from the master mould and bonded to a glass slide through oxygen plasma treatment. Inlet and outlet ports of the device were manually punched with a mechanical puncher.
Microtubing for microfluidic device
To test the hypothesis of improving single zoospore encapsulation rate by using small inner diameter microtubings, six different fused silica microtubings (inner diameters: 20, 50, 75, 100, 150, and 1000 μm; length: 2 m; Upchurch Scientific, WA, USA) were, respectively, used to connect between a syringe and a zoospore inlet port (see inlet 1 in Fig. 1a) of the device. Inside the syringe was only culture media without any zoospores. The outlet and other inlet ports of the device were inserted by 1000 μm-inner diameter microbore tubings (Cole-Parmer, IL, USA). Instead of loading a zoospore suspension into the syringe, we preloaded a suspension solution directly into one of the aforementioned six microtubings. The zoospore suspension was pushed by a syringe pump flowing directly from the microtubing into the device. Here, the microtubings were placed horizontally, minimizing zoospore aggression that generally would deteriorate the homogeneity of zoospores.
Figure 1.
(a) Schematic microfluidic setup for droplet encapsulation of highly motile P. sojae zoospores. A zoospore suspension was preloaded directly into a microtubing with a reduced inner diameter. Flow rate of each flow was controlled by a syringe pump. (b) Time-lapse images extracted from a movie showing a typical process of droplet formation and zoospore encapsulation. Blue, red, and white circles indicate three different zoospores, respectively. The time interval for trapping these zoospores was not uniform because zoospores were not exactly uniformly distributed inside a microtubing and thus they arrived at a T-junction at different time instants. t is time in seconds. (c) Droplets containing both zoospores and a chemical were collected in a collecting reservoir. (d) Encapsulated zoospores in droplets. Zoospores are highlighted with white circles. Scale bars in (b) and (d) represent 100 μm.
P. sojae zoospore suspension and zoospore-chemical mixture preparation
Phytophthora sojae zoospores were prepared according to a published protocol.24 Briefly, the pathogen was grown on V8 solid medium for 5 days at 22 °C and soaked overnight with sterile double distilled water. Following day, P. sojae plates were washed repeatedly to deplete nutrients of the medium so that sporangia were formed for production of zoospores in the following morning. Zoospores were counted and used at a concentration of 104–106 zoospores/ml. An aqueous phase mixture of a zoospore suspension and an antioomycete candidate chemical, with a specific concentration of the chemical, was prepared by adding an appropriate amount of the chemical into a zoospore suspension. In this study, metalaxyl, a chemical toxic to P. Sojae,15 was used as a model chemical.
Microscopic recording
In order to observe and record zoospore germination and growth of germlings within droplets, we used a Leica M205FA stereo microscope. The microscope has a 1× and 2× objective lens, enabling 7.1× to 230× magnification which was adequate for the designed experiments. The microscope was coupled with a digital camera (QICam 12-bit color Fast 1394) that enabled the capture of digital images at a specified time interval. These images were compressed into the Audio Video Interleave (.avi) video format. The.avi video was then post-processed by a zoospore growth tracking software briefly described below.
Tracking program for zoospore growth in droplets
The zoospore growth tracking program was able to simultaneously extract the germinating filament length of zoospores in about 25–30 droplets. The number of the droplets being processed was actually determined by not only the magnification factor of the microscope but also, more importantly, the image resolution that allowed the program to differentiate a growing zoospore from its nearest pixels. The program was able to (i) identify the initial location of a spherical zoospore in each droplet to set up a dynamic origin in the coordinate plane; (ii) track developing germlings of multiple zoospores using an articulated model; and (iii) calculate germinating filament lengths of multiple zoospores. The current version of the software allowed us to analyze the.avi video at a fixed rate of five frames per minute and at a fixed resolution of 800 × 600 pixels with a ∼12% error in germination length caused by rounding to the nearest pixels. It was possible to simultaneously process a larger number of droplets with a less calculation error by optimizing the microscopic video system and the tracking program.
