Abstract
We have used Brownian dynamics-finite element method to examine two conformational preconditioning approaches for improving DNA stretching in a microcontraction for the purpose of direct gene analysis. The newly proposed “pre-stretching” strategy is found to significantly improve the degree of DNA extension at the exit of the contraction. On the other hand, applying an oscillating extensional field to DNA yields no preconditioning effect. Detailed analysis of the evolution of DNA extension and conformation reveals that the success of our “pre-stretching” strategy relies on the “non-local” effect that cannot be predicted using simple kinematics analysis. In other words, accurate prediction can only be obtained using detailed simulations. Comparing to the existing preconditioning strategies, our “pre-stretching” method is easy to implement while still providing a very good performance. We hope that the insight gained from this study can be useful for future design of biomicrofluidic devices for DNA manipulation.
INTRODUCTION
Manipulation of DNA has been an active research field for several decades, and many applications have been created from the developed knowledge.1 Among related studies, a topic received much attention is to find an efficient and reliable way for performing DNA genome analysis.2, 3 For some applications, a high resolution of DNA sequence is not necessary, while a low resolution of gene map is sufficient. Examples include personal identification, national defense, and disease examinations. However, current methods for these applications usually involve slow and expensive processes such as polymerase chain reaction and gel electrophoresis.4 To overcome this bottleneck, a new technology called “direct linear analysis (DLA)”5, 6, 7 has been proposed, and it promises faster and cheaper operation than the traditional methods. The basic idea of DLA is to mark targeted DNA sequences with specifically designed fluorescent tags, and then to stretch DNA so that the locations of specific genes can be optically determined. Comparing to the traditional methods, DLA not only avoids time-consuming biochemical processes and separation steps but also provides a direct way to examine the targeted sequences.
Realizing DLA requires two core technologies: the creation of the adequate sequence markers and the linearization of DNA for direct identification. The former can be readily achieved by various biomarkers,5, 8 while the latter is yet to be fulfilled by an efficient and reliable way. Several methods have the potential to meet this need and can be roughly divided into four categories: molecular combing,9, 10 confinement,8, 11, 12 tethered DNA,13, 14 and dynamic stretching.7, 15 Molecular combing uses the receding meniscus to stretch DNA on a substrate.10 Using confinement to stretch DNA takes advantage of the fact that DNA as a self-avoiding chain will extend to a significant degree when confined in a space with at least one of the dimensions smaller than DNA persistence length.16 Tethered DNA has one of its ends fixed on a substrate, and therefore, it can be easily stretched out by a uniform flow or electric field. Some applications attach a bead to one end of DNA and stretch DNA using magnetic17 or optical tweezers.18 Dynamic stretching uses hydrodynamic flow gradient or electric field gradient19 to stretch DNA. Among these methods for DLA, dynamic stretching has received more attention because of its low cost and high throughput. However, DNA extension obtained using dynamic stretching is only moderate and somewhat difficult to control.
To dynamically stretch DNA to near its contour length, two basic requirements have to be met. First, the strain rate must be high enough so that the Deborah number (De) of DNA is larger than 0.5.20 Secondly, the strain imposed to DNA must be adequate. Insufficient strain leads to partially stretched DNA, and this has become the primary limiting factor of the current design.21 A plausible solution is just to increase the strain the device can provide. However, implementing this idea is practically difficult. For example, a typical device to perform dynamic DNA stretching is a microcontraction. However, large strain implies that DNA will have a very high velocity when reaching its maximum extension. This high velocity of DNA usually leads to difficulty in the detection of targeted biomarkers. Moreover, a contraction capable to provide large strain will have a very large aspect ratio that is limited by the strength of the material used to construct the device. Therefore, to improve the method of dynamic stretching, it is necessary to keep both the strain and the strain rate in an adequate range. Constrained by these criteria, researchers have focused on creating various “pre-conditioning” strategies that can tune DNA configuration to a state more suitable for the successive stretching, usually by an extensional flow.
Different conformational preconditioning strategies found in literature include pre-shearing DNA,22 passing DNA through a gel matrix7 or obstacle arrays,23, 24 and exposing DNA to an oscillating extensional flow.22 “Pre-shearing” is to apply a shear flow to DNA before it is being stretched by an extensional flow. Pre-shearing has been shown to effectively reduce the “folded” DNA population, and in general helpful to improve DNA stretching. However, the “shear” component of the flow also causes DNA to rotate and thus prevent DNA from being highly stretched.15 The gel matrix set in the flow cell acting as tethering points that can hook DNA and make DNA partially stretched. DNA pre-conditioned by this way has been shown to have excellent final extension, but setting up the gel matrix in a flow cell is complicated and easy to fail. Obstacle array is a more controllable version of gel matrix, while the resulting benefit is also compromised. Finally, exposing DNA to an oscillating extensional flow in prior to the successive extensional flow has been predicted to facilitate DNA stretching.22 However, the prediction has not yet been tested. We denote it as a “massage” approach, and it is to be examined in this study. To summarize, the currently available conformation pre-conditioning methods have not yet met the needs for practical applications.
The goal of this study is to use the quantitative Brownian dynamics-finite element method (BD-FEM) simulations to explore new preconditioning approaches that are more effective to stretch DNA and easier to implement into real devices. The BD-FEM simulation technique21, 24, 25, 26, 27, 28, 29 models the behavior of macromolecules using Brownian dynamics simulations30, 31, 32 with the flow or electric field solved by the finite element method.33 The method that combines the power of the coarse-grained molecular simulations and continuum calculations has provided researchers an accurate but affordable way to quantitatively predict the behavior of DNA in microfluidic devices. Despite its power, BD-FEM technique has often been used as a complementary tool for explaining experimental results.24, 26, 34 Here, we want to use BD-FEM simulation as a simulation-guided design (SGD) tool that goes before the experiments, just like the role of computational fluid dynamics for engineering design.21 In this study, we use BD-FEM technique to exam two DNA pre-conditioning strategies. The first is a “pre-stretching” approach proposed first time by the current study, and the second is the implementation of the “massage” method that exposes DNA to an oscillating extensional field for conformational preconditioning. A microcontraction is used to stretch preconditioned DNA. We perform most of our simulations at De = 23 to evaluate these preconditioning strategies.
