Abstract
Background
MTG16, MTGR1 and ETO are nuclear transcriptional corepressors of the human ETO protein family. MTG16 is implicated in hematopoietic development and in controlling erythropoiesis/megakaryopoiesis. Furthermore, ETO homologue genes are 3'participants in leukemia fusions generated by chromosomal translocations responsible of hematopoietic dysregulation. We tried to identify structural and functional promoter elements of MTG16 and MTGR1 genes in order to find associations between their regulation and hematopoiesis.
Results
5' deletion examinations and luciferase reporter gene studies indicated that a 492 bp sequence upstream of the transcription start site is essential for transcriptional activity by the MTG16 promoter. The TATA- and CCAAT-less promoter with a GC box close to the start site showed strong reporter activity when examined in erythroid/megakaryocytic cells. Mutation of an evolutionary conserved GATA -301 consensus binding site repressed promoter function. Furthermore, results from in vitro antibody-enhanced electrophoretic mobility shift assay and in vivo chromatin immunoprecipitation indicated binding of GATA-1 to the GATA -301 site. A role of GATA-1 was also supported by transfection of small interfering RNA, which diminished MTG16 expression. Furthermore, expression of the transcription factor HERP2, which represses GATA-1, produced strong inhibition of the MTG16 promoter reporter consistent with a role of GATA-1 in transcriptional activation. The TATA-less and CCAAT-less MTGR1 promoter retained most of the transcriptional activity within a -308 to -207 bp region with a GC-box-rich sequence containing multiple SP1 binding sites reminiscent of a housekeeping gene with constitutive expression. However, mutations of individual SP1 binding sites did not repress promoter function; multiple active SP1 binding sites may be required to safeguard constitutive MTGR1 transcriptional activity. The observed repression of MTG16/MTGR1 promoters by the leukemia associated AML1-ETO fusion gene may have a role in hematopoietic dysfunction of leukemia.
Conclusions
An evolutionary conserved GATA binding site is critical in transcriptional regulation of the MTG16 promoter. In contrast, the MTGR1 gene depends on a GC-box-rich sequence for transcriptional regulation and possible ubiquitous expression. Our results demonstrate that the ETO homologue promoters are regulated differently consistent with hematopoietic cell-type- specific expression and function.
Background
The highly conserved ETO (Eight-Twenty-One) corepressor gene family traced to Drosophila ETO homologue Nervy [1] contains the myeloid translocation gene (MTG) 16 (MTG16) (gene name CBFA2T3) or murine ETO-2 (gene name cbfa2t3), MTG8 (gene name CBFA2T1, RUNXT1, ETO) and MTG-related protein 1 (MTGR1) (gene name CBFA2T2). The ETO homologues do not bind directly to DNA but rather function as protein scaffolds and bring about gene repression indirectly through an interplay with multiple transcriptional regulatory proteins [2,3]. MTG16 has been shown to interact with canonical transcription factors such as PLZF, BCL6, TAL1/SCL, Gfi1and Heb [4-9], and MTGR1 with TAL1/SCL [10]. The ETO homologues also recruite chromatin regulating proteins such as nuclear corepressors [3,11,12] and histone deacetylases (HDACs) that catalyze chromatin modifications, resulting in transcriptional repression. Importantly, ETO homologue genes are involved in chromosomal translocations of acute leukemia, MTG16 in the generation of the AML1- MTG16 fusion gene of t(16;21) [13] in patients with therapy-induced leukemia, ETO in the generation of the AML1-ETO fusion gene of t(8;21) [14,15] and MTGR1 in the generation of AML1-MTGR1 fusion gene of t(20;21) [16]. The leukemic fusion proteins so encoded disrupt normal function of transcriptional regulators, to promote dysregulation of hematopoietic cell differentiation [17].
Expression of the MTG16 (murine ETO-2) corepressor is confined to early hematopoiesis whereas MTGR1 is present throughout hematopoietic maturation [18]. Thus, ETO-2 increases during differentiation of murine embryonic stem cells into hematopoietic cells [19] suggesting a role in the development of the blood cell compartment. This is consistent with MTG16 being the most highly expressed ETO homologue gene in the stem/progenitor cell compartment [18]. Furthermore, MTG16/ETO-2 has a role in controlling erythropoiesis and megakaryopoiesis. In this context, MTG16/ETO-2 is incorporated in Ldb1 (LIM domain- binding protein 1) and TAL1 (T-cell acute lymphocytic leukemia protein 1) containing transcription factor complexes [5,8,20,21], mediating transcriptional suppression. As a heteromer with MTG16, also MTGR1 complexes with TAL1 in erythroid cells, enhancing transcriptional repression [10]. The ETO-2 recruitment within nuclear complexes may determine the onset of terminal erythroid/megakaryocytic differentiation [5], while the downmodulation of MTG16/ETO-2 at late stages of erythropoiesis suggests that decreased transcriptional repression is necessary for late, terminal erythroid differentiation to occur. For example, in erythroid MEL cells, induction of differentiation is accompanied by dissociation of nuclear protein complexes concomitant with a decrease in the relative level of ETO-2 [8]. Moreover, shedding the negative regulator ETO-2 [21] transforms the Ldb complex into a positive regulator of final erythroid differentiation, supporting the notion that multi-protein complexes may keep erythroid target genes suppressed until corepressor ETO-2 is eliminated at final differentiation [21]. Similarly, in megakaryopoiesis the ETO-2 expression is restricted to immature cells consonant with a function in repression of genes that should be only terminally expressed [22]. Finally, a physiological hematopoietic role for MTG16 has been proven by results from targeted disruption of murine MTG16 (ETO-2) [23]. Although no complete block in hematopoietic differentiation was discovered by ETO-2 knockout, a requirement was demonstrated for ETO-2 in fate decisions, proliferation and stress- dependent hematopoiesis. As a corroboration of functional differences between ETO homologues, targeted disruption of MTGR1 showed no phenotypic consequences for hematopoiesis [24].
Endogenous cell-type-specific hematopoietic expression of the ETO homologues [18] suggests differences in gene regulation. The ETO promoter is regulated by the GATA transcription factor in erythroid/megakaryocytic cells [25]. We now examined functional promoter elements upstream of the coding sequences for MTG16 and MTGR1. A critical role in transcriptional regulation was shown for an evolutionary conserved GATA binding site of the MTG16 promoter. In contrast, MTGR1 has a strong TATA-less promoter with multiple GC box-rich regions of possible importance for constitutive expression. Our results show that the ETO homologue promoters are regulated differently, consistent with cell-type- specific expression.
Results
Identification of transcriptional start site and sequence analysis of proximal promoters of MTG16 and MTGR1
The MTG16 corepressor is associated with developing hematopoiesis and control of erythropoiesis/megakaryopoiesis. However, transcriptional factors regulating the expression of the MTG16 and MTGR1 genes have not been identified. Therefore, we tried to discover the important cis-acting regions and trans-acting factors regulating expression in hematopoietic cells. First, to find the position of the transcriptional start site for MTG16, we performed 5'- rapid amplification of cDNA-ends (RACE). MTG16-cDNA amplified from erythropoietic HEL cell mRNA showed a complete sequence match with a region of MTG16a transcript (NCBI Ref. Seq: NT_010542.15). The transcription start site (TSS) was determined at -176 bp upstream of the translation start codon ATG at +1 (Figure 1A). The translation initiation in MTG16b is located further downstream to that of MTG16a as determined by alignment of mRNA sequence of MTG16a (NM_005187.5) and MTG16b (NM_175931.2). Inspection of the proximal promoter upstream of the transcription start site start (-176 bp) revealed several consensus binding elements (Figure 1A) such as potential ETS1 binding sites at -491 and at -419; SP1 binding sites at -425 and at -316; GATA binding sites at -356 and at -301; MZF1 sites at -251 and at -191; TFAP2A at -389 (Figure 1A). No canonical TATA or CCAAT boxes were detected in the region near the transcription start.
Figure 1.
Localization and structure of MTG16 promoter. (A) Nucleotide sequence of the MTG16 promoter. Identification of transcription start site (TSS) and amplification of the promoter was performed as described in Methods. Nucleotide +1 is the translational start site (ATG). Putative consensus binding sites for transcription factors identified with MatInspector http://www.genomatix.de//matinspector.html and http://jaspar.genereg.net/ are marked. The promoter lacks TATA and CCAAT boxes, but a GC box (underlined) close to the TSS may be important for start site utilization. (B) Homology search was performed for a stretch of sequence from -251 to -499 human sequencewith NCBI nucleotide blast (Blastn) programme in database for "whole-genome shotgun reads" (wgs) and an evolutionary conserved GATA site was identified (marked between straight lines). The following accession numbers appeared containing homologus sequences, Chimpanzee, b| AACZ03106631.1|; Orangutan gb| ABGA01087101.1|; Gorilla, emb| CABD02143420.1|; Rhesus Monkey, gb|AANU01214553.1|; Marmoset, gb|ACFV01163109.1|; Mouse, ref|NT_078575.6|Mm8_78640_37; Rat, ref|NW_047536.2|Rn19_WGA2058_4. The homology sequences were also found at http://genome.ucsc.edu/cgi-bin/hgBlat.
However, a conserved GC box close to the transcriptional start site (Figure 1A) may regulate transcriptional initiation. Sequence comparisons showed the GATA binding site at -301 to be conserved between eight species (Figure 1B).