DROPLET ENCAPSULATION OF RANDOM FAST MOVING PATHOGENIC ZOOSPORES
Droplet encapsulation of zoospores
Figure 1a illustrates a microfluidic setup for the droplet encapsulation of P. sojae zoospores. A premixed aqueous zoospore-chemical mixture (the chemical was metalaxyl as mentioned earlier) was flowed directly from a microtubing into a vertical channel through inlet 1. A continuous oil phase solution (mineral oil, #0121-1, Fisher Scientific, NJ, USA) was also flowed into a horizontal channel through inlet 2. The zoospore-chemical flow was then cut into fine droplets at the T-shaped junction of the two channels by the oil flow. Thus, the zoospores were trapped into the droplets. The oil flow also carried the droplets to a collecting reservoir (Figs. 1a, 1c). Surfactant (Span 80, 2 wt. %, Sigma-Aldrich, MO, USA) was pre-added into the mineral oil to prevent droplet coalescence.25 After the reservoir received enough droplets, all syringe pumps were turned off and the microtubings were all unplugged. Consequently, a static droplet distribution would be achieved in the collecting reservoir in several minutes when the internal fluid pressure dropped to the atmospheric pressure. Actually, several existing droplet trapping and stabilization techniques could be applied to immobilize the droplets.10, 26, 27, 28 But, the droplet stabilization was not the topic of the present work. Here we conducted the experiment on a vibration free table to maintain a static distribution during testing. Figure 1b shows a typical process for the droplet formation and zoospore encapsulation, where three zoospores were being trapped in succession by three droplets.
We needed to be concerned about three important factors, in order to realize high-efficiency trapping of single zoospores into droplets. First, an appropriate hydrodynamic force in the vertical channel was required to overcome the random fast self-motion of highly motile zoospores, while posing a minimal shear stress on zoospore surface (discussed in Sec. 3B). Second, increasing the fraction of zoospore diameter in the inner diameter of the microtubing would, to some extent, increase the order of zoospores in the longitudinal direction of the microtubing (discussed in Sec. 3C). Last, a sufficient amount of zoospores was needed to supply to the front of the zoospore-chemical flow (discussed in Sec. 3D).
To figure out influences of the three factors, i.e., zoospore-chemical flow rate, microtubing’s inner diameter, and zoospore suspension density (ZSD), on zoospore encapsulation rate, a thorough, very large experimental matrix would be required. But we could reduce the experimental matrix by fixing two of the three parameters for the following two reasons. First, it was obvious that the higher the ZSD, the higher the encapsulation rate, given other conditions the same during trapping. The highest ZSD available was 106 zoospores/ml. Second, as mentioned early, it was reasonable to think that using a smaller inner diameter microtubing to deliver zoospores would add more to the degree of zoospore ordering in the longitudinal direction of the microtubing, enabling us to obtain a higher encapsulation rate. The smallest inner diameter for a microtubing was 20 μm commercially available. Therefore, in Sec. 3B, we have chosen the ZSD of 106 zoospores/ml and the inner diameter of 20 μm to study influence of zoospore-chemical flow rate on zoospore encapsulation rate; in Sec. 3C we have chosen the ZSD of 106 zoospores/ml and an optimized flow rate obtained in Sec. 3B, to find out quantitatively the relationship between microtubing size and encapsulation rate; and in Sec. 3D we have chosen the inner diameter of 20 μm and the optimized flow rate, to study quantitatively how different ZSDs could impact zoospore encapsulation rate.
Influence of zoospore-chemical flow rate on zoospore encapsulation rate
When the zoospore-chemical solution was flowed slowly in the vertical channel, achieving a high zoospore encapsulation rate was difficult because zoospores had a high chance of moving away from the front of the zoospore-chemical flow just before encapsulation. Here, the encapsulate rate here referred to the success rate of trapping a single zoospore in droplet. The data were obtained by dividing the number of single zoospore-containing droplets by the total number of the droplets (∼110) collected in the reservoir. In this experiment, the microtubing containing a zoospore suspension was 20 μm in diameter. The ZSD was 1 × 106 zoospores/ml and the oil flow rate was 0.02 ml/h. It was found that at a relatively high zoospore-chemical flow rate no obvious random motion of zoospores was observed. Most likely hydrodynamic force of laminar flows suppressed the zoospores’ self-motion. As shown in Fig. 2, the single zoospore encapsulation rate increased as the zoospore-chemical flow rate was increased up to 0.008 ml/h. At 0.008 ml/h the droplet diameter was 110.2 ± 5.4 μm. About 60.5% of droplets contained one zoospore. However, further increase of zoospore-chemical flow rate resulted in decreasing the single zoospore encapsulation rate. This was because the droplet size was also increased with increasing zoospore-chemical flow rate and one droplet could hold more than one zoospore, limiting the single zoospore encapsulation rate. We should point out that although the zoospores’ self-motion was significantly reduced during the encapsulation process, their motility were returned to normal after droplets became still.