THE PRE-CONDITIONING STRATEGIES AND DEVICE DESIGN
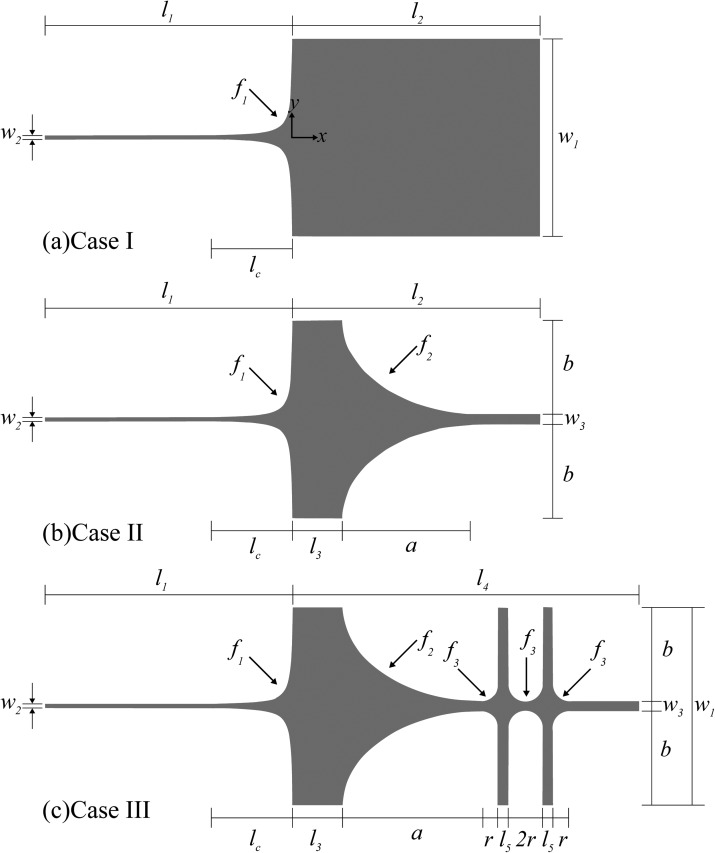
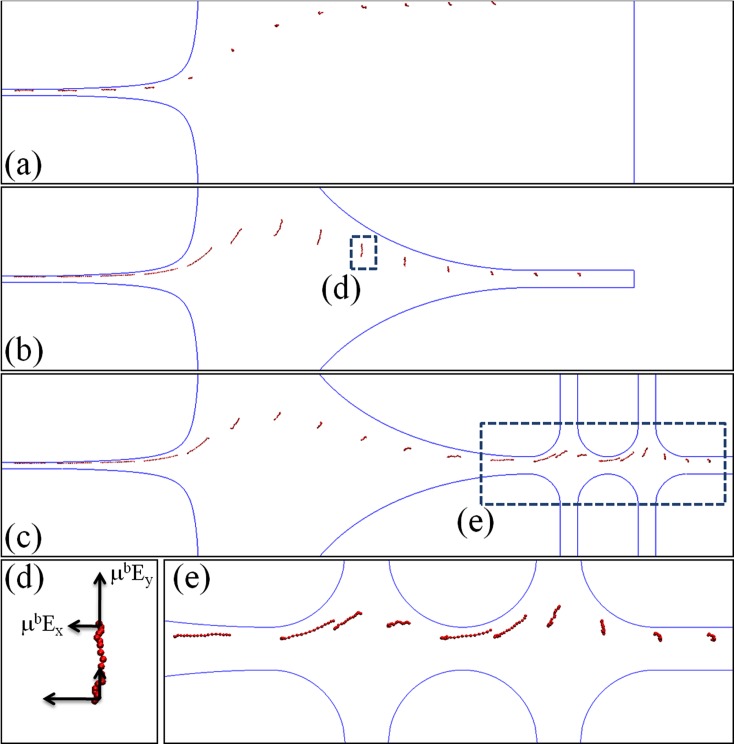
The reference device chosen to test the pre-conditioning strategies is based on the design of Kim and Doyle.21 The device is a microcontraction and DNA in buffer solutions are driven through the device by electrophoresis. DNA chains are stretched when passing through the contraction funnel due to the presence of the strong electric field gradient. Since the funnel shape has been shown to have a little effect on the degree of DNA stretching, we choose the case IV of Kim and Doyle21 as the reference device through this study. We denote the reference device to be the case I of the current study. The schematic diagram of case I is presented in Fig 1a. The geometrical parameters w1, w2, l1, l2 and lc are set to 200 μm, 3.8 μm, 250 μm, 250 μm and 80 μm, respectively.21 The shape of the contraction funnel is defined by a hyperbolic function,
| (1) |
We denote the inlet and the outlet as the rightmost and leftmost of the device. The height of the devices (not shown in Fig. 1) is set to be 2 μm through this study.
Figure 1.
The schematic diagram of the devices implemented with different pre-conditioning approaches. (a) Contraction only, (b) contraction with an expansion for pre-stretching DNA, (c) contraction with two cross-slots for pre-conditioning DNA with an oscillating extensional field. The expansion connecting the contraction and the cross-slots is identical to that of case II. The numerical values of the design parameters are listed in Table TABLE I..
Two different preconditioning approaches are examined in this study. For the first approach, we propose to “pre-stretch” DNA in the direction perpendicular to the funnel orientation, and then to turn the DNA chain to align with the funnel direction for the successive stretching. We denote this approach to be the “pre-stretching” approach. Whether the approach will succeed or fail depends on whether the DNA extension achieved in y direction can be preserved when DNA turns to x direction. The “pre-stretching” approach is implemented in the design of case II shown in Fig. 1b. Case II is different from case I by having an expansion between the inlet of the device and the contraction. In case II, the geometry parameters a, b, w3, and l3 are set to be 140 μm, 95 μm, 10 μm, and 50 μm, respectively. The shape of the expansion is defined by the function .
The second preconditioning approach is the “massage” method based on the prediction of Larson.22 It is implemented in the design of case III shown in Fig. 1c by appending two cross-slots to the design of case II. The cross-slots are responsible for generating an oscillating extensional field to pre-condition DNA. The design parameter l4 and l5 are chosen to be 350 μm and 10 μm, respectively. The boundary of the cross-slot is defined by the function with r = 17.5 μm. We choose to add the cross-slots at the end of case II rather than that of case I because there must be an expansion at the junction between the cross-slots and the contraction. Thus, the design of case III is not only reasonable but also helpful for determining the net effect of the “massage” approach by comparing with case II. All the numerical values of the design parameters are summarized in Table TABLE I..
TABLE I.
Design parameters of the three testing devices.
| w1 | 200 μm | l1 | 250 μm | l4 | 350 μm | a | 140 μm |
| w2 | 3.8 μm | l2 | 250 μm | l5 | 10 μm | b | 95 μm |
| w3 | 10 μm | l3 | 50 μm | lc | 80 μm | r | 17.5 μm |
SIMULATION METHODS
Brownian dynamics
We use bead-spring model31 that contains Nb beads and Ns (= Nb-1) springs to simulate DNA behavior. In this study, λ-DNA with a contour length L = 20.5 μm is chosen to be the representative molecule, and it is modeled by 19 beads connected with 18 springs. The BD simulation is reproduced from Doyle and coworkers.21, 24, 25 The force balance for i–th bead is described as below
| (2) |
where ri is the position of i-th bead, μb is the electrophoretic mobility of a bead, E(ri) is the electric field at ri, and ζ is the drag coefficient of a bead. FiB is the Brownian force, FiS is the sum of the spring forces acting on i-th bead, FiEV is the sum of the excluded volume force between beads, and FiEV,wall results from the excluded volume interaction between bead i and the wall. In Eq. 2, the theory of electro-hydrodynamic equivalence35 is employed so the effect of the electric field on DNA can be modeled by a hydrodynamic flow field equal to μbE.
For studies about the dynamic properties of macromolecules such as DNA, it is always a concern for the influence of hydrodynamic interactions (HI). It has been shown by theory36, 37 that velocity disturbance decays as 1/r2 in slit-like confinement, implying the existence of long-ranged HI effect. However, the scaling analysis given by Balducci et al.38 shows that the combination of the algebraic decay of the velocity disturbance and the DNA segment distribution in good solvent results in hydrodynamic screening in the transverse directions in slit-like confinement. In addition to the theoretical analysis, recent experimental studies38, 39, 40, 41 have also supported the scenario of HI screening in such condition. As a result, the assumption of negligible HI has been used in many studies involving DNA in confinement.21, 24, 42 In the current study, because the equilibrium size of λ-DNA is close to the channel height, and the DNA contour length is much larger than the channel height, we also assume that the hydrodynamic interaction can be neglected in our simulations.
To perform the simulations, we non-dimensionalize Eq. 2 using
| (3) |
where l is the maximum extension of a spring (= L/Ns), kBT is the thermal energy, and is set to be the electric field at the inlet of case I. The dimensionless force balance equation becomes
| (4) |
where is the bead Peclet number, D is the diffusivity of the bead (). is the dimensionless Brownian force term which is Gaussian in reality. This term must have a zero mean and its second moment must satisfy the fluctuation-dissipation theorem.43, 44 However, it is difficult to generate the force term obeying Gaussian distribution in simulation. On the other hand, for the purpose of BD simulations a Gaussian distribution and a uniformly distributed random number with zero mean are statistically indistinguishable as long as their second moments are the same. Thus, it is more convenient to represent the term in the simulation with the following function:43, 44
| (5) |
where is a vector with each of the three component uniformly distributed between 1 and −1. The spring force between bead i and j, denoted as , is described by the Marko-Siggia spring law45
| (6) |
where is the number of the true persistence length Ap per spring, is the ratio of the effective persistence length Aeff to the true DNA persistence length. We set Ap to be 0.053 μm, and thus ν is 21.488. Aeff is set to be 0.0735 μm for λ-DNA simulated with 19 beads.25, 46 is the dimensionless distance between bead i and j. The sum of the spring force acting on bead i is calculated from
| (7) |
The dimensionless excluded volume force between beads is modeled by a soft potential expressed as follows:47
| (8) |
where is the dimensionless excluded volume parameter. υEV,p is set to 0.0004 μm.25 In addition to the excluded volume effect between beads, it is also important to consider the excluded volume between the beads and the wall. Here, we model the bead-wall excluded volume effect by the modified Heyes-Melrose algorithm that moves penetrated beads to the nearest wall.25, 48 The displacement vector for the penetrated beads can be expressed as25
| (9) |
where Δpi is the vector from the penetrated bead to the nearest point of the device boundary, Δpi is the magnitude of Δpi, and H denotes the Heaviside step function that is nonzero only for penetrated beads.