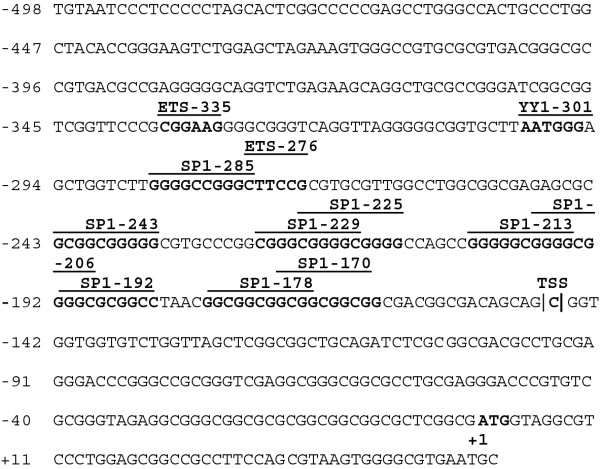
The transcription start site for the MTGR1 gene (NCBI Ref. Seq: NM_001032999.2) is present at -146 bp upstream of the translation start codon [16]. As for MTG16, the proximal promoter of MTGR1 lacks canonical TATA and CCAAT boxes. However, the region between -285 to -170 bp contains multiple GC motifs showing strong homology with the classical GC box, 5GGGGCGGGG3' (Figure 2). These boxes are predicted to compose multiple binding sites for SP1. In addition, ETS1 sites are noted at -335 and -276 as well as a YY1 potential binding sites at -301 bp. The different composition of the proximal promoters of MTG16 and MTGR1 suggests that they are under distinct regulatory mechanisms. While the promoters of both genes lack classical TATA or CCAAT boxes, the presence in the MTGR1 promoter of multiple SP1 sites and a CpG island from -450 to -1 bp covering the transcriptional start site (data not shown) indicate characteristics of a housekeeping gene.
Figure 2.
Localization and structure of MTGR1 promoter. Nucleotide sequence of the MTGR1 promoter. The promoter of MTGR1 was amplified by PCR as described in Methods. Nucleotide +1 is the translational start site (ATG) and transcription start site at -176 is marked (TSS). The MTGR1 promoter lacks TATA and CCAAT boxes but is GC-rich with multiple SP1 binding sites. Putative consensus binding sites for transcription factors identified with MatInspector http://www.genomatix.de//matinspector.html and with http://jaspar.genereg.net/ are marked.
Functional evaluation of MTG16 promoter
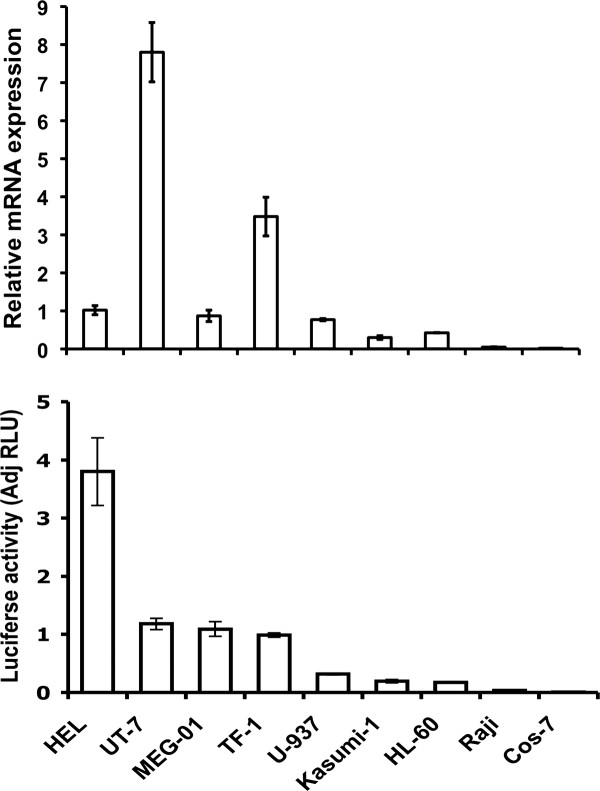
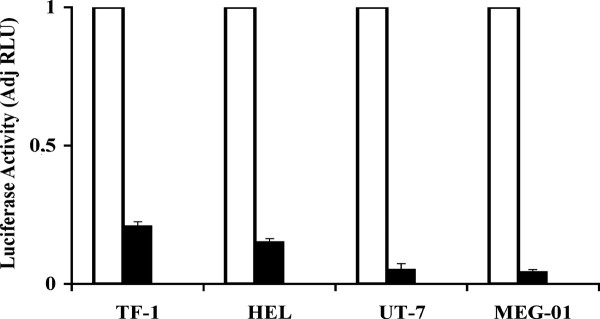
We cloned a -820 to -57 bp region (translational start codon at +1) of the sequence upstream of the transcription start of the MTG16 for determination of transcriptional activity. The cloned region was inserted upstream of the luciferase reporter gene in promoterless pGL3/Basic vector (Promega) to give the pGL3/-820 to -57 plasmid, which was transfected into hematopoietic cell lines and luciferase activity was determined and normalized as described in Methods. pGL3/-820 to -57 showed an approximately 4-fold increased reporter signal as compared to pGL3/SV40-promoter in erythroid HEL cells (Figure 3) indicating strong transcriptional activity in cells expressing the endogenous MTG16 gene. The MTG16 luciferase reporter also showed a strong signal in other erythroid and in megakaryocytic cell lines investigated (Figure 3). However, no direct correlation was observed between luciferase activity and MTG16 transcripts in erythroid and megakaryocytic cell lines. Differences in cellular environment and regulatory elements outside the cloned promoter that modulate endogenous gene expression could explain the lack of direct correlation between promoter activity and gene expression. Promyelocytic HL-60 and acute myeloid leukemia Kasumi-1 cells showed only low luciferase activity; lymphoblastoid Raji cells and monkey kidney COS cells lacked reporter activity. The latter results correlate with low MTG16 transcript levels.
Figure 3.
Reporter gene analysis of the MTG16 promoter. Promoter activity in different cell lines. pGL3 -820-57 (-820) was transfected to indicated cell lines and luciferase activity was determined as described in Methods. The relative MTG16 mRNA expression of the cell lines is also given. Kasumi, HL-60, Raji and COS cells show both the lowest reporter signals and the lowest MTG16 mRNAs.
A GATA consensus binding site is critical for MTG16 promoter function in erythroid/megakaryocytic cells
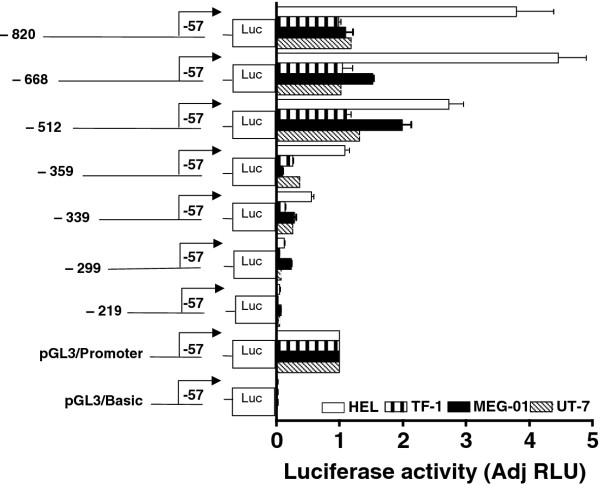
In order to identify important cis regulatory elements, a series of constructs was produced by sequential deletions from the 5' end of the -820 to -57 bp region shown in Figure 4. The deletion constructs were incorporated upstream of the luciferase reporter gene in promoterless pGL3/Basic resulting in the pGL3 -820-57, pGL3 -668-57, pGL3 -512-57, pGL3 -359-57, pGL3 -339-57, pGL3 -299-57, and pGL3 -219-57 reporter constructs, which were transfected into erythroid HEL/TF-1, erythroid/megakaryocytic UT-7 and megakaryocytic MEG-01 cells. Deletion of the region from -820 to -668 did not reduce the transcriptional activity of the promoter, while extended 5'-deletions caused a prominent decrease in luciferase activity (Figure 4). We conclude that the region between -668 and -57 bp is likely to contain the major regulatory cis-elements of the proximal MTG16 promoter, as it was the smallest fragment created with retention of full transcriptional activity (Figure 4).
Figure 4.
Functional analysis of the MTG16 promoter by sequential 5'-deletion. The following 5'deleted reporter constructs were generated from the pGL3 -820-57 (-820) construct and examined after expression in erythroid HEL/TF-1, erythroid/megakaryocytic UT-7 and megakaryocytic MEG-01cells: pGL3 -668-57 (-668), pGL3 -512-57 (-512), pGL3 -359-57 (-359), pGL3 -339-57 (-339), pGL3 -299-57 (-299) and pGL3 -219-57 (-219). Promoterless pGL3/basic and the pGL3/SV40-promoter are used as negative and positive control, respectively. The luciferase activity was normalized against pGL3/SV40- promoter activity. Results are shown for 3 to 5 separate transfections; bars represent the mean and the error bars show SEM.