Figure 2.
Single zoospore encapsulation rate (left y-axis) and droplet diameter (right y-axis) with respect to zoospore-chemical flow rate. For all different zoospore-chemical flow rates, ZSD = 1 × 106/ml. Here the microtubing was 20 μm in diameter and the oil flow rate was 0.02 ml/h.
Influence of microtubing’s inner diameter on zoospore encapsulation rate
In this experiment, the zoospore suspension with the ZSD of 1 × 106 zoospores/ml was preloaded into the aforementioned six different microtubings. The zoospore-chemical flow rate used here was 0.008 ml/h according to the result obtained in the previous section. The oil flow rate was still 0.02 ml/h. Figure 3a demonstrates that the large reduction to the microtubing’s inner diameter was beneficial to increase the single zoospore encapsulation rate up to 60.5%. Specifically, when the inner diameter was changed between 75 and 1000 μm, the single zoospore encapsulation rate was little affected, falling between 32.2% and 35.3%. By converting the ZSD of 1 × 106 zoospores/ml to an average 0.7 zoospores per droplet (110.2 ± 5.4 μm in diameter) and substituting λ with 0.7 in the Poisson distribution equation listed above, interestingly, we found about 34.8% of droplets would contain a single cell, whereas 15.6% would contain more than one cell. This indicated that when the microtubing’s inner diameter was equal to or above 75 μm, the delivery of zoospores into the inlet port of the microfluidic device was still a stochastic process that followed Poisson distribution. However, when the 50 μm-inner diameter microtubing was used, the single-zoospore encapsulation rate was slightly increased to about 43.4 ± 6.9%. Further, a significant increase to about 60.5 ± 4.7% was achieved at the microtubing’s inner diameter of 20 μm (the minimum inner diameter of microtubing commercially available that moment). Although a quantitative full explanation for this result is still lacking, the plausible explanation is as follows. The zoospores studied here had a diameter of about 5 μm, a quarter or a significant fraction of the microtubing’s inner diameter (20 μm). This might lead to adding an additional degree of zoospore ordering in the longitudinal direction of the microtubing. Thus the stochastic loading of zoospores from the microtubing into the channel and thus into droplets was somehow influenced. Also, it would possibly happen that further decreasing the microtubing’s inner diameter would increase the single zoospore encapsulation rate.
Figure 3.
(a) Encapsulation rate for 0, 1, and ≥ 2 zoospores in a droplet with respect to the inner diameter of the microtubing containing a zoospore suspension with ZSD = 1 × 106 zoospores/ml. The zoospore-chemical flow rate used here was 0.008 ml/h and the oil flow rate was 0.02 ml/h. (b) Encapsulation rate for 0, 1, and ≥ 2 zoospores in a droplet as a function of the ZSD. The inner diameter of the microtubing was 20 μm and the oil flow rate was 0.02 ml/h. For comparison, the Poisson distribution of the number of zoospores in droplets were calculated. The value of λ shown in this figure represents an average number of zoospores per droplet if zoospores are uniformly distributed among all droplets.
Influence of ZSD on single zoospore encapsulation rate
The single zoospore encapsulation rate related significantly to the ZSD used in the experiments. We tested zoospore encapsulation rates at four different ZSDs (0.2, 0.5, 0.7, and 1 × 106 zoospores/ml), using the 20 μm-inner diameter microtubing to deliver zoospore suspensions. The zoospore-chemical flow rate was 0.008 ml/h and the oil flow rate was 0.02 ml/h in this experiment. Figure 3b plots the zoospore encapsulation rate as a function of the ZSD. At the low ZSD of 0.2 × 106 zoospores/ml or λ = 0.14, the single zoospore encapsulation rate was low mainly due to the insufficient zoospore supply, and the use of the 20 μm-inner diameter microtubing did not change the Poisson distribution of the number of zoospores per droplet. However, as the ZSD was increased, the single zoospore encapsulation rate was increased significantly. By comparing between the measured actual distribution and the calculated Poisson distribution shown in Fig. 3b, it was clear that the 20 μm-inner diameter microtubing developed a higher level disturbance to the Poisson distribution with a high ZSD zoospore supply than it did with a low ZSD one.