To perform BD simulations, we use the explicit Euler’s method to integrate Eq. 4. The dimensionless time step is fixed to be for De ≤ 10. The time step size is adjusted by a factor 10/De for De >10. The simulated radius of gyration of λ-DNA in bulk condition is 0.70 , consistent with experimental value of 0.69 μm. To define the Deborah number in thesimulations, we obtain DNA relaxation time in the 2 μm height channel by fitting the time evolution of the ensemble averaged mean square end-to-end distance of an initially stretched chain at no field condition to a single exponential function25
| (10) |
where is the dimensionless end-to-end distance of a chain, represents an ensemble average of the results from 300 independent runs, is the equilibrium value of , and A is a fitting parameter. In our simulations, we first stretch a DNA chain to 70% of its contour length and let it relax. When the end-to-end distance of the chain becomes smaller than 30% of its contour length, we record as a function of time, and fit its value to Eq. 10 to obtain . The dimensionless relaxation time is 0.52, in very good agreement with the results in literature.25
In our simulations, we define the characteristic De to be the De in the contraction so that the results of all three cases can be compared on the same basis. Since the electric field at the inlet of the contraction is much smaller than that at the outlet, we calculate the characteristic De as , where is electrophoretic velocity at the outlet of the contraction (x/lc = −1).21 The extension of DNA denotes to that is defined to be the distance between the maximum coordinates and the minimum coordinates of the DNA chain.21 We note that all the simulation results presented in the rest of the sections are the ensemble average of 300 independent runs. Each run starts with a different initial configuration in order to include the effect of “molecular individualism.”49 The initial x position of the center of mass of the chains are set to be 168 μm, 205 μm, and 300 μm for case I, case II, and case III, respectively. The initial y position of the center of mass of each chain is evenly distributed along the y-axis at the initial x position so that the inhomogeneity of the electric field is also accounted for in the simulation results.
Finite element method
The electric field in the testing devices is obtained by solving the Laplace equation of the electric potential using finite element method (FEM).33 In this study, the commercial software COMSOL is used to perform the task. The governing equation and boundary conditions are listed as follows:21
| (11) |
| (12) |
where Ω is the problem domain, denotes the boundary, and n denotes the normal vector along the boundary. The boundary condition is imposed at the inlet and the outlet of the device, and is the insulating condition set on the wall of the device. Since the height of the device is constant and the wall of the device is insulated, this problem can be treated 2-dimensionally and triangular elements are used in FEM. The boundary condition at the exit of the device is always set to be zero, while the value of at the inlet of the device is chosen so that the electric field at the exit is equal to . The solved electric field E is stored in a database for later use in BD simulations.
Connecting FEM with BD
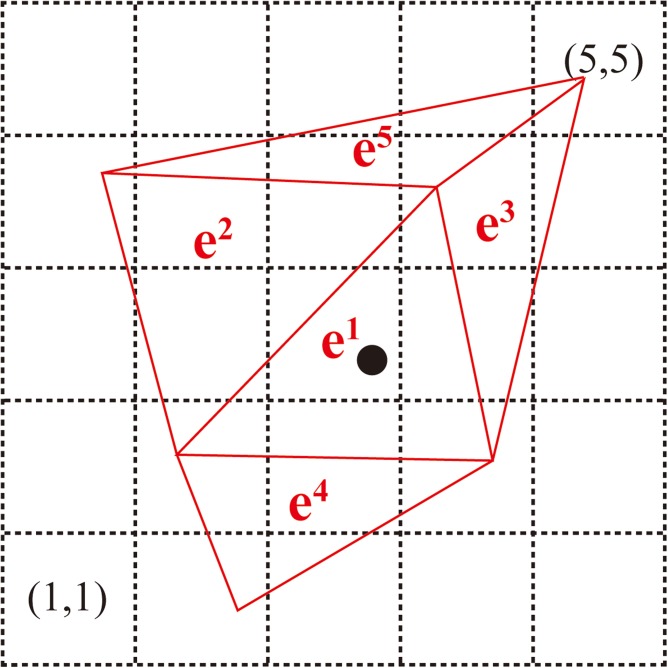
In order to use the electric field solved by FEM in BD simulations, we need to extract the electric field at the position of each bead from the FEM results in every time step. This is trivial if the meshes are rectangular since locating the bead in which element only involves algebra. However, it becomes extremely time-consuming to locate even just one bead from irregular meshes because an intensive searching might be required. Given about 105 elements used in this study, we might need to scan an order of elements in each time step if an explicit searching algorithm is employed. Kim and Doyle25 have developed an efficient “target-induced” searching algorithm based on graph theory. However, it is complicate to implement. Thus, we choose to develop our own searching algorithm that is not only efficient to use but also easy to implement. Our searching algorithm is illustrated in Fig. 2 and described as follows:
-
1.
We first create virtual square meshes over the given device, and construct a map between the square meshes and the irregular elements.
-
2.
To locate a given bead from irregular elements, we first locate the square mesh that contains the bead. This step requires only algebra but no searching.
-
3.
After the square mesh is determined, we consult the map built in the first step and only search the irregular elements that overlap with the square mesh.
-
4.
Once the element that contains the bead is found, we use a 3-node linear shape function to interpolate the electric field at the bead position.
Figure 2.
Demonstration of the method used to locate a given bead from irregular meshes. To use this method, we first create virtual square meshes over the problem domain, and then build a map connecting the square meshes and the irregular meshes. For example, the square mesh (3, 3) contains part of the triangular elements e1 and e2, and the square mesh (4, 4) contains part of the triangular elements e1, e2, e3, and e5. To locate a given bead (black point) in which triangular element, we first locate the bead in the square mesh (3, 3). This step is trivial because it only involves algebra. Since the square mesh (3, 3) only contains the triangular elements e1 and e2, we can determine which triangular element contains the bead within two calculations.
Since we use very small time steps, the beads stay in the same element over many steps. Thus, we only have to do the search once in many steps, and on average we check fewer than 5 elements in each search. This method can also be used for 3-dimensional problems.
Validation of the simulation scheme
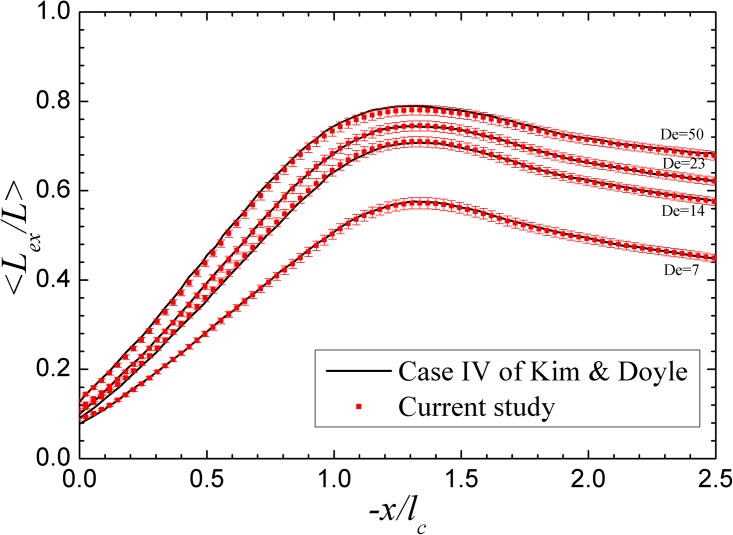
Our simulation scheme is different from that of Kim and Doyle25 in the methods of integration and BD-FEM connection scheme. We verify our simulation scheme by comparing our results with those of case IV in Kim and Doyle.21 Except the time step size, we use the identical parameters. The comparison is shown in Fig. 3 for various values of De, and it is clear that our results agree very well with the original results. All of our simulations are performed on a desktop workstation with an Intel® Core2 Q9400 CPU. The typical run time for one chain passing through the device is several minutes, while it strongly depends on the length of the device and the initial position of the chain.