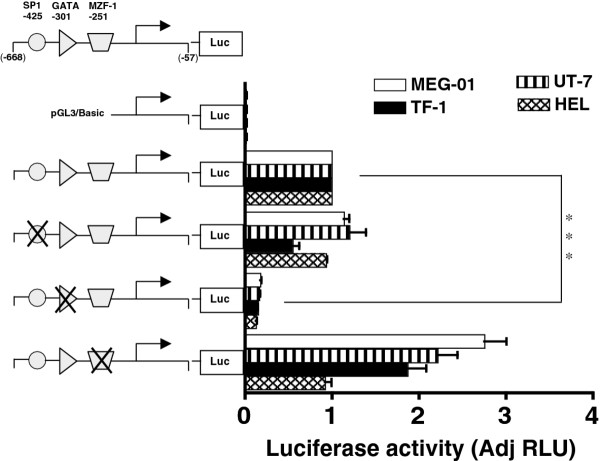
As noted in Figure 1A potential ETS1, SP1, GATA and MZF1 binding sites are detected in the proximal MTG16 promoter. We examined whether these binding sites contribute to promoter transactivation using site-directed mutagenesis. Most notably, disruption of the GATA-301 site led to a 4 to 10-fold reduction in luciferase reporter gene activity in either HEL, TF1, UT-7 or MEG-01 cells relative to intact MTG16 promoter (P < 0.0001) (Figure 5), indicating this GATA site as an important cis-factor for MTG16 gene expression. Furthermore, the GATA -301 site is highly conserved among species (Figure 1B) supporting its functional importance. Moreover, GATA factors are known to play an important role in erythroid/megakaryocytic differentiation [26-28]. Our results suggest that the GATA -301 site may be involved in transcriptional activation in erythroid/megakaryocytic cells.
Figure 5.
Functional analysis of putative transcription factor binding sites in the MTG16 promoter. Putative Sp1 (-425), GATA (-301) and MZF-1 (-251) sites in the -668-57 (-668) promoter were individually destroyed by site directed mutagenesis as decribed in Methods. Erythroid HEL/TF-1, erythroid/megakaryocytic UT-7 and megakaryocytic MEG-01cells were transfected and luciferase activity was determined. The pGL3/basic and pGL3/SV40-promoter are used as negative and positive control, respectively. The luciferase activity of mutated MTG16 promoter is normalized against luciferase activity of wildtype MTG16 promoter.
The GATA-301 site is necessary but not sufficient for transactivation of the MTG16 promoter as some deletion constructs (-359, -339) that retain the GATA -301 site do not retain transcriptional activity (Figure 4). However, all mutation experiments were done with the longer -668 construct and its promoter activity was blocked completely by mutation of the GATA-301 site. An element upstream of the GATA-301 site that cooperates with GATA bound to -301 might explain our results. However, no such element was identified as disruption of all upstream potential binding sites that were identified e. g. the SP1 -425, ETS1 -491, ETS1 -419, SP1 -316, and GATA -356 sites did not affect the reporter signal (data not shown). Disruption of the MZF1 -191 sites downstream of the GATA-301 site did not affect the luciferase signal either (data not shown). Mutation of the downstream MZF1 - 251 increased the signal except in HEL cells (data not shown), suggesting that this site possibly mediates binding to some transcriptional repressor.
GATA-1 binds in vitro/in vivo to the -301 consensus site in the MTG16 promoter
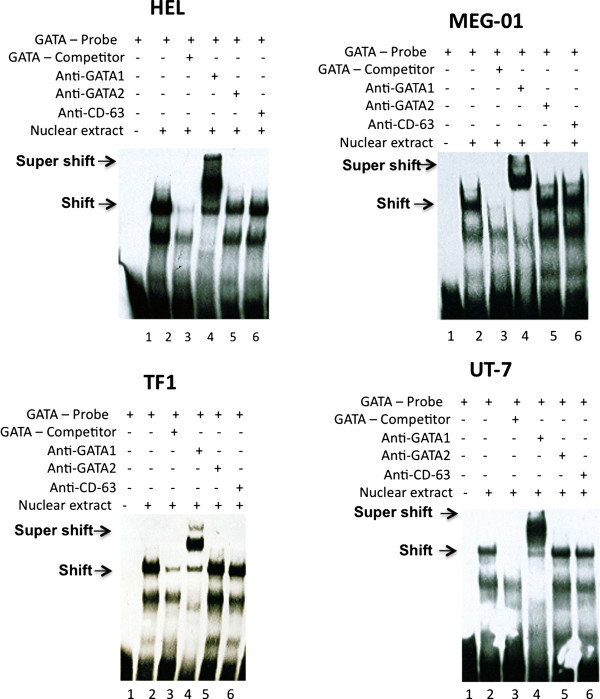
The finding that destruction of the conserved GATA (-301) site reduced gene promoter activity (Figure 5) suggested binding of trans-acting factors. Therefore, Electrophoretic Mobility Shift Assays (EMSA) and antibody supershift assays were performed to examine whether the -301 site binds GATA protein in vitro. A double-stranded biotin-labeled oligonucleotide probe (see Methods) was prepared for the core consensus GATA -301 site, the sequence and location of which is shown in Figure 6. The oligonucleotide probe was incubated with nuclear extracts prepared from HEL, TF-1, UT-7 or MEG-01 cells to examine interactions between probe and proteins in vitro. Nuclear extract proteins from all cell types showed a shift, indicating binding of protein to the biotinylated probe (Figure 6). Binding was specific, as shown by absence of shift in the presence of excess unlabeled competitor probe. Protein bound to the GATA-301 probe was "super-shifted" by anti-GATA-1 antibody, but not with control anti-CD63 antibody (Figure 6). Thus, our results demonstrate that GATA-1 can bind to the consensus GATA-binding site (-301) within the MTG16 promoter. No supershift was observed by anti-GATA-2 antibodies (Figure 6) despite testing of several different antibodies (data not shown), suggesting lack of strong binding of GATA-2 to the consensus site. This finding is similar to previous results from the characterization of the ETO promoter, in which a GATA-site binds GATA-1 but not GATA-2 [25].
Figure 6.
Detection of DNA-protein interactions using electrophoretic mobility shift/supershift assays in vitro of consensus binding sequences in the 5'promoter of MTG16 and nuclear extracts. The following sequence of the oligonucleotide probe was used in EMSA for the conserved GATA (-301) core consensus site (red): GATA-301 probe 5'- CCCGGCATTATCACGGGGACAC. Results are given for HEL/MEG-01/TF-1/UT-7 cells. Primary DNA-nuclear protein interactions are shown by arrows marked shift; DNA- nuclear protein-antibody interactions are shown by arrows marked supershift. All cell lines tested showed identical. A shift is shown for the GATA -301 probe that is competed for by excess unlabelled probe (competitor) (3) indicating binding of nuclear extract protein to the biotinylated probe that containing the the GATA -301 sequence. Proteins bound to the probe were "super-shifted" by antibody to GATA-1 (4) but not with antibody to GATA-2 (5). All experiments were repeated at least twice.
In addition to the specific shift, another band is clearly visible in the EMSA (Figure 6). This band-shift is unspecific since it is unaffected or only weakly affected by the competitor probe. However, it obviously contains GATA-1 protein as it is lost by supershifting with anti GATA-1 (Figure 6).
The SP1 -425 and the MZF1 -251 binding sites of the MTG16 promoter were also examined by EMSA. No supershifting was observed by appropriate antibodies suggesting lack of binding of these transcription factors to the respective consensus binding sites (data not shown).
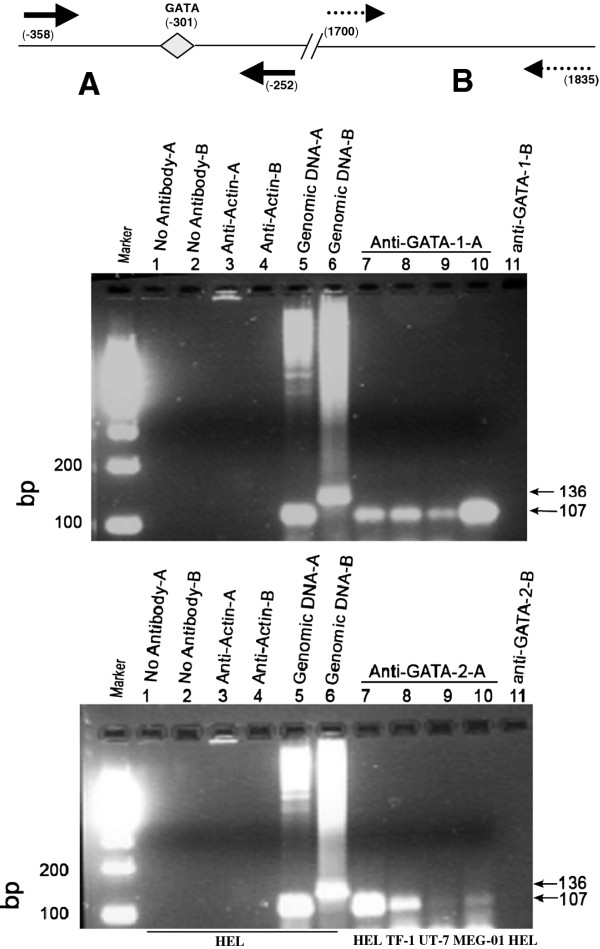
Chromatin immunoprecipitation (ChIP) was used to determine whether GATA-1-2 bound to the putative MTG16 gene promoter in vivo. The assays were carried out with chromatin isolated from HEL, TF-1, UT-7 or MEG-01 cells and antibody towards chromatin-bound GATA-1 or GATA-2. Precipitated DNA was identified by PCR amplification of the MTG16 promoter fragment with gene specific oligonucleotides. By the use of primers specific for the -358 to -252 region, which flank the conserved GATA (-301) site, a 107 bp PCR product was generated using precipitation with either anti-GATA-1 or anti-GATA-2 (but not with anti-actin antibody or with no antibody, serving as negative controls) of all the cell lines examined (Figure 7). Control precipitation with anti-GATA-1 or anti-GATA-2 and amplification of a downstream control region was negative. Thus, specific amplification was achieved after precipitation with both anti-GATA-1 or anti-GATA-2.
Figure 7.