It seemed rational to think that increasing the ZSD could raise the single zoospore encapsulation rate further. Unfortunately, obtaining a ZSD higher than 106 zoospores/ml was practically hard. A centrifuge method is often used to increase the suspension density of biological cells. This method, however, was not suitable for zoospores. The zoospores lose their motility even after exposure to very low centrifugal forces because they encyst rapidly. For instance, the zoospores were fully encysted after centrifugation at ∼300 rpm for just 2 min. Thus, unavailability of a higher ZSD suspension posed a major limitation in obtaining a higher encapsulation rate above 60.5%.
ZOOSPORES’ PHENOTYPIC RESPONSES TO AN ANTIOOMYCETE MODEL CHEMICAL
The purpose of creating multiple isolated droplet compartments was to screen antioomycete candidate chemicals by analyzing phenotypic changes of zoospores (e.g., germination, and germling growth) in response to different chemical environments. Using multiple small droplets would allow us to obtain a medium-to-large-sized data set, thus a reliable statistic. Germination and germling growth of a single zoospore in a droplet were independent and consumed a low dose chemical, but not affected by germinating zoospores in neighboring droplets, thus a high spatial resolution of zoospores. In contrast, a Petri dish-based approach for phenotypic responses of zoospores to drugs is often associated with a network-like appearance of germlings making it difficult to grasp actual drug affects on individual germinating zoospores.
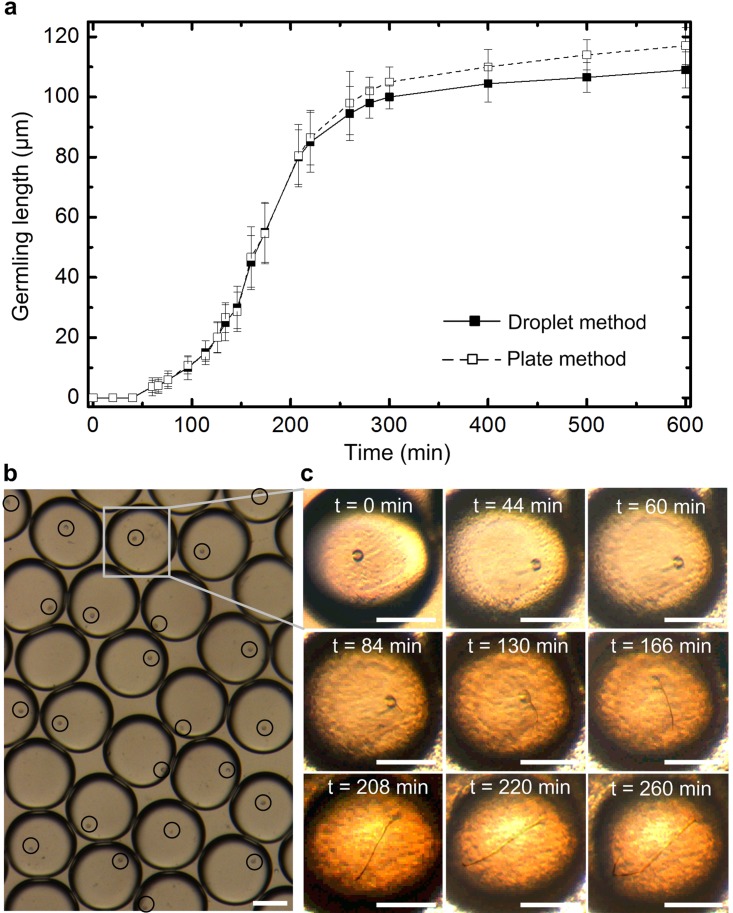
A control experiment was first conducted by encapsulating single zoospores into droplets with no any chemical. When all droplets were stabilized in the collecting reservoir, the microscopic recording system and the tracking program mentioned above started to simultaneously record and analyze zoospore germination and germling growth inside 25–30 droplets. Figure 4a shows the average germling length of zoospores in the multiple droplets, over a 600 min period. In a typical case (Fig. 4c), a zoospore first moved actively in the droplet and then settled down. About 1 h later, the zoospore began germinating to form a protrusion at the zoospore surface. Then, the tiny germinating filament was extended from the surface protrusions. The germling grew almost along the direction of the initial protrusion. As the germinating filament reached the water-oil interface of the droplet, the germling continued growing within the droplet by curling itself to fit into the droplet rather than breaking through the droplet wall into the oil phase or any of the neighboring droplets. We did not see the zoospore moving out of a droplet in the course of the experiment. The viability of the encapsulated zoospores was also examined. Almost all of the encapsulated zoospores were viable inside the droplets (with no antioomycete chemical) after the 600 min. This was because of the low zoospore-chemical flow rate used during the encapsulation process that posed a minimal shear stress on zoospore surface. In order to compare the results obtained in the droplets with those obtained on traditional plates, we monitored zoospore germination in a 1-in. plastic Petri dish (Corning, NY) containing the same culture medium. The monitoring time was also 600 min. It was found that the zoospore germination in droplets were almost the same as that in the Petri dish in the first 260 min; afterwards, the Petri dish environment allowed zoospores to grow slightly faster than the droplet environment (Fig. 4a). This was probably because the mineral oil used for generating droplets was not gas permeable that might affect zoospore germination in droplets.