Figure 3.
Comparison between the simulation results obtained using the method of this study (red symbols) and those of case IV of Kim and Doyle (black solid line).
RESULTS AND DISCUSSIONS
Field kinematics and the evolution of DNA configuration
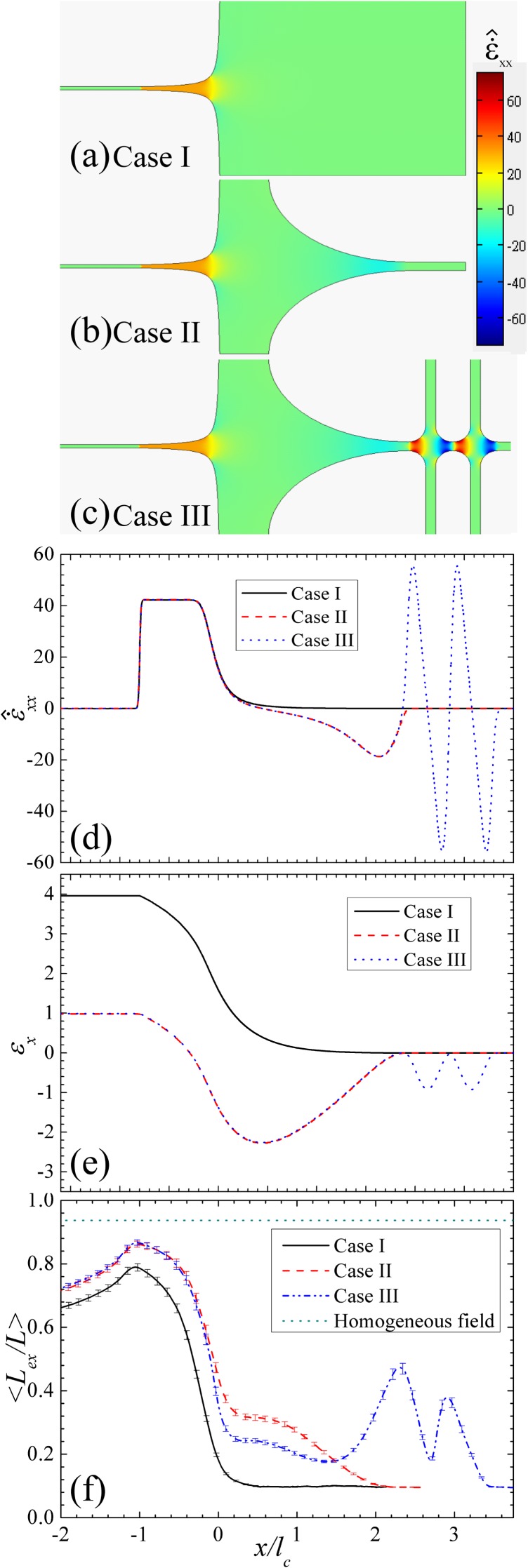
In Fig. 4, we present the analysis of the field kinematics and the evolution of the averaged relative DNA extension in three devices for De = 23. We note that all six subfigures of Fig. 4 share the same x axis so they can be easily crossreferred. Figs. 4a, 4b, 4c are the contour plots of the xx-component of the dimensionless electrophoretic strain rate () in the three devices. Here, is defined by . We are particularly interested in because our primary concern is to improve DNA extension in the contraction that aligns with x direction. From Figs. 4a, 4b, 4c, is zero at the inlets and becomes highly positive in the contraction for all cases. For case II (Fig. 4b), becomes moderately negative at the expansion but soon gets back to zero at the end of the expansion. For case III, while oscillates about zero in the cross-slot region, it is identical to that of case II in the rest of the device. From the field kinematics, the positive value of indicates that DNA will be stretched in x direction and compressed in y direction and vice versa. Thus, we can qualitatively predict the conformation change of DNA in the devices using the field kinematics.
Figure 4.
The field kinematics and the evolution of the average DNA extension in three devices for De = 23. All the subfigures share the same x-axis. (a), (b), and (c) the contour plots of the xx-component of strain rate for case I, case II, and case III, respectively. (d) and (e) and the accumulated x-component of strain calculated along the axis of three devices as function of x. (f) the ensemble averaged relative extension of DNA as a function of x in three cases. The results of homogeneous field are obtained using a dumbbell model by balancing the drag force and the worm-like chain spring force with all other force neglected.
Figs. 4d, 4e display the variation of and the accumulated x-component of the electrophoretic strain along the x-axis of the three devices as a function of x. Here, is defined by . Figs. 4d, 4e provide the electrophoretic strain rate and the electrophoretic strain that a non-Brownian deformable object will experience through the device if it is released at the inlet of the device at y = 0. Although and also vary with y, their sign is only a function of x. Therefore, we choose and calculated along the axis of the devices as the representative quantities for the following discussion. The value of shown in Fig. 4d matches the qualitative analysis based on Figs. 4a, 4b, 4c. Moreover, Figs. 4a, 4b, 4c, 4d show that the field kinematics at the contraction, the expansion and the cross-slots is nearly independent. Thus, the influence of each part of the device to DNA behavior can be independently identified.
From Fig. 4e, we have found that the accumulated electrophoretic strain at the end of the contraction of case II and case III is the same but is less than that of case I. This implies that case II and case III actually provide less strain to stretch DNA than case I does. In other words, when a deformable object passes through either case II or case III, it will experience less deformation in x direction than that in case I. At this point, the two devices equipped with the new pre-conditioning design do not seem able to improve DNA stretching over the original device. However, the prediction based on the kinematics analysis is only accurate when DNA is small comparing to the dimension of the device. The analysis does not consider any “non-local” effect,21 meaning that different parts of DNA could experience qualitatively different field kinematics. This “non-local” effect often has a significant influence on DNA behavior and can only be quantitatively captured using detailed simulations. As we will show in a later section, the “non-local” effect is the key to the preconditioning effect of these devices.