Chromatin immunoprecipitation (ChIP) assay for examining interactions in vivo of consensus binding sequences in the 5'promoter of MTG16. The forward and reverse primers used to amplify the proximal promoter region from -358 to -252 (primers A, solid arrows) and forward and reverse primers for a downstream region from 1700 to 1835 as control (primers B, dashed arrows) are shown. ChIP assays were carried out as described in Methods using chromatin isolated from erythroid HEL/TF-1, erythroid/megakaryocytic UT-7 and megakaryocytic MEG-01cells. PCR products were separated on a 2% gel and representative results are shown. Lane 1-2, no antibody and primers A or B; lanes 3-4, actin antibody and primers A or B; lanes 5-6, genomic DNA and primers A or B; lanes 7-11, GATA-1(top) or GATA-2 (bottom) antibody and primers A or B (lane 11). By use of the specific primers, a PCR product is generated both from the anti-GATA-1 and the anti- GATA-2 immunoprecipitated chromatin. No amplification is seen without antibody or in the presence of anti-actin. All experiments were repeated at least twice.
Taken together, the results from EMSA/supershift assays demonstrate GATA-1 binding in vitro to the GATA -301 binding site and results from ChIP assays demonstrate binding in vivo of both GATA-1 and GATA-2 to the putative MTG16 promoter. This result further supports a functional role at least for GATA-1 in activation of the MTG16 promoter, consistent with the results of the mutagenesis studies shown in Figure 5.
Overexpression of GATA-1
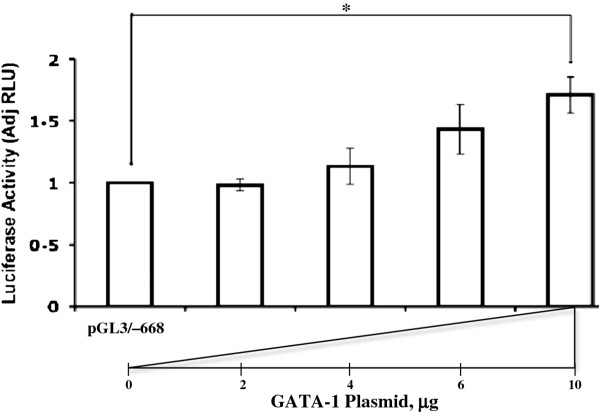
GATA-1 was transiently overexpressed to examine its effect on co-transfected MTG16 promoter activity in erythroid/megakaryocytic cells. The reporter signal was increased by overexpression in megakaryocytic MEG-01 cells (p < 0.05) (Figure 8). The specificity of this result is supported by lack of impact by GATA-1 overexpression on the Renilla control (data not shown).
Figure 8.
Effects on the MTG16 promoter activity of GATA-1 overexpression. MEG-01 cells were co-transfected with 15 μg MTG16 -668 to -57 bp promoter plasmid and increasing amounts of GATA-1 plasmid. The luciferase activity is normalized against Renilla and the activity of the MTG16 -668 to -57 bp promoter. The MTG16 promoter is activated by GATA-1 overexpression. The data are from 3 separate experiments. Bars represent the mean and the error bars show SEM. Statistically significant differences are marked by star.
Repression of GATA-1 by siRNA and inhibition of GATA-1 transcriptional activity with HERP2 leads to decreased MTG16 promoter activity
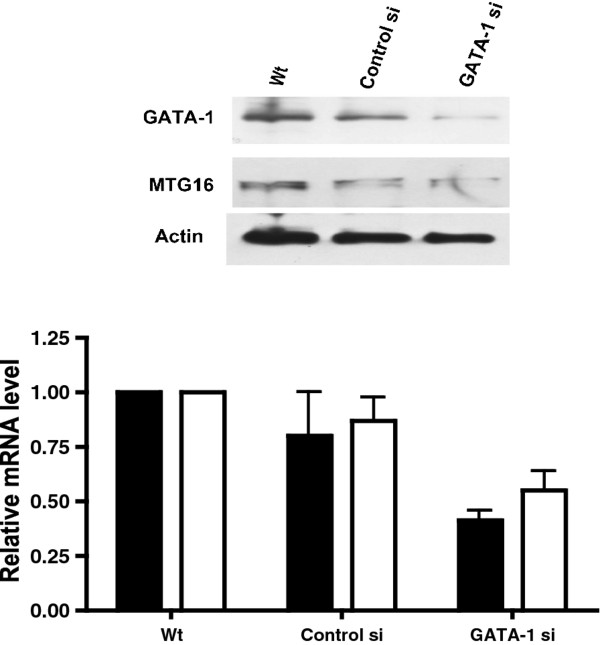
To verify a role of GATA-1 in MTG16 expression, siRNA was transfected into HEL cells as described in Methods. Introduction of GATA-1 siRNA led to a decrease by approximately 50% of both GATA-1 and MTG16 mRNA levels and the results were confirmed on the protein levels (Figure 9), indicating a role of GATA-1 in MTG16 transcriptional regulation. Three different siRNA constructs, all bioinformatically (Rosetta algorithm, SIGMA) designed to minimize off-target effects were used. However, only one resulted in a robust suppression of GATA-1 protein. Off-target effects can therefore not be excluded. However, in combination with our results from reporter experiments, EMSA, ChIP, and overexpression, we believe that we can draw the conclusion that GATA-1 is a positive trans-acting factor on the promoter. Additional support for a role of GATA-1 was obtained by use of the basic helix-loop-helix transcription factor HERP2, which shows physical interaction with GATA-1 and represses GATA-1-mediated transcriptional activation [29]. Both erythroid and megakaryocytic cell lines cotransfected with HERP2 showed strong inhibition of the MTG16 promoter reporter (Figure 10) consistent with a role of GATA-1 in transcriptional activation of MTG16.
Figure 9.
Effects on the MTG16 promoter activity of GATA-1 siRNA. Transfection was as described in Methods. The relative mRNA expression of GATA-1 (black bar) and MTG16 (white bar) is given for HEL cells (wt), HEL cells transfected with non silencing scrambled control siRNA (control si) and HEL cells transfected with siRNA for GATA-1 (GATA-1 si). The protein expression of GATA-1 and MTG16 are shown with GAPDH as acontrol of equal loading. The experiments were performed in triplicate. Bars represent the mean and the error bars show SEM.
Figure 10.
Inhibition of the MTG16 promoter activity through inhibition of GATA-1 by HERP2. TF-1, HEL, UT-7, or MEG-01 cells were co-transfected with 10 μg MTG16 -668 to -57 bp promoter and the HERP2 plasmid construct. White bars show MTG16 -668 to -57 bp promoter activity without HERP2. Black bars show activity after co-transfection with HERP2, relative to the luciferase activity of the MTG16 -668 to -57 bp promoter in the absence of HERP2 (white bars). All cell lines examined demonstrated repression of GATA-1 driven MTG16 luciferase promoter. Renilla was not affetected by HERP2. The experiments were performed in triplicate. Bars represent the mean and the error bars show SEM.
Functional evaluation of the MTGR1 promoter
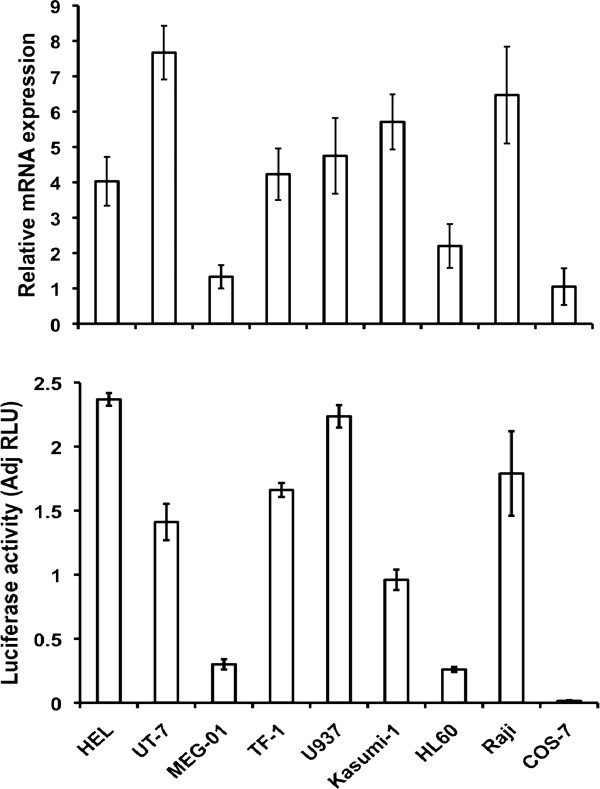
We cloned a -989 to +52 bp region (translational start codon at +1) for examination of transcriptional activity. The cloned region was inserted upstream of the luciferase reporter gene in promoterless pGL3/Basic vector to give the pGL3/-989 to +57 plasmid, which was transfected into hematopoietic cell lines and luciferase activity was determined and normalized as described in Methods. An approximately 2-fold luciferase signal in HEL cells, as compared to the positive control pGL3/promoter, indicated strong transcriptional activity of the cloned region (data not shown). Upon transfection of the shortest luciferase construct with full promoter activity (pGL3 -499 to +52 bp) (see Figure 11 below) to a panel of hematopoietic cell lines, different activities were revealed (Figure 12). As with the MTG16 promoter, the strongest activity was found in HEL cells, but the activity patterns between the two promoters were not uniform (compare Figure 3 to Figure 12). The MTGR1 promoter construct produced a signal in all cell lines examined except for COS cells. All cell lines examined expressed endogeneous MTGR1 mRNA supporting ubiquitous expression (Figure 12).