Figure 4.
(a) Germling length of P. sojae zoospores in droplets and a Petri dish as a function of time, without any antioomycete chemical. (b) 25-30 Droplets simultaneously imaged and analyzed to track the zoospore germination and germling growth. Black circles indicate the zoospores. (c) Time-lapse images extracted showing a typical growth trajectory of a germling. Scale bars in (b) and (c) represent 50 μm.
The model antioomycete candidate chemical metalaxyl was used as in the following experiments. This chemical essentially poses significant interference to the infection process and metabolic pathway of oomycete pathogens by blocking rRNA synthesis.29 Zoospore-metalaxyl mixtures with 14 different metalaxyl concentrations (from 0 to 10 mg/ml) were prepared. Zoospore-metalaxyl droplets with a specific metalaxyl concentration were formed and the zoospore growth data were tracked by using the same procedures as for the previous control experiment. As shown in Fig. 4a, germination of P. sojae zoospores in both droplets and Petri dish slowed down significantly after 260 min. Therefore, in the next set of experiments we monitored responses of zoospores to antioomycete candidate chemicals over a 260 min period.
Figure 5 summarizes the final length of zoospore germlings inside droplets at different metalaxyl concentrations at the recording time of t = 260 min. Each data point was obtained by averaging the zoospore growth data from 25 to 30 droplets. At the metalaxyl concentrations lower than 0.5 mg/ml, the zoospore germination and germling growth was not seriously inhibited. An abrupt drop in germling length was observed when the metalaxyl concentration was 0.75 mg/ml. At metalaxyl concentration 1.0 mg/ml, the germling length was reduced significantly to 18% of the germling length in the metalaxyl-free control. At concentration above 2.0 mg/ml, the zoospore germination was fully inhibited and no germlings were detected during the entire 260 min period. Figure 6 displays the germling length as a function of time at different metalaxyl concentrations over the 260 min period. The germling length results shown in Figs. 56 revealed that the droplet-based screening was reliable to screen antioomycete chemicals in that chemical responses of zoospores in droplets were almost the same as those in a Petri dish over the 260 min period. We also would like to point out that the relative standard deviation of uniformity of droplet diameter was about 5.2% (the mean droplet diameter: 110.2 μm). We believe that this size inconsistency level had little influence on the consistency of environments for zoospore germination because the zoospore is more than three orders of magnitude smaller than the droplet in volume.
Figure 5.
(a) Final germling length of P. sojae zoospores in droplets and a Petri dish as a function of metalaxyl concentration after 260 min. (b) Images showing the final germlings at different metalaxyl concentrations. Scale bars represent 50 μm.
Figure 6.
Germling length of P. sojae zoospores in droplets and a Petri dish as a function of time at various metalaxyl concentrations.
CONCLUSIONS
We have demonstrated to encapsulate highly motile P. sojae zoospores and a model chemical metalaxyl in microfluidic droplets. By preloading a zoospore suspension directly into a microtubing with a reduced inner diameter, an additional degree of zoospore ordering in the longitudinal direction of the microtubing was possibly obtained to change the inherent limitation of stochastic encapsulation of zoospores. This led to a relatively high single zoospore encapsulation rate of 60.5% on a simple T-junction droplet generator. No any special channel geometry was employed. Zoospore germination and germling growth at different chemical concentrations inside multiple droplets were measured and analyzed in parallel. A medium throughput data set for the zoospore germination and germling growth was achieved at a single zoospore spatial resolution, showing promise of development of a reliable screening assay for chemicals against highly motile pathogenic zoospores.