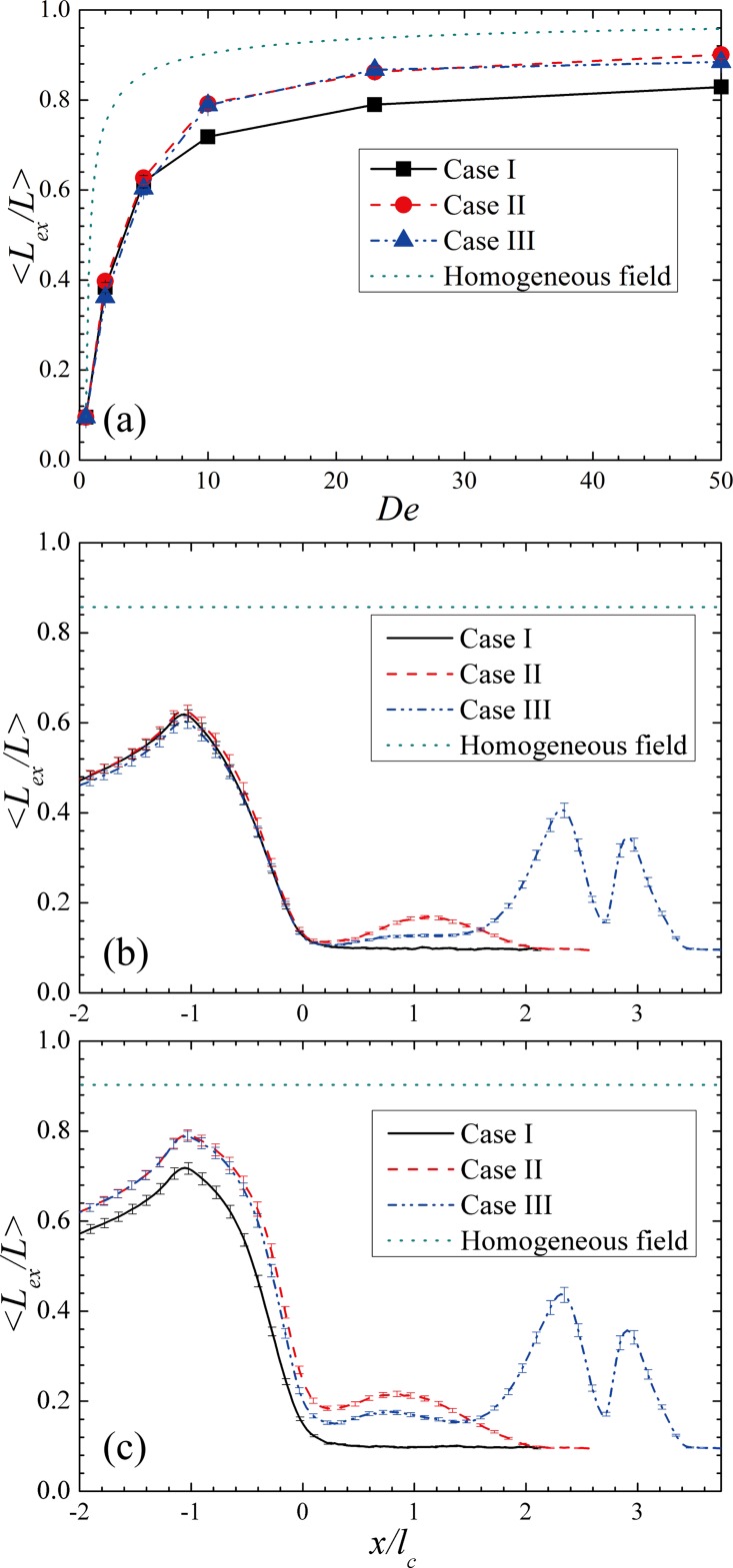
Fig. 4f compares the evolution of the ensemble averaged relative extension of DNA in the three cases for De = 23. We note that the extension of DNA presented in this Fig. 4f is not just the projection of the DNA extension in x direction, but the true DNA extension Lex defined previously. The results of homogeneous field (shown by the dotted line) are obtained using a dumbbell model by balancing the drag force and the worm-like chain spring force with all other force neglected.24 The estimation is obtained under the steady-state assumption. Therefore, it represents the theoretical limit of the DNA extension at the corresponding De. For case I, DNA is found only being stretched in the contraction, consistent with the kinematics of the field. On the other hand, DNA in case II is pre-stretched at the expansion and then being stretched again to a higher degree in the contraction. For case III, the relative extension of DNA first oscillates as they pass the cross-slot region, but then decreases after entering the expansion. The averaged DNA extension rises again from the middle of expansion (around x/lc ≈ 1.5). Finally, DNA enters the contraction and reaches the same maximum degree of extension as DNA in Case II does. For De = 23, the maximum values of the ensemble averaged relative DNA extension reached in case I, case II, case III and homogeneous field are 0.78, 0.86, 0.86, and 0.92, respectively. The improvement of case II and case III over case I is about 8% of DNA contour length. It represents a significant improvement because the maximum possible extension is only 6% away. Moreover, the testing devices could have better performance if they are optimized.
From the results shown in Fig. 4f, it is obvious that case II and case III outperform case I. Based on our earlier analysis, this improvement should be attributed to the preconditioning effects of the new design. However, there are still several questions to be addressed. Firstly, how exactly does the new design precondition DNA? Secondly, why does DNA in case III adopt less extended conformation than those in case II at the inlet of the contraction (x/lc = 0)? Thirdly and more strangely, why is there no difference between the maximum DNA extension achieved in case II and case III?
To understand the pre-conditioning mechanism, we collect the snapshots of DNA conformation in the three devices and present them in Figs. 5a, 5b, 5c. To generate these snapshots, DNA with the same initial configuration is released from the same y position near the inlet of each device. For case I (Fig. 5a), DNA is not stretched until very close to the inlet of the contraction. For case II (Fig. 5b), DNA is first being stretched in y direction at the expansion. However, when DNA is about to leave the expansion, it starts to rotate and gradually aligns with the funnel direction. Before DNA relaxes, it enters the contraction and is stretched again to reach the maximum extension at the outlet of the contraction. For case III (Fig. 5c), DNA is found to be stretched and compressed periodically in the cross-slot region. As DNA leaves the cross-slots and enters the expansion, it is already partially stretched and aligned with x direction. However, the value of is negative in the expansion, and thus DNA has to be compressed before it can be stretched in y direction. This also explains the decrease of DNA extension seen in Fig. 4f between x/lc = 2.5 and x/lc = 1.25. When DNA proceeds further, it shows the same rotational motion as observed in case II. However, DNA in case III generally adopts less stretched conformation at the inlet of the contraction than those in case II.
Figure 5.
The snapshots of DNA conformation in case I(a), case II (b), and case III (c) at De = 23. (d) The magnified view of the rectangular region in (b). The relative magnitude of the electrophoretic velocity components at different parts of DNA is also displayed as using the arrows. The difference in the velocity at the top and the bottom of DNA results in DNA stretching in y direction and rotating clockwise. (e) The magnified view of the rectangular region in (c).
Combining the information presented in Figs. 45, we find that the rotational motion of DNA observed between the expansion and the contraction for both case II and case III is the key to the preconditioning effect. This rotational motion is caused by the difference in the electrophoretic velocity at different parts of DNA. It might be a little confusing that an irrotational filed50 can generate rotational motion of DNA. However, we note that the field is only locally irrotational, while the non-local electrophoretic velocity difference is still capable to cause the rotational motion of a large object, such as an extended DNA. To illustrate this point, we plot in Fig. 5d, the components of the electrophoretic velocity of DNA in the rectangular region of case II. As can be seen, the difference of the y-component of the electrophoretic velocity over the DNA chain results in DNA stretched in y direction. Moreover, DNA segments closer to the center axis of the device experience a stronger electric field in x direction than those farther from the axis. This velocity difference causes DNA to rotate clockwise, and lets the DNA segments closest to the center axis always enter the contraction earlier than the rest segments. This rotational motion allows DNA to preserve the extension originally achieved in y direction to x direction and leads to a higher degree of final DNA extension at the end of the contraction. Without this rotational motion, there will be no preconditioning effect at all. Fig. 5e is the magnified view of the rectangular region in Fig. 5c. It shows the detailed evolution of DNA conformation in the cross-slots region. We can clearly see that the DNA conformation changes periodically and becomes partially stretched and aligned with the channel at the entrance of the expansion.