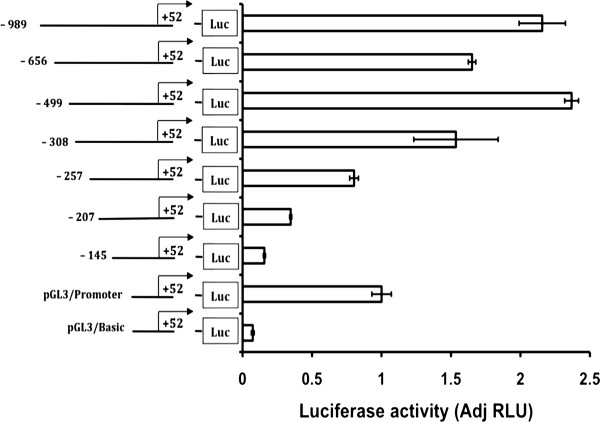
Figure 11.
Reporter gene analysis of the MTGR1 promoter. Functional analysis by sequential 5'-deletion. The 5'deleted MTGR1 reporter constructs -989 to +52 (-989), -656 to +52 (-656), -499 to +52 (-499), -308 to +52 (-302), -257 to +52 (-257), -207 to +52 (-257), and -145 to +52 (-145) were examined after expression in HEL cells. The transcriptional activity is retained by the -499 to +52 bp region indicating the location of the proximal MTGR1 promoter. The pGL3/basic and pGL3/SV40-promoter are used as negative and positive control, respectively in (A) and (B). The luciferase activity was normalized against pGL3/SV40-promoter activity and is shown for 3 to 5 separate transfections; bars represent the mean and the error bars show SEM.
Figure 12.
MTGR1 promoter activity in various cell lines. pGL3/-499 to +52 (the shortest construct with full promoter activity) was transfected to indicated cell lines and luciferase activity was determined as described in Methods. The relative MTGR1 mRNA expression of the cell lines is also given. The promoter shows a 1 to 2.5-fold increased reporter signal in most cell lines. MEG-01, HL-60 and COS cells show both the lowest reporter signals and the lowest MTGR1 mRNAs.
To define the promoter in more detail, a series of constructs was generated by sequential deletions from the 5'end of the -989 +52 bp region after which transcriptional activity was analyzed in HEL cells (Figure 11). Deletion to -499 did not affect transcriptional activity, but further deletion reduced the luciferase reporter expression. Thus the sequence from -499 to +52 bp retained full transcriptional activity and therefore is likely to contain the core proximal MTGR1 promoter. Further sequential 5' deletions of the -499 to +52 bp region resulted in a step by step decreasing luciferase reporter signal (Figure 11) suggesting cooperation between factors binding to different regions of the promoter.
Site-directed mutagenesis was used to examine whether potential transcription factor binding sites contributed to promoter transactivation. The -308 to +52 bp region retains approximately two thirds of the transcriptional activity of the fully active promoter as was shown in Figure 11. This region is GC box-rich with 9 SP1 consensus binding sites (Figure 2) located immediately upstream of the transcription start site at -146 bp consistent with the housekeeping gene character of MTGR1 with ubiquitous expression. However, disruption of individual SP1 sites; SP1-285, SP1-243, SP1-229, SP1-225, SP1-213, SP1-206, SP1-192, SP1-178, SP1-170; did not affect reporter gene activity relative to intact MTGR1 promoter (data not shown), suggesting that SP1 is not important for promoter activity. However, it is possible that multiple active SP1 binding creates a redundancy in SP1 binding and elimination of several SP1 binding sites is needed to reveal their functional relevance. Disruption of the -335 ETS1, the -276 ETS1 and the -301 YY1 sites did also not affect promoter activity (data not shown). Overall, our results indicate that a GC-box-rich sequence with multiple SP1 sites is important for transcriptional regulation of MTGR1 promoter.
AML1-ETO represses MTG16 and MTGR1 gene reporters
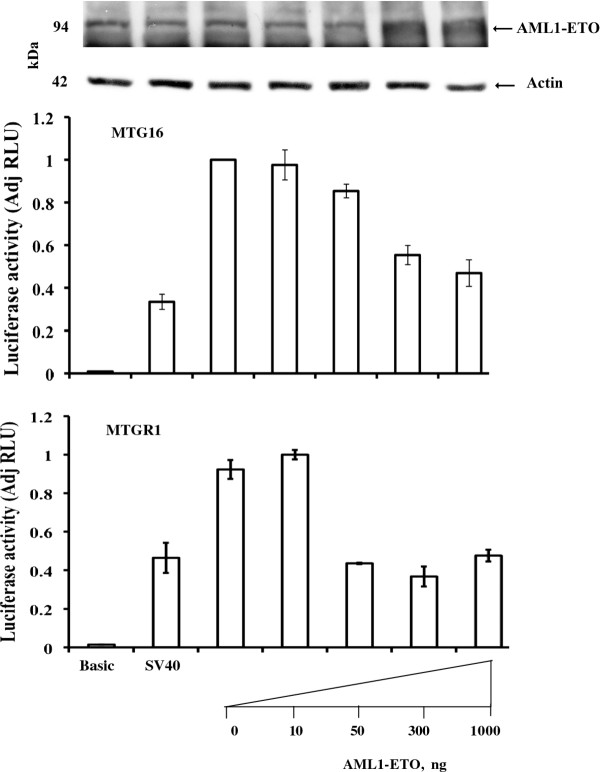
The AML1-ETO fusion protein, which is a gene product of t(8;21) of acute leukemia [14,15] binds the promoter region of many genes causing transcriptional suppression [30]. AML1- ETO has been shown to suppress an ETO promoter reporter in erythroid/megakaryocytic cells [25]. Therefore, AML1-ETO was transiently expressed in HEL cells to examine also the effect on co-transfected MTG16 and MTGR1 promoter reporters. In both cases the reporter was strongly repressed in a dose-dependent manner by AML1-ETO (Figure 13). This finding is not directly consistent with the inhibition observed in vivo in AML1-ETO-positive Kasumi cells of only MTG16 (Figure 3) but not MTGR1 transcripts (Figure 12) suggesting influence of cell-specific contexts.
Figure 13.
Effects of AML1-ETO on the MTG16 and MTGR1 promoter reporter. Erythroid HEL cells were co-transfected with 15 μg MTG16 -668 to -57 bp promoter plasmid (-668) or MTGR1 -499 to +52 bp promoter plasmid (-499) and 0 to 1 μg of AML1- ETO plasmid. pGL3/Basic (Basic). pGL3/SV40 (SV40). The luciferase activity is normalized against MTG16 -668 to -57 bp or MTGR1 -499 to +52 bp promoter. Both promoters are repressed in a dose-dependent manner by AML-ETO. Western blotting shows exogeneous AML1-ETO expression detected with anti-MTG. The experiments were repeated three times with similar results.
Discussion
The negative transcriptional regulator ETO was originally discovered as the 3' participant in the leukemia chromosomal translocation t(8;21) [31]. Now, ETO and its homologues MTG16 and MTGR1 are known to be involved in transcriptional regulation of genome-wide targets. To explain cell-type-specificity of ETO homologue expression and function [18] exploration of promoter regulation is warranted. The human ETO proximal promoter is regulated by the GATA-1 transcription factor in an erythroid/megakaryocytic-specific manner [25]. Here, we continued to identify essential cis-acting elements and trans-acting factors also of the promoters of MTG16 and MTGR1 within human hematopoietic cells.
MTG16 promoter regulation
TATA and CAAT-like promoter motifs facilitate interactions with general transcription factors and are thus involved in initiation of transcription. The proximal MTG16 promoter investigated lacks these motifs. GC-boxes are known to regulate the utilization of start sites in TATA-less promoters [32-35]. Therefore, the conserved GC-box situated close to start site (Figure 1A) may be involved in transcriptional initiation. Among hematopoietic cell lines, the promoter activity was strongest in the erythroid HEL/TF-1, erythroid/megakaryocytic UT-7 and megakaryocytic MEG-01 cells consistent with a role of MTG16 in erythropoiesis/megakaryopoiesis [5,8,20,21]. Deletion analysis of the proximal MTG16 promoter defined a 492 bp region upstream of the transcription start to be required for maximal promoter activity. This region includes a potential GATA binding site (-301), which is strongly conserved between species. Disruption of the GATA -301 binding site, repressed transactivation of the MTG16 promoter. The importance of this cis-acting binding site was confirmed by results from examination in vitro with EMSA, which revealed specific GATA-1 binding. Further, in vivo binding of both GATA-1 and GATA-2 to this region of the promoter was confirmed by ChIP/PCR analysis. Taken together these results demonstrate binding of the GATA-1 transcription factor to the MTG16 promoter and strongly support a role for GATA-1 in transcriptional regulation of the MTG16 gene in hematopoietic cells.
MTGR1 promoter regulation
A 499 bp region upstream of the translation start site of MTGR1 was defined as sufficient for maximal activity in erythroid HEL cells. Upon analysis using different cell lines, we found that the promoter is active in most of them, which is in agreement with ubiquitous presence of native MTGR1 gene product. Furthermore, the lack of classical TATA or CCAAT boxes, the presence of multiple SP1 sites and a CpG island covering the transcriptional initiation site of the MTGR1 promoter (Figure 2) are similar to characteristics of a housekeeping gene [36], expected to be generally active in a non-tissue specific manner. Few potential binding sites for known transcription factors were found in the -499 promoter, rather a GC-box-rich sequence stretch with multiple SP1 binding sites is a prominent part of the promoter. Importantly, the GC-box-rich region was responsible for most of the transcriptional activity. However, we were not able to inhibit promoter activity by the elimination of individual SP1 binding sites. Possibly, individual SP1 binding sites may be equally involved in guaranteeing constitutive MTGR1 expression. Further 5'deletion of the -499 promoter region showed gradually decreasing promoter activity observed upon successive deletions, indicating transcriptional cooperation between promoter sequences consistent with cooperating SP1- binding sites.