Many further studies remain to be done to understand theoretically the influence of the microtubing’s inner diameter on ordering zoospores inside the microtubing. Besides, from the experimental perspective, many existing microfluidic droplet techniques can be adopted to achieve a truly high throughput phenotypic screening for antioomycete chemicals toxic to zoospores. Through microfluidic tuning, flexible control over droplet size, chemical concentration and composition, and generation speed can provide versatility in designing additional experiments for other biological studies. In the present work, each concentration of chemicals was prepared and tested independently, which somehow diminished throughput and user convenience. Several clever methods have been demonstrated by other group to generate concentration gradients inside droplets, including by varying flow rates of constituting streams of liquid,30 by on chip-dilution,31 by using aspiration technique,32 by droplet-on-demand technology,33 or by use of droplet libraries.34 Single cell screen coupled with generation of concentration gradient on the same chip was demonstrated recently.35 Also, as mentioned previously, the present reservoir was simple and droplets were prone to slight vibrations, requiring a vibration-free table. There have been several methods of droplet trapping and stabilization, including creating droplet spots and using various trapping geometries.26, 27, 28, 10 In addition, some excellent droplet generation mechanisms can also be applied to simplify droplet formation process.36 Lastly, by creating gas-surrounding droplets on chip,37 it is possible to eliminate the possible influence of non-gas permeable oil on zoospore germinations in droplets, making the droplet system more reliable in screening antioomycete chemicals.
ACKNOWLEDGMENTS
The authors thank Mr. Rishi Sumit for the generous zoospore supply and Santosh Pandey for his helpful discussion. This work was supported in part by the National Science Foundation (ECCS-1102354), by the McGee-Wagner Interdisciplinary Research Fund Award, and by the Iowa State Dean’s Research Initiative for High Throughput Computational Biology.
References
- Whitesides G. M., Nature (London) 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Teh S. Y., Lin R., Hung L. H., and Lee A. P., Lab Chip 8, 198 (2008). 10.1039/b715524g [DOI] [PubMed] [Google Scholar]
- McClain M. A., Culbertson C. T., Jacobson S. C., Allbritton N. J., Sims C. E., and Ramsey J. M., Anal. Chem. 75, 5646 (2003). 10.1021/ac0346510 [DOI] [PubMed] [Google Scholar]
- Weibel D. B. and Whitesides G. M., Curr. Opin. Chem. Biol. 10, 584 (2006). 10.1016/j.cbpa.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Song Y., Hormes J., and Kumar C. S. S. R., Small 4, 698 (2008). 10.1002/smll.200701029 [DOI] [PubMed] [Google Scholar]
- Nie Z., Li W., Seo M., Xu S., and Kumacheva E., J. Am. Chem. Soc. 128, 9408 (2006). 10.1021/ja060882n [DOI] [PubMed] [Google Scholar]
- Kang L., Chung B. G., Langer R., and Khhademhosseini A., Drug Discover Today 13, 1 (2008). 10.1016/j.drudis.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich P. S. and Manz A., Nature Rev. Drug Discov. 5, 210 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- Wheeler A. R., Throndset W. R., Whelan R. J., Leach A. M., Zare R. N., Liao Y. H., Farrell K., Manger I. D., and Daridon A., Anal. Chem. 75, 3581 (2003). 10.1021/ac0340758 [DOI] [PubMed] [Google Scholar]
- Shi W., Qin J., Ye N., and Lin B., Lab Chip 8, 1432 (2008). 10.1039/b808753a [DOI] [PubMed] [Google Scholar]
- Song H., Chen D. L., and Ismagilov R. F., Angew. Chem., Int. Ed. 45, 7336 (2006). 10.1002/anie.v45:44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke D. E. L., Drenth A., Duncan J. M., Wagels G., and Brasier C. M., Fungal Genet. Biol. 30, 17 (2000). 10.1006/fgbi.2000.1202 [DOI] [PubMed] [Google Scholar]