Probability distribution of DNA extension
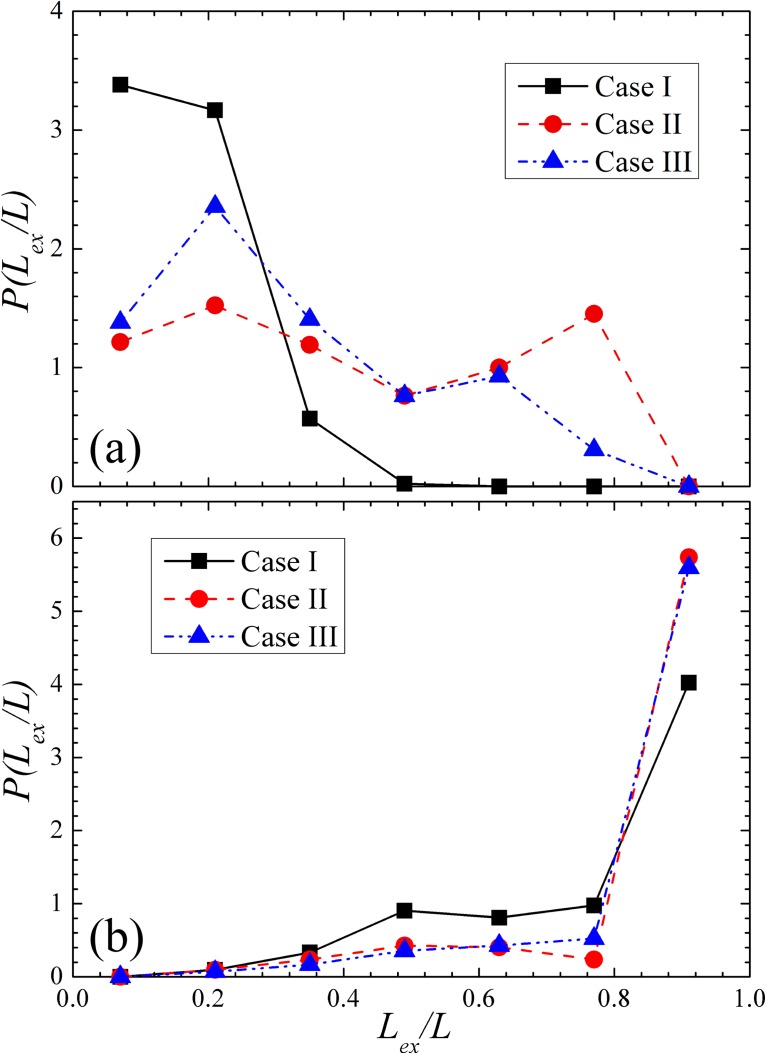
To further understand the preconditioning mechanism, we compare the probability density of the relative DNA extension at De = 23 for all three cases at the inlet (x/lc = 0) and the outlet of the contraction (x/lc = −1) in Figs. 6a, 6b, respectively. Consistent with the results shown in Fig. 4f, Fig. 6a shows that more DNA molecules that undergo pre-conditioning adopt partially extended conformations at the inlet of the contraction. For case II, about half of the DNA population has been stretched over 50% of their contour length. For case III, about one third of DNA population was over 50% stretched. However, Fig. 6b shows that the distribution of DNA extension in case II and case III is undistinguishable at the outlet of the contraction. This is somewhat strange because the field kinematics is the same in the contraction for all three cases. Given that the probability distribution of DNA extension is different at the inlet of the contraction, why would it become identical at the outlet?
Figure 6.
The probability density of relative DNA extension at (a) the inlet and (b) the outlet of the contraction for case I, case II and case III. Data are collected at De = 23.
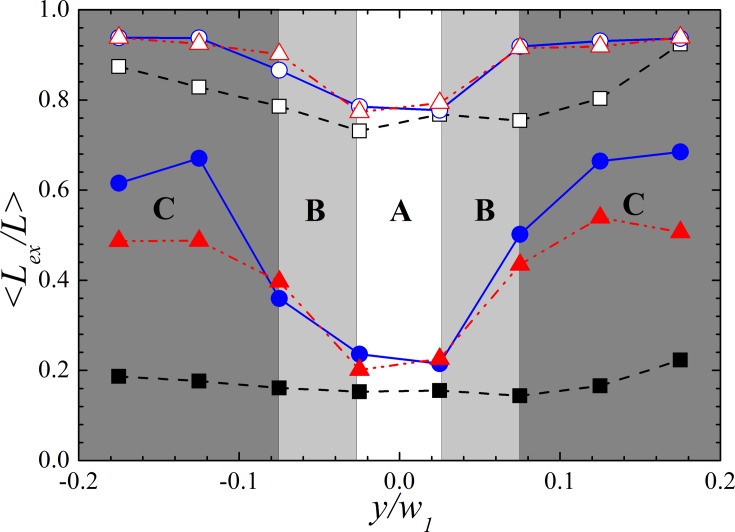
To answer this question, we plot the averaged relative DNA extension at the inlet and the outlet of the contraction as a function of their y-component of the center of mass in Fig. 7. The data shown in the figure are only plotted in the range from y/w1 =− 0.2 to 0.2 because only about 5% of total chain populations pass the rest region and the data collected there suffer from poor statistics. To compare the difference between the three cases, we define three regimes A, B, and C as shown in the Fig. 7. We first focus on the results obtained at the inlet of the contraction. Regime A is around the center axis of the devices. DNA passing this regime has similar extension in all three devices, indicating that little or no preconditioning effect presents. This makes sense because DNA passing this regime is likely to have its segments on both sides of the axis. Due to the symmetry of the field about the device axis, the rotational motion of DNA illustrated in Figs. 5b, 5c will not happen, and consequently case II and case III perform similarly to case I. Regime B is a little farther from the axis. In regime B, DNA extension for case II and case III is very close but significantly higher than that for case I. Regime C is even farther from the axis where DNA extension is highest in case II, middle in case III, and lowest in case I. DNA passing regime B and C has experienced prominent preconditioning effect. When DNA in case III pass the cross-slot regime, those far away from the center axis will be stretched to a higher degree than others close to the axis due to inhomogeneity of the field.21 However, as shown in Figs. 5c, 5e, DNA stretched in the cross-slot will align with x-axis and therefore become more difficult to extend in y direction at the expansion. As a result, these DNA will gain less pre-conditioning effect comparing with those passing the same off-axis region in case II.
Figure 7.
Averaged relative DNA extension as a function of the y-component of their center of mass at (a) the inlet of the contraction (x/lc = 0) and (b) the outlet of the contraction (x/lc =− 1) at De = 23 for case I (▪), case II (•) and case III (▴). The filled and hollow symbols represent the results obtained at the inlet and the outlet of the contraction, respectively. Three regimes A, B, and C are defined for discussion in the context.
We now turn our attention to DNA conformation at the outlet of the contraction. The average extension of DNA in regime A and B is much higher but qualitatively very similar to that at the inlet of the contraction. It is expected because regime A and B are strain-limited regime where the strain provided is not enough for the chains to be fully stretched. In regime C, however, it is found that DNA of case II and case III reach the same degree of extension despite the difference of their states at the inlet of the contraction. This is because DNA chains passing off the contraction axis experience more effective “strain” for stretching.21 Therefore, regime C is actually the strain-abundant regime where preconditioned DNA chains receive more strain than they need to be fully stretched. As a result, DNA molecules passing this regime reach the extension limit regardless the difference in their conformation at the inlet of the contraction. This explains why DNA in case II and case III achieve the same maximum average extension at the outlet of the contraction.
Efficiency of the pre-conditioning strategies at different De number
Fig. 8a compares the maximum relative DNA extension at different De in the three devices. The results of homogeneous field represent the theoretical limit of DNA extension at the corresponding De. From Fig. 8a, we find that whether case II and case III outperform case I depends on De. When De ≤ 5, all three cases yield the identical maximum DNA extension, indicating that the preconditioning effect does not exist at all. When De ≧ 10, the preconditioning effect is prominent but is only weakly dependent on De. To clarify why a threshold De exists for the presence of preconditioning effect, we plot the ensemble averaged DNA extension versus the x-component of the center of mass of DNA for De = 5 and De = 10 in Figs. 8b, 8c, respectively. As can be seen in Fig. 8b, DNA extension in all three devices is identical in the contraction for De = 5. It is because the local De at the expansion is too low to stretch DNA despite that the De is still much higher than one in the contraction. Consequently, the preconditioning effect vanishes. When De is larger than 5, the preconditioning effect presents. However, the improvement from case I to case II and case III changes very little with increasing De. This is because the ability of a device to stretch DNA depends strongly on the effective strain provided by the device but depends only weakly on De once De exceeds a threshold value. Therefore, it is not surprising to find the efficiency of preconditioning does not change much with De for De > 10.
Figure 8.
(a) The maximum ensemble averaged relative DNA extension obtained in the three cases and under homogeneous field at De = 2, 5, 10, 23, and 50. (b) the ensemble averaged relative extension of DNA as a function of x in the three cases at De = 5. (c) the ensemble averaged relative extension of DNA as a function of x in the three cases at De = 10.