ETO homologues in hematopoiesis
The regulation of both the ETO [25] and the MTG16 promoter (this work) by GATA-1 is consistent with a role of human ETO/MTG16-mediated gene suppression at a phase of erythropoiesis/megakaryopoiesis when GATA-1 levels are high [37,38]. However, despite shared trans-activation, MTG16 and ETO are differently expressed in human hematopoiesis [18]; MTG16 is downmodulated at late stages of erythropoiesis whereas ETO is expressed transiently in (human) erythropoiesis. Therefore, cell-type-specific expression of these ETO homologues may be governed also by trans-activating factors other than GATA. Although both MTG16 and ETO are expressed in human erythroid cells [18] yet the role of ETO in murine erythropoiesis is not yet supported [10].
GATA-1 and GATA-2 are expressed reciprocally during development of the erythropoietic lineage, GATA-2 decrease is followed by GATA-1 increase [39]. GATA-2 seems to be essential for the development of early and definitive erythropoiesis/megakaryopoiesis and GATA-1 for terminal erythroid/megakaryocytic maturation [26-28]. The present results are consistent with upregulation of MTG16 as a result of rising GATA-1 levels. Furthermore, results from genome-wide analyses [40] are consistent with early GATA-2 induction of murine ETO-2 during erythropoiesis. These analyses also showed that most occupied sites are located away from promoters indicating that GATA factors may be involved in long- range control by occupancy at non-promoter regions.
Our finding of in vitro binding of GATA-1 but lack of GATA-2 binding to the GATA -301 binding site raises questions about the relative importance of GATA-1/GATA-2 in MTG16 promoter regulation. GATA-2 is reported to contribute in hematopoietic stem cell and progenitor survival [41] and GATA-1 in erythroid cell survival [42,43]. Furthermore, genetic complementation studies show that GATA-1 can replace GATA-2 [44,45]. A lack of binding in EMSA experiments suggests that GATA-2 may not be a primary transcription factor to initiate nucleation of the transcription complex at the MTG16 promoter. Possibly, GATA-2 has a weaker binding affinity than GATA-1. However, our finding in ChIP assay of the binding of both GATA-1 and GATA-2 to MTG16 promoter suggests that GATA-1 and GATA-2 cooperate. This is supported by results from in vivo studies by ChIP-seq with human K562 erythroleukemia cells [40] showing chromatin occupancy by both GATA-1 and GATA-2 at the conserved GATA motif corresponding to the -301 site of the MTG16 promoter. In addition, a role of GATA-2 in stimulating murine ETO-2 expression was also suggested [40]. Thus, GATA-2 was found to occupy a conserved GATA binding site in the murine erythroid G1E cell, which lack GATA-1 expression; the occupied murine site corresponds to the human -301 GATA binding site of MTG16 promoter. Overall, available data suggest a role of both GATA-1 and GATA-2 in MTG16 promoter activation.
Murine MTG16 (ETO2) is involved in erythroid progenitor expansion [5], which is coupled to inhibition of differentiation. This is also the case in megakaryopoiesis where ETO2 represses inappropriate early expression of terminal genes [22] coupled to restrained differentiation of immature megakaryocytes. MTG16 and ETO show reciprocal expression during in vitro-induced human erythropoiesis; MTG16 decreases and ETO increases during the peak of erythropoiesis [18]. As both promoters are regulated by GATA-1, specific negative autoregulation of MTG16 [40] may explain the difference. This is consonant with a role for MTG16 in repressing genes associated with terminal erythroid differentiation thereby inhibiting differentiation and a role for ETO in repressing genes associated with self-renewal and proliferation thereby supporting erythroid differentiation.
Suppression of MTG16/MTGR1 promoter by AML1-ETO
The leukemic AML1-ETO fusion protein [14,15] binds to the promoter region of a number of genes resulting in transcriptional suppression [30]. However, some genes are affected by AML1-ETO in an indirect manner and do not bind the fusion protein directly [30,46]. The observed suppression of the MTG16 and the MTGR1 promoter by AML1-ETO is likely to be indirect as binding sites for AML1 were not detectable on the promoters. Furthermore, a potential mechanism for the MTG16 promoter inhibition by AML1-ETO would be suppression of GATA-1 through impaired acetylation [47]. Suppression of MTG16 is consistent with the low MTG16 promoter reporter signal observed in AML1-ETO-positive Kasumi cells, which showed very low MTG16 transcripts (Figure 3), whereas MTGR1 transcripts were high in this cell line (Figure 12). AML1-ETO-mediated suppression of the ETO promoter has also been reported [25] and suggested to facilitate the AML1-ETO- induced block of erythroid lineage commitment [47,48]. AML1-ETO-mediated repression of MTG16 may have a similar effect because of blocking MTG16/ETO2-mediated erythroid progenitor expansion [5]. The repression of all ETO homologue genes by AML1-ETO may have a general role in inhibition of hematopoietic differentiation in t(8;21) leukemia.
Conclusions
This work shows differences in promoter regulation between MTG16 and MTGR1 that may explain differences in hematopoietic expression of these genes. Evolutionary conserved GATA binding sites are critical in transcriptional regulation of both MTG16 (this work) and ETO [25] promoters. The MTGR1 promoter has a GC box-rich sequence with multiple SP1 binding sites, which may secure transcriptional regulation and ubiquitous expression of this gene.
Methods
Cell Culture
Myelomonocytic U-937, erythroid HEL and TF-1, megakaryocytic MEG-01 cells were grown in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS) (Gibco BRL, Life Technologies, Rockville, MD, USA); TF-1 cells also received 20 ng/ml GM-CSF (R&D Systems, MN, USA). The erythroid/megakaryocytic UT-7 cells were grown in MEM Alpha modification medium with L-Glutamine and Nucleosides (PAA Laboratories GmBh, Austria) supplemented with 20% FBS and 30 ng/ml GM-CSF. The acute myeloid leukemia Kasumi-1, Burkitt's lymphoma Raji, and promyelocytic HL-60 cells were supplemented with 20% FBS. Monkey kidney COS cells were grown in DMEM medium with 10% FBS supplemented with high glucose (4.5 g/L) and L-glutamine.
Identification of the transcription start site of the MTG16 gene
The 5'-end (transcription start) of the mRNA was identified by 5'-RACE carried out with mRNA prepared from HEL cells using Oligotex Direct mRNA mini Kit (Qiagen, Hilden, Germany), using the First choice RNA Ligase mediated (RLM)-RACE kit (Ambion Inc., TX, USA). Nested PCR of RACE reactions was performed with adapter primer 5'-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3' and nested gene specific primer 5'-GCCTTAGCTTTCCTGTCCACTGG-3'. RACE- products were cloned into pGEM-T Easy Vector system (Promega Corporation, WI, USA) and sequenced.
Amplification of MTG16 promoter region
A 1208 bp region upstream of the translation start (from -1264 to -57 bp) was amplified from human genomic DNA by PCR. The forward and reverse primers used were 5'-TGAGGCGGTACCACCTCCAGCCATGGCATC -3' and 5'-TGCCTAGATCTACCTCCTGCAGCCTTGAGG -3', respectively. Regions corresponding to -905 to -57 and -820 to -57 bp were amplified from the 1208 bp fragment by nested PCR with forward primers 5'- TATGGTACCGTGGGGGAGCCCTGCTGTCTCCACA -3' (KpnI restriction site underlined),5'- AGGCGGGAGGTACCTTGAGGACAGGTCAGG -3' (KpnI restriction site underlined), and a common reverse primer 5'-TGCCTAGATCTACCTCCTGCAGCCTTGAGG -3' (BglII restriction site underlined).
Sequential 5' deletions of the -820 to -57 bp promoter region were generated by PCR from the cloned genomic DNA as template to generate -668 to -57, the -512 to -57, the -359 to -57 bp, the -339 to -57, the -299 to -57 and the -219 to -57 regions. Forward primers were 5'-TAAGGTACCCTGGGGAAAGCTCGGCTGAC-3', 5'-TATGGTACCCCTGCCCTGGGGCCGAGG -3', 5'-CCAGGTACCTCCTGTCAGGGAAGTGGCGG-3', 5'-TATGGTACCGGGCGCAGCTGCAGGCC
-3', 5'- TATGGTACCTCACGGGGACACAGCTGGC-3' and 5'-TATGGTACCATGAGCCCCAGGGCTCCCACCC -3' with KpnI restricton sites underlined. The common reverse primer is the same as used for amplification of the -1208 to -57 bp region. All sequences were verified by sequencing.
Amplification of MTGR1 promoter region
A 5' flanking region of 1965 bp from -1913 to +52 bp was amplified from human genomic DNA by PCR. The forward and reverse primers used were 5'-TATTGGTACCGCAACACCATGTCTGGCTAA -3' (KpnI restriction site underlined) and 5'- CTACAGATCTGCATTCACGCCCCACTTAC -3' (BglII restriction site underlined), respectively. Regions corresponding to -989 to +52, -656 to +52 and -499 to +52 bp were amplified from the 1965 bp fragment by nested PCR with forward primers 5'-TTGCGGTACCCTTCAAACTCCTGACCTCGTGA -3' (KpnI restriction site underlined), 5'- TTGCGGTACCCCCGGCCAACAGTTATTAAA -3' (KpnI restriction site underlined), 5'- TTGCGGTACCTGTAATCCCTCCCCCTAGCACT -3' (KpnI restriction site underlined), and a common reverse primer 5'-CTACAGATCTGCATTCACGCCCCACTTAC -3' (BglII restriction site underlined).