- Hahn M. G., Bonhoff A., and Grisebach H., Plant Physiol. 77, 591 (1985) 10.1104/pp.77.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrather J. A. and Koenning S. A., J. Nematol. 39, 173 (2006). [PMC free article] [PubMed] [Google Scholar]
- Guy S. O., Oplinger E. S., and Grau C. R., Agron. J. 81, 529 (1989). 10.2134/agronj1989.00021962008100030027x [DOI] [Google Scholar]
- Yamaguchi H., Uchida K., Nagino K., and Matsunaga T., J. Infect. Chemother. 8, 374 (2002). 10.1007/s10156-002-0201-y [DOI] [PubMed] [Google Scholar]
- Espinel-Ingroff A., C. W.Kish, Jr., Kerkering T. M., Fromtling R. A., Bartizal K., Galgiani J. N., Villareal K., Pfaller M. A., Gerarden T., and Rinaldi M. G., J. Clin. Microbiol. 30, 3138 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Nat. Biotechnol. 19, 212 (2001). 10.1038/85634 [DOI] [PubMed] [Google Scholar]
- Inglese J., Auld D. S., Jadhav A., Johnson R. L., Simeonov A., Yasgar A., Zheng W., and Austin C. P., Proc. Natl. Acad. Sci. U.S.A. 103, 11473 (2006) 10.1073/pnas.0604348103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edd J. F., Carlo D. D., Humphry K. J., Koster S., Irimia D., Weitz D. A., and Toner M., Lab Chip 8, 1262 (2008). 10.1039/b805456h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anna S. L., Bontoux N., and Stone H. A., Appl. Phys. Lett. 82, 364 (2003). 10.1063/1.1537519 [DOI] [Google Scholar]
- Chabert M. and Viovy J.-L., Proc. Natl. Acad. Sci. U.S.A. 105, 3191(2008). 10.1073/pnas.0708321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- Ward E. W. B., Lazarovits G., Unwin C. H., and Buzzell R. I., Phytopathology 69, 951 (1979). 10.1094/Phyto-69-951 [DOI] [Google Scholar]
- Thorsen T., Roberts R. W., Arnold F. H., and Quake S. R., Phys. Rev. Lett. 86, 4163 (2001). 10.1103/PhysRevLett.86.4163 [DOI] [PubMed] [Google Scholar]
- Schmitz C. H. J., Rowat A. C., Koster S., and Weitz D. A., Lab Chip 9, 44 (2009). 10.1039/b809670h [DOI] [PubMed] [Google Scholar]
- Huebner A., Bratton D., Whyte G., Yang M., deMello A. J., Abell C., and Hollfelder F., Lab Chip 9, 692 (2009). 10.1039/b813709a [DOI] [PubMed] [Google Scholar]
- Bai Y., He X., Liu D., Patil S. N., Bratton D., Huebner A., Hollfelder F., Abell C., and Huck T. S. H., Lab Chip 10, 1281 (2010). 10.1039/b925133b [DOI] [PubMed] [Google Scholar]
- Fisher D. J. and Hayes A. L., Crop Prot. 3, 177 (1984). 10.1016/0261-2194(84)90052-8 [DOI] [Google Scholar]
- Song H. and Ismagilov R. F., J. Am. Chem. Soc. 125, 14613 (2003). 10.1021/ja0354566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Gielen F., Edel J. B., and deMello A. J., Nat. Chem. 3, 437 (2011). 10.1038/nchem.1046 [DOI] [PubMed] [Google Scholar]
- Clausell-Tormos J., Griffiths A. D., and Merten C. A., Lab Chip 10, 1302 (2010). 10.1039/b921754a [DOI] [PubMed] [Google Scholar]
- Churski K., Korczyk P., and Garstecki P., Lab Chip 10, 816 (2010). 10.1039/b925500a [DOI] [PubMed] [Google Scholar]
- Brouzes E., Medkova M., Savenelli N., Marran D., Twardowski M., Hutchison J. B., Rothberg J. M., Link D. R., Perrimon N., and Samuels M. L., Proc. Natl. Acad. Sci. U.S.A. 106, 14195 (2009). 10.1073/pnas.0903542106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J.-C., Beck Y., Billas-Massobrio I., Moras D., and Griffiths A. D., Chem. Biol. 17, 528 (2010). 10.1016/j.chembiol.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Gu H., Murade C. U., Duits M. H. G., and Mugele F., Biomicrofluidics 5, 011101 (2011). 10.1063/1.3570666.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zeng S., Qin J., and Lin B., Electrophoresis 30, 3181 (2009). 10.1002/elps.200900331 [DOI] [PubMed] [Google Scholar]