Comments on the failure of the “massage” preconditioning approach
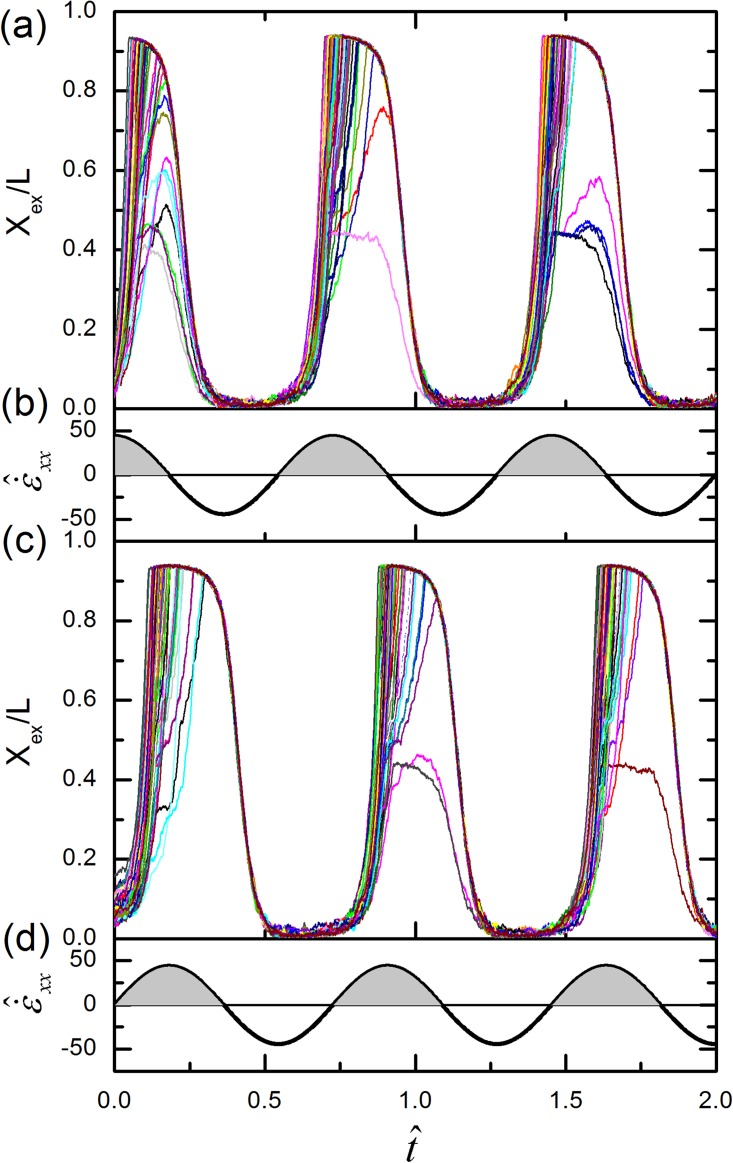
From the discussion of the previous sections, we find that applying an oscillating extensional field to DNA does not yield a net preconditioning effect, at least in the device tested in this study. We were not sure if the discrepancy between our results and the previous prediction22 is due to the specific design of our device or due to other causes. To clarify this issue, we perform the simulations of the original study and present the results in Fig. 9a. The figure resembles Fig. 2 of the original study that shows the relative DNA stretch (Xex/L) versus time for 60 DNA chains under an oscillating planar extensional flow. Slightly different from Lex, Xex is defined by the distance in the flow direction between the downstream-most and upstream-most parts of the molecule. The simulation parameters for DNA are from the current study while the function of the strain rate is taken from the original study. The strain rate of the fluid is described by
| (13) |
Here, , , and are the xx-, yy-, and zz-components of the dimensionless rate of strain tensor, respectively. is the dimensionless amplitude of the oscillatory extension rate, and is set to be 5 as the amplitude of the imposed strain. The results in Fig. 9a are obtained by setting . As shown in the figure, DNA stretch oscillates with time, in accordance with the variation of plotted in Fig. 9b. It can be easily seen that more DNA chains are fully stretched in the second and the third cycles than in the first cycle. This observation leads to the original conclusion that an oscillating planar extensional flow can precondition DNA so that they are less prone to form folds that resist stretching. However, from the strain rate variation presented in Fig. 9b, we notice that DNA chains experience a shorter period of positive strain rate in the first cycle than in the following cycles. More quantitatively, the effective strain provided to DNA in the first cycle is only half of that in the following cycles. Here, the effective strain is estimated by for positive in each cycle, and it is presented graphically in Fig. 9b as the shaded area. Therefore, the preconditioning effect observed in Fig. 9a is actually due to the less strain provided to DNA in the first cycle. To support our point, we redo the simulations with , and display the results in Fig. 9c. The strain rate versus time is presented in Fig. 9d, and the strain for DNA stretching (shaded area) becomes the same in all cycles. As expected, the preconditioning effect no longer exists in Fig. 9c. Thus, we conclude that in general an oscillating extensional flow (or field) does not have the preconditioning effect for DNA stretching.
Figure 9.
(a) DNA extension versus time for 60 chains in an oscillating planar extension flow. (b) the strain rate used to produce the results in (a). The shaded area represents the strain for stretching DNA in each cycle of strain rate variation. (c) and (d) are the same as (a) and (b) with .
CONCLUSIONS
Brownian dynamics-finite element method is employed to evaluate two different conformational preconditioning approaches for improving DNA stretching in a microcontraction. An efficient algorithm connecting BD with FEM has been developed to improve the performance of the simulations. The numerical accuracy of the simulation techniques is verified by comparing our data with the existing simulation results. The “pre-stretching” approach that stretches DNA in one direction and turns it to another direction for a second stretching is found to significantly improve the degree of DNA extension at the exit of the contraction. The success of this approach relies on the rotational motion of DNA caused by the “non-local” effect that is tricky to predict from simple kinematics analysis. On the other hand, the “massage” approach that applies an oscillating extensional field to DNA prior to stretching is found not effective for preconditioning DNA. We identify that the ineffectiveness of the “massage” approach is due to an error made in the original study.
Our group is performing experiments to verify the principal prediction given in this study. We notice that the proposed devices are only the prototypes for testing our ideas, and optimization is still needed to maximize the efficiency. The insight gained from this study can be useful for future design of biomicrofluidic devices for DNA manipulation. We also hope that the current work would be one of the pioneering studies that take advantage of the recent development of simulation techniques to facilitate the design of biomicrofluidic devices for novel applications.
ACKNOWLEDGMENTS
The authors thank the National Science Council of Taiwan for supporting this research under the grant of NSC 99-2221-E-002 -013.