Sequential 5' deletions of the -499 to +52 bp promoter region were generated by PCR from the cloned genomic DNA as template to generate -308 to +52, the -257 to +52, the -207 to +52 bp and the -145 to +52 regions. Forward primers were 5'-TTGCGGTACCGTGCTTAATGGGAGCTGGTCT.-3', 5'-TTGCGGTACCGCGGCGAGAGCGCGCGGCGGGGGCG -3', 5'-TTGCGGTACCGCCGGGGGCGGGGCGGGGCGCGGCC -3' and 5'-TTGCGGTACCGTGGTGGTGTCTGGTTAGCTC-3', with KpnI restricton sites underlined. The common reverse primer is the same as used for amplification of the -499 to +52 bp region. All sequences were verified by sequencing.
Site-directed mutagenesis of transcription factor-binding sites
Oligonucleotide primers including the desired mutations were synthesized and used in two-step spliced overhang extension PCR. The following potential MTG16 promoter transcription factor binding sites were mutated: ETS1/PU.1 sites at positions -491, 5'-AGGAAA-3' changed to 5'- ATTCAA-3'; SP1 sites at positions -425, 5'-GGGAGG-3' changed to 5'-GTTCGG-3'; ETS1/SPI1 sites at positions -419, 5'-AGGAAGT-3' changed to 5' - ATTCAGT-3' TFAP2A sites at positions -389, 5'-GCCCGAGGG-3' changed to 5'-GCCTTCGGG-3'; GATA/YY1 sites at positions -356, 5'-GATGGT-3' changed to 5'-GACTTT-3'; SP1 sites at positions -316, 5'-CTCCGGCCCC-3' changed to 5'-CTCCTTACCC-3'; GATA sites at positions -301, 5'-TATCA-3' changed to 5'-TGGTA-3' and MZF1 sites at positions -251 & -191, 5'-TCCCCA-3' changed to 5'-TAAACA-3'.
The following potential MTGR1 promoter transcription factor binding sites were mutated: ETS1 sites at positions -335, 5'-CTTCCG-3' changed to 5'-CGTATG-3'; YY1 sites at positions -301, 5'-CCCATT-3'changed to 5'-CTTCTT-3'; SP1 sites at positions -285, 5'-CCCGGCCCCA-3' changed to 5'- CACTGTCCCA -3'; ETS1 sites at positions -276, 5'-CTTCCG-3' changed to 5'- CG TATG -3' SP1 sites at positions -243, 5'-CCCCCGCCGC-3' changed to 5'- CCCTTGCAGC -3'; SP1 sites at positions -229, 5'-CCCCGCCCGC-3' changed to 5'- CCTTGCACGC -3'; SP1 sites at positions -225, 5'-CCCCGCCCCG-3' changed to 5'- CCTTGCACCG -3'; SP1 sites at positions -213, 5'-CCCCGCCCCC-3' changed to 5'- CCTTGCACCC -3'; SP1 sites at positions -206, 5'-CCCCGCCCCG-3' changed to 5'- CCTTGCACCG -3'; SP1 sites at positions -192, 5'-CGCCGCCGCC-3' changed to 5'- CGTTGCAGCC -3'; SP1 sites at positions -178, 5'-CGCCGCCGCC-3' changed to 5'- CGTTGCAGCC -3' and SP1 sites at positions -170, 5'-CGCCGCCGCC-3' changed to 5'- CGTTGCAGCC -3'. After subcloning into promoterless pGL3/Basic reporter plasmid, all mutations were verified by sequencing.
Luciferase reporter assays of MTG16 and MTGR1 promoter
MTG16 promoter was cloned into promoterless pGL3/Basic reporter plasmid using firefly luciferase as reporter to generate pGL3 -905-57, pGL3 -820-57, pGL3 -668-57, pGL3 - 512-57, pGL3 -359-57, pGL3 -339-57, pGL3 -299-57 and pGL3 -219-57 reporter constructs. Mutants were cloned into the same reporter plasmid. Transient transfections of hematopoietic cell lines were performed by electroporation as previously described by [49]. The pGL3/SV40-promoter vector served as positive control and promoterless pGL3/Basic vector as negative control. Renilla luciferase, pRL/SV40 vector (Promega Corporation, WI, USA) was used as internal control for transfection efficiency. Thirtyfive μg pGL3 DNA were used for HL-60 target cells and 15 μg for HEL, TF-1, U937, UT-7, MEG-01, Raji and Kasumi-1 target cells. COS cells were transfected with 1.5 μg of DNA by use of Polyfect (Qiagen, Hilden, Germany). Twentyfour hours after transfection, cells were lysed and subjected to luciferase dermination using the Dual luciferase reporter assay kit (Promega Corporation, WI, USA). Firefly and Renilla luminiscense were quantified (Run Promega Protocol, DLR-0-INJ) in a GLOMAX 20/20 Luminometer (Promega Corporation, WI, USA). Firefly was normalized to Renilla luciferase as internal control for transfection efficiency. Results are given as adjusted Relative Luciferase Units (AdjRLU) normalized to pGL3/promoter (set to 1). Three to five independent transfections were performed in each case and each sample was measured in triplicate.
Similarly, MTGR1 pGL3 -1913 + 52, pGL3 -989 + 52, pGL3 -656 + 52, pGL3 -499 + 52, pGL3 -308 + 52, pGL3 -257 + 52, pGL3 -207 + 52 and pGL3 -145 + 52 reporters were generated, transfected into cells and assayed as described above.
Transfection of GATA-1 small interfering RNA
Twentyfour-well Falcon plates (Becton Dickinson, New Jersey 07417, USA) were coated with 1 mg/ml retronectin (Takara Bio inc, Verviers, Belgium) in PBS for two h at room temperature. Excess retronectine was removed and wells were blocked with 2% BSA in PBS for 30 min. After removal of the supernatant, 50,000 HEL cells were added per well in 600 μl RPMI medium containing 10% FBS and 20 nM phorbol 12-myristate 13-acetate (PMA) to generate adherent cells by incubation at 37°C for 24 h. After washing with PBS transfection was carried out with 5 μM siRNA using the N-TER transfection reagent (Sigma-Aldrich Corp. St. Louis, MO USA) in RPMI without FBS. After 4 h, 300 μl of RPMI with 20% FBS was gently added and cells were incubated for 48 h. Eventually, gently rinsed with PBS, tripsinized and examined by Western blotting. The GATA-1 siRNA sequence was 5'-AGUUGAGGCAGGGUAGAGC-3' (Oligo SASI_Hs02_00333089, Sigma).
Inhibition of GATA-1 transcriptional activity by HERP2
The transcription factor HERP2 represses transcriptional activation by GATA-1 [29] by physical interaction. To get support for a role of GATA-1 in gene activation of MTG16, HEL, TF-1, UT-7, and MEG-01cells were co-transfected with 10 μg MTG16 pGL3 -668 luciferase reporter and 10 μg HERP2 plasmid (kindly provided by Dr N Goldfarb, Charlottesville, Virginia) and the reporter signal was measured after 24 h as described above for luciferase reporter assays. The results were normalized to the luciferase activity of pGL3 -668 luciferase reporter.
Electrophoretic Mobility Shift Assay (EMSA) of MTG16 promoter
EMSA was performed as described previously [25]. The potential GATA-1 sites at -301 was examined. The probe sequence was biotin-5-CCCGGCATTATCACGGGGACAC -3'. Nuclear extracts from HEL, TF-1, UT-7 and MEG-01 cells were prepared as described by Andrews and Faller [50]. One to two μl polyclonal anti-GATA-1 (Active Motif, Carlsbad, CA, USA), monoclonal anti-GATA-2 (Santa Cruz Biotechnology Inc., CA, USA, sc-9008) or polyclonal anti-CD63 (sc-7080, Santa Cruz Biotechnology Inc., CA, USA) antibodies were added to the reaction mixtures.
Chromatin Immunoprecipitation (ChIP) assay of MTG16 promoter
ChIP was performed as described previously [25]. For IP, 4 μl of polyclonal anti-GATA-1 (Active Motif, Carlsbad, CA, USA) or 0.8 μg monoclonal anti-GATA-2 antibodies (R&D Systems, MN, USA) were used. Forward and reverse primers for GATA sites were 5'-CAGATGGTTCCTGTCAGGGAAGTGGCG-3' and 5'-AGGTCTCCCTGCAGCCTGCGGGTGAG-3'. Control forward and reverse primers were: 5'-TAACACAGAGTACCCAGCCACTGTGC-3 ' and 5'-TGGTTGCACGGACAGAAGCCCCT-3'. Three different chromatin preparations were used for each IP.
Quantitative real-time PCR
Real-time PCR was performed as described previously [18]. Transcript levels were calculated from a standard curve based on the Ct values of the samples,. Relative quantification based on the ΔCt method [51] was used. Normalization: ΔCt = Ct (sample) - Ct (HEL corresponding DNA concentration). Relative quantification = 2 - ΔCt .
Immunoprecipitation (IP) and Western blotting
IP and Western blotting were performed as described previously [52]. The following antibodies were used: polyclonal anti-GATA-1 (Active Motif, Carlsbad, CA, USA) and polyclonal anti-GATA-2 (R&D Systems, MN, USA).
Bioinformatics
The cDNA sequences were analyzed by use of the NCBI Blast program http://www.ncbi.nlm.nih.gov/BLAST/. Conserved regions were searched by multiple alignment to genomic sequences using ClustalW http://www.ebi.ac.uk/Tools/clustalw2/index.html.
Potential transcription factor binding sites were examined with the MatInspector http://www.genomatix.de//matinspector.html and the Jaspar database (Jaspar.genereg.net).
Statistical analysis
The statistical significance between two samples was determined by unpaired t-test in Prism Programme. Triple and single asterisk represents P < 0.0001 and P < 0.05, respectively.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RA carried out experiments, analyzed data, supervised experimental design/data analysis and drafted the manuscript. PK carried out experiments, analyzed data and drafted the manuscript. RSD supervised experimental design/data analysis. UG supervised experimental design/data analysis. IO supervised experimental design/data analysis and drafted the manuscript. All authors critically revised and approved the final manuscript.
Contributor Information
Ram Ajore, Email: Ram.Ajore@med.lu.se.
Parveen Kumar, Email: Parveen.Kumar@med.lu.se.
Rakesh Singh Dhanda, Email: singh.rakesh@pgimer.edu.in.
Urban Gullberg, Email: Urban.Gullberg@med.lu.se.
Inge Olsson, Email: Inge.olsson@med.lu.se.
Acknowledgements
This work was supported by the Swedish Cancer Foundation and the Swedish Childhood Cancer Foundation. We are grateful to Ann-Maj Persson for kind help and expert guidance.
References
- Feinstein PG, Kornfeld K, Hogness DS, Mann RS. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140(2):573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18(12):7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG. et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18(12):7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier N, Corcoran CM, Lennon C, Hyjek E, Chadburn A, Bardwell VJ, Licht JD, Melnick A. ETO protein of t(8;21) AML is a corepressor for Bcl-6 B-cell lymphoma oncoprotein. Blood. 2004;103(4):1454–1463. doi: 10.1182/blood-2003-06-2081. [DOI] [PubMed] [Google Scholar]
- Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25(2):357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee L, Bryan J, Elliott L, Grimes HL, Kazanjian A, Davis JN, Meyers S. Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism. J Cell Biochem. 2003;89(5):1005–1018. doi: 10.1002/jcb.10548. [DOI] [PubMed] [Google Scholar]
- Melnick AM, Westendorf JJ, Polinger A, Carlile GW, Arai S, Ball HJ, Lutterbach B, Hiebert SW, Licht JD. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2000;20(6):2075–2086. doi: 10.1128/MCB.20.6.2075-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P. et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005;25(23):10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305(5688):1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- Cai Y, Xu Z, Xie J, Ham AJ, Koury MJ, Hiebert SW, Brandt SJ. Eto2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem Biophys Res Commun. 2009;390(2):295–301. doi: 10.1016/j.bbrc.2009.09.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95(18):10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand D, Tiefenbach J, Heinzel T, Grez M, Maurer AB. Multiple regions of ETO cooperate in transcriptional repression. J Biol Chem. 2001;276(13):9889–9895. doi: 10.1074/jbc.M010582200. [DOI] [PubMed] [Google Scholar]
- Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91(11):4028–4037. [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80(7):1825–1831. [PubMed] [Google Scholar]
- Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12(7):2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastadisegni MC, Lonoce A, Impera L, Di Terlizzi F, Fugazza G, Aliano S, Grasso R, Cluzeau T, Raynaud S, Rocchi M. et al. CBFA2T2 and C20orf112: two novel fusion partners of RUNX1 in acute myeloid leukemia. Leukemia. 2010;24(8):1516–1519. doi: 10.1038/leu.2010.106. [DOI] [PubMed] [Google Scholar]
- Nimer SD, Moore MA. Effects of the leukemia-associated AML1-ETO protein on hematopoietic stem and progenitor cells. Oncogene. 2004;23(24):4249–4254. doi: 10.1038/sj.onc.1207673. [DOI] [PubMed] [Google Scholar]
- Lindberg SR, Olsson A, Persson AM, Olsson I. The Leukemia-associated ETO homologues are differently expressed during hematopoietic differentiation. Exp Hematol. 2005;33(2):189–198. doi: 10.1016/j.exphem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Okumura AJ, Peterson LF, Lo MC, Zhang DE. Expression of AML/Runx and ETO/MTG family members during hematopoietic differentiation of embryonic stem cells. Exp Hematol. 2007;35(6):978–988. doi: 10.1016/j.exphem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133(24):4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W. et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24(3):277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlett I, Draper J, Strouboulis J, Iborra F, Porcher C, Vyas P. Characterization of megakaryocyte GATA1-interacting proteins: the corepressor ETO2 and GATA1 interact to regulate terminal megakaryocyte maturation. Blood. 2008;112(7):2738–2749. doi: 10.1182/blood-2008-03-146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, Hiebert SW. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol Cell Biol. 2008;28(20):6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann JM, Chyla BJ, Ellis TC, Martinez A, Moore AC, Franklin JL, McGhee L, Meyers S, Ohm JE, Luce KS. et al. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol Cell Biol. 2005;25(21):9576–9585. doi: 10.1128/MCB.25.21.9576-9585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajore R, Dhanda RS, Gullberg U, Olsson I. The leukemia associated ETO nuclear repressor gene is regulated by the GATA-1 transcription factor in erythroid/megakaryocytic cells. BMC Mol Biol. 2010;11:38. doi: 10.1186/1471-2199-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16(13):3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P, Ault K, Jackson CW, Orkin SH, Shivdasani RA. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 1999;93(9):2867–2875. [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8(10):1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Elagib KE, Xiao M, Hussaini IM, Delehanty LL, Palmer LA, Racke FK, Birrer MJ, Shanmugasundaram G, McDevitt MA, Goldfarb AN. Jun blockade of erythropoiesis: role for repression of GATA-1 by HERP2. Mol Cell Biol. 2004;24(17):7779–7794. doi: 10.1128/MCB.24.17.7779-7794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Sun D, Schuetz J, Hiebert SW. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol Cell Biol. 1998;18(6):3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PF, Robinson M, Owens G, Drabkin HA. The ETO portion of acute myeloid leukemia t(8;21) fusion transcript encodes a highly evolutionarily conserved, putative transcription factor. Cancer Res. 1994;54(7):1782–1786. [PubMed] [Google Scholar]
- Lederfein D, Yaffe D, Nudel U. A housekeeping type promoter, located in the 3 region of the Duchenne muscular dystrophy gene, controls the expression of Dp71, a major product of the gene. Hum Mol Genet. 1993;2(11):1883–1888. doi: 10.1093/hmg/2.11.1883. [DOI] [PubMed] [Google Scholar]
- Fraizer GC, Wu YJ, Hewitt SM, Maity T, Ton CC, Huff V, Saunders GF. Transcriptional regulation of the human Wilms' tumor gene (WT1). Cell type-specific enhancer and promiscuous promoter. J Biol Chem. 1994;269(12):8892–8900. [PubMed] [Google Scholar]
- Pinte S, Guerardel C, Deltour-Balerdi S, Godwin AK, Leprince D. Identification of a second G-C-rich promoter conserved in the human, murine and rat tumor suppressor genes HIC1. Oncogene. 2004;23(22):4023–4031. doi: 10.1038/sj.onc.1207504. [DOI] [PubMed] [Google Scholar]
- Zhao C, He X, Tian C, Meng A. Two GC-rich boxes in huC promoter play distinct roles in controlling its neuronal specific expression in zebrafish embryos. Biochem Biophys Res Commun. 2006;342(1):214–220. doi: 10.1016/j.bbrc.2006.01.134. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Sazer S, Tjian R, Schimke RT. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature. 1986;319(6050):246–248. doi: 10.1038/319246a0. [DOI] [PubMed] [Google Scholar]
- Martin DI, Zon LI, Mutter G, Orkin SH. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344(6265):444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- Shimizu R, Yamamoto M. Gene expression regulation and domain function of hematopoietic GATA factors. Semin Cell Dev Biol. 2005;16(1):129–136. doi: 10.1016/j.semcdb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Leonard M, Brice M, Engel JD, Papayannopoulou T. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood. 1993;82(4):1071–1079. [PubMed] [Google Scholar]
- Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371(6494):221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Simon MC, Pevny L, Wiles MV, Keller G, Costantini F, Orkin SH. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat Genet. 1992;1(2):92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA. 2003;100(15):8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA. 2004;101(4):980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, Venturini E, Zhang DE, Pelicci PG, Alcalay M. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4(11):e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Elagib KE, Delehanty LL, Goldfarb AN. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1. Cancer Res. 2006;66(6):2990–2996. doi: 10.1158/0008-5472.CAN-05-2944. [DOI] [PubMed] [Google Scholar]
- Tonks A, Pearn L, Tonks AJ, Pearce L, Hoy T, Phillips S, Fisher J, Downing JR, Burnett AK, Darley RL. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2003;101(2):624–632. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- Lennartsson A, Pieters K, Ullmark T, Vidovic K, Gullberg U. AML-1, PU.1, and Sp3 regulate expression of human bactericidal/permeability-increasing protein. Biochem Biophys Res Commun. 2003;311(4):853–863. doi: 10.1016/j.bbrc.2003.10.067. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19(9):2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30(6):503–512. doi: 10.1016/S0301-472X(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Dhanda RS, Lindberg SR, Olsson I. The human SIN3B corepressor forms a nucleolar complex with leukemia-associated ETO homologues. BMC Mol Biol. 2008;9:8. doi: 10.1186/1471-2199-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]