References
- Craighead H., Nature (London) 442(7101), 387 (2006). 10.1038/nature05061 [DOI] [PubMed] [Google Scholar]
- Shendure J. and Ji H. L., Nat. Biotechnol. 26(10), 1135 (2008). 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- Riehn R., Lu M. C., Wang Y. M., Lim S. F., Cox E. C., and Austin R. H., Proc. Natl. Acad. Sci. U. S. A. 102(29), 10012 (2005). 10.1073/pnas.0503809102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viovy J. L., Rev. Mod. Phys. 72(3), 813 (2000). 10.1103/RevModPhys.72.813 [DOI] [Google Scholar]
- Chan E. Y., Goncalves N. M., Haeusler R. A., Hatch A. J., Larson J. W., Maletta A. M., Yantz G. R., Carstea E. D., Fuchs M., Wong G. G., Gullans S. R., and Gilmanshin R., Genome Res. 14(6), 1137 (2004). 10.1101/gr.1635204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K. M., Larson J. W., Yantz G. R., D’Antoni C. M., Gallo M. V., Gillis K. A., Goncalves N. M., Neely L. A., Gullans S. R., and Gilmanshin R., Nucleic Acids Res. 33(18), 5829 (2005). 10.1093/nar/gki895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G. C., Schultz K. M., and Doyle P. S., Lab Chip 6(4), 516 (2006). 10.1039/b515326c [DOI] [PubMed] [Google Scholar]
- Lim S. F., Karpusenko A., Sakon J. J., Hook J. A., Lamar T. A., and Riehn R., Biomicrofluidics 5(3), 034106 (2011). 10.1063/1.3613671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X., Ekong R., Fougerousse F., Rousseaux S., Schurra C., Hornigold N., vanSlegtenhorst M., Wolfe J., Povey S., Beckmann J. S., and Bensimon A., Science 277(5331), 1518 (1997). 10.1126/science.277.5331.1518 [DOI] [PubMed] [Google Scholar]
- Bensimon A., Simon A., Chiffaudel A., Croquette V., Heslot F. and Bensimon D., Science 265(5181), 2096 (1994). 10.1126/science.7522347 [DOI] [PubMed] [Google Scholar]
- Reisner W., Morton K. J., Riehn R., Wang Y. M., Yu Z. N., Rosen M., Sturm J. C., Chou S. Y., Frey E., and Austin R. H., Phys. Rev. Lett. 94(19), 4 (2005). 10.1103/PhysRevLett.94.196101 [DOI] [PubMed] [Google Scholar]
- Tegenfeldt J. O., Prinz C., Cao H., Chou S., Reisner W. W., Riehn R., Wang Y. M., Cox E. C., Sturm J. C., Silberzan P., and Austin R. H., Proc. Natl. Acad. Sci. U. S. A. 101(30), 10979 (2004). 10.1073/pnas.0403849101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle P. S., Ladoux B., and Viovy J. L., Phys. Rev. Lett. 84(20), 4769–4772 (2000). 10.1103/PhysRevLett.84.4769 [DOI] [PubMed] [Google Scholar]
- Ferree S. and Blanch H. W., Biophys. J. 85 (4), 2539–2546 (2003). 10.1016/S0006-3495(03)74676-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. W., Yantz G. R., Zhong Q., Charnas R., D’Antoni C. M., Gallo M. V., Gillis K. A., Neely L. A., Phillips K. M., Wong G. G., Gullans S. R., and Gilmanshin R., Lab Chip 6(9), 1187 (2006). 10.1039/b602845d [DOI] [PubMed] [Google Scholar]
- Hsieh C. C. and Doyle P. S., Korea-Aust. Rheol. J. 20(3), 127 (2008). [Google Scholar]
- Strick T. R., Allemand J. F., Bensimon D., Bensimon A., and Croquette V., Science 271(5257), 1835 (1996). 10.1126/science.271.5257.1835 [DOI] [PubMed] [Google Scholar]
- Perkins T. T., Quake S. R., Smith D. E., and Chu S., Science 264(5160), 822 (1994). 10.1126/science.8171336 [DOI] [PubMed] [Google Scholar]
- Tang J. and Doyle P. S., Appl. Phys. Lett. 90(22), 3 (2007). [Google Scholar]
- Hsieh C. C. and Larson R. G., J. Rheol. 49(5), 1081 (2005). 10.1122/1.2000971 [DOI] [Google Scholar]
- Kim J. M. and Doyle P. S., Lab Chip 7(2), 213 (2007). 10.1039/b612021k [DOI] [PubMed] [Google Scholar]
- Larson R. G., J. Non-Newtonian Fluid Mech. 94(1), 37 (2000). 10.1016/S0377-0257(00)00125-7 [DOI] [Google Scholar]
- Balducci A. and Doyle P. S., Macromolecules 41(14), 5485 (2008). 10.1021/ma8010087 [DOI] [Google Scholar]
- Trahan D. W. and Doyle P. S., Biomicrofluidics 3(1), 012803 (2009). 10.1063/1.3055275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M. and Doyle P. S., J. Chem. Phys. 125(7), 17 (2006). [DOI] [PubMed] [Google Scholar]
- Hsieh C. C., Park S. J., and Larson R. G., Macromolecules 38(4), 1456 (2005). 10.1021/ma0491255 [DOI] [Google Scholar]
- Chun-Cheng Z., Feng J., Qian-Qian C., and Jing-Song Y., Polymer 49(3), 809 (2008). 10.1016/j.polymer.2007.12.013 [DOI] [Google Scholar]
- Hu X., Wang S. N., and Lee L. J., Phys. Rev. E 79(4), 041911 (2009). 10.1103/PhysRevE.79.041911 [DOI] [PubMed] [Google Scholar]
- Zuo C. C., Ji F., and Cao Q. Q., Polymer 50(22), 5326 (2009). 10.1016/j.polymer.2009.09.043 [DOI] [Google Scholar]
- Hsieh C. C. and Larson R. G., J. Rheol. 48(5), 995 (2004). 10.1122/1.1781171 [DOI] [Google Scholar]
- Hsieh C. C., Li L., and Larson R. G., J. Non-Newtonian Fluid Mech. 113(2–3), 147 (2003). 10.1016/S0377-0257(03)00107-1 [DOI] [Google Scholar]
- Wei Y. F. and Hsiao P. Y., Biomicrofluidics 3(2), 022410 (2009). 10.1063/1.3129563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerlind L. J., Applied Finite Element Analysis, 2nd ed. (Wiley, New York, 1984). [Google Scholar]
- Ou J., Carpenter S. J., and Dorfman K. D., Biomicrofluidics 4(1), 013203 (2010). 10.1063/1.3283903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D., Dobrynin A. V., Rubinstein M., and Ajdari A., J. Chem. Phys. 108(3), 1234 (1998). 10.1063/1.475485 [DOI] [Google Scholar]
- Liron N. and Mochon S., J. Eng. Math. 10(4), 287 (1976). 10.1007/BF01535565 [DOI] [Google Scholar]
- Staben E., Zinchenko A. Z., and Davis R. H., Phys. Fluids 15(6), 1711 (2003). 10.1063/1.1568341 [DOI] [Google Scholar]
- Balducci A., Mao P., Han J. Y., and Doyle P. S., Macromolecules 39(18), 6273 (2006). 10.1021/ma061047t [DOI] [Google Scholar]
- Hsieh C. C., Balducci A., and Doyle P. S., Macromolecules 40(14), 5196 (2007). 10.1021/ma070570k [DOI] [Google Scholar]
- Lin P. K., Fu C. C., Chen Y. L., Chen Y. R., Wei P. K., Kuan C. H., and Fann W. S., Phys. Rev. E 76(1), 011806 (2007). 10.1103/PhysRevE.76.011806 [DOI] [PubMed] [Google Scholar]
- Tang J., Levy S. L., Trahan D. W., Jones J. J., Craighead H. G., and Doyle P. S., Macromolecules 43(17), 7368 (2010). 10.1021/ma101157x [DOI] [Google Scholar]
- Trahan D. W. and Doyle P. S., Macromolecules 44(2), 383 (2011). 10.1021/ma1020837 [DOI] [Google Scholar]
- Grassia P. and Hinch E. J., J. Fluid Mech. 308, 255 (1996). 10.1017/S0022112096001474 [DOI] [Google Scholar]
- Larson R. G., J. Rheol. 49(1), 1 (2005). 10.1122/1.1835336 [DOI] [Google Scholar]
- Marko J. F. and Siggia E. D., Macromolecules 28(26), 8759 (1995). 10.1021/ma00130a008 [DOI] [Google Scholar]
- Underhill P. T. and Doyle P. S., J. Rheol. 49(5), 963 (2005). 10.1122/1.2008294 [DOI] [Google Scholar]
- Jendrejack R. M., Schwartz D. C., de Pablo J. J., and Graham M. D., J. Chem. Phys. 120(5), 2513 (2004). 10.1063/1.1637331 [DOI] [PubMed] [Google Scholar]
- Heyes D. M. and Melrose J. R., J. Non-Newtonian Fluid Mech. 46(1), 1 (1993). 10.1016/0377-0257(93)80001-R [DOI] [Google Scholar]
- deGennes P. G., Science 276(5321), 1999 (1997). 10.1126/science.276.5321.1999 [DOI] [PubMed] [Google Scholar]
- Randall G. C. and Doyle P. S., Phys. Rev. Lett. 93(5), 4 (2004). 10.1103/PhysRevLett.93.058102 [DOI] [PubMed] [Google Scholar]