Abstract
Background
Enzybiotics are becoming increasingly recognized as potential alternative therapies for drug-resistant bacteria. Although only a few enzybiotics are currently well characterized, much information is still missing or is unavailable for researchers. The construction of an enzybiotics database would therefore increase efficiency and convenience in investigating these bioactive proteins and thus help reduce or delay the recent increase in antibiotic resistance.
Description
In the present manuscript, we describe the development of a novel and original database called EnzyBase, which contains 1144 enzybiotics from 216 natural sources. To ensure data quality, we limited the source of information to authoritative public databases and published scientific literature. The interface of EnzyBase is easy to use and allows users to rapidly retrieve data according to their desired search criteria and blast the database for homologous sequences. We also describe examples of database-aided enzybiotics discovery and design.
Conclusion
EnzyBase serves as a unique tool for enzybiotic studies. It has several potential applications, e.g. in silico enzybiotic combination as cocktails, and novel enzybiotic design, in response to continuously emerging drug-resistant pathogens. This database is a valuable platform for researchers who are interested in enzybiotic studies. EnzyBase is available online at http://biotechlab.fudan.edu.cn/database/EnzyBase/home.php.
Background
Antibiotic abuse is, in part, responsible for the dramatic increase in the resistance of pathogens to traditional antibiotics [1]. Superbugs, such as MRSA and NDM-1, frequently and seriously threaten public safety [2,3]. Consequently, the need to develop new classes of antibiotics with novel mechanisms of action against drug-resistant pathogens is becoming very urgent. Enzybiotics [4-8] and antimicrobial peptides (AMPs)[9] have attracted much attention as potential substitutes for conventional antibiotics.
In the present manuscript, enzybiotics are referred to as bacterial cell wall-degrading enzymes, including lysins, bacteriocins, autolysins, and lysozymes. The most important characteristics of enzybiotics are their novel mechanisms of antibacterial action and capacity to kill antibiotic-resistant bacteria [10]. Another significant feature of certain enzybiotics is their low probability of developing bacterial resistance [11]. Compared with AMPs, enzybiotics are large, heat-labile, and narrow-spectrum types of antimicrobial proteins. Consequently, enzybiotics are not always suitable antimicrobial agents. Despite this, certain enzybiotics have been well characterized and widely used. Lysostaphin [12-15] and lysozymes [16-18] are the most studied enzybiotics in regards to their clinical or food applications. Furthermore, despite their apparent limitations in medicine, their potency against multi-drug-resistant pathogens should not be ignored. Therefore, an enzybiotic specific database that not only mobilizes research on enzybiotics, but also makes it more efficient and convenient, needs to be constructed.
Over the past decade, many databases have been developed for AMPs. These databases, including APD [19,20], ANTIMIC [21], CAMP [22], BACTIBASE [23,24], PhytAMP [25], PenBase [26], Defensins [27], CyBase [28], and peptaibols Peptaibol [29], contain AMP sequences from diverse origins or specific families and accordingly have accelerated and stimulated research on AMPs. Conversely, the majority of the sequenced enzybiotics are stored in the manually annotated UniProt/Swiss-Prot [30] database or scattered in the scientific literature. As a result, it is difficult to find information on enzybiotics for recent users. Developing a central database that stores information on enzybiotics is warranted by investigators to promote their research on enzybiotics discovery and design.
The idea of constructing a database that stores information on enzybiotics arose from our own research experience. We found that we constantly had to query information on enzybiotics from public databases, such as UniProt, and scientific literature. Thus, we decided to construct a database that simplified our research efforts, and comprehensively collected this information. EnzyBase, a novel and original database for enzybiotics studies, was developed and currently contains 1144 enzybiotics from 216 natural sources. This database provides a platform for current users to comprehensively and conveniently research enzybiotics and can be useful for exploring and designing novel enzybiotics for medical use.
Construction and content
EnzyBase was built on an Apache HTTP Server (V2.2.14) with PHP (V5.2.13) and MySQL Server (V5.1.40) as the back-end, and Personal Home Page (PHP), HyperText Markup Language (HTML) and Cascading Style Sheets (CSS) as the front-end. Apache, MySQL, and PHP were preferred as they are open-source software and platform independent, respectively, making them suitable for academic use. The web server and all parts of the database are hosted at Information Office of Fudan University, Shanghai, China.
All enzybiotic sequences were collected manually from the annotated UniProt/Swiss-Prot database or scientific literature. Each enzybiotic without the UniProt link had been excluded. The enzybiotics collected in EnzyBase database are primarily from natural sources, with the exception of genetically-modified sequences. Additional physicochemical data of each enzybiotic was either calculated via Bioperl programs or identified from scientific literature via a PubMed search. All of the collected information was classified and filled into six relational tables in MySQL. For each enzybiotic, a unique identification number (i.e., enzy id) was assigned, beginning with the prefix EN. Each entry also contains general data, such as protein name, protein full name, producer organism, simple function annotation and protein sequence, domains, 3D structure, and relevant references. For all proteins that already exist in the UniProt, Interpro [31], and/or PDB [32] databases, hyperlinks to these databases were created in EnzyBase. Additional physicochemical data, including calculated isoelectric point (pI) and charge at pI, are also provided. Moreover, minimal inhibitory concentrations (MICs) are included, if data are available. The BlastP program (BLASTP V2.2.25+) [33,34] was used for sequence homology searches against EnzyBase.
Utility and discussion
Database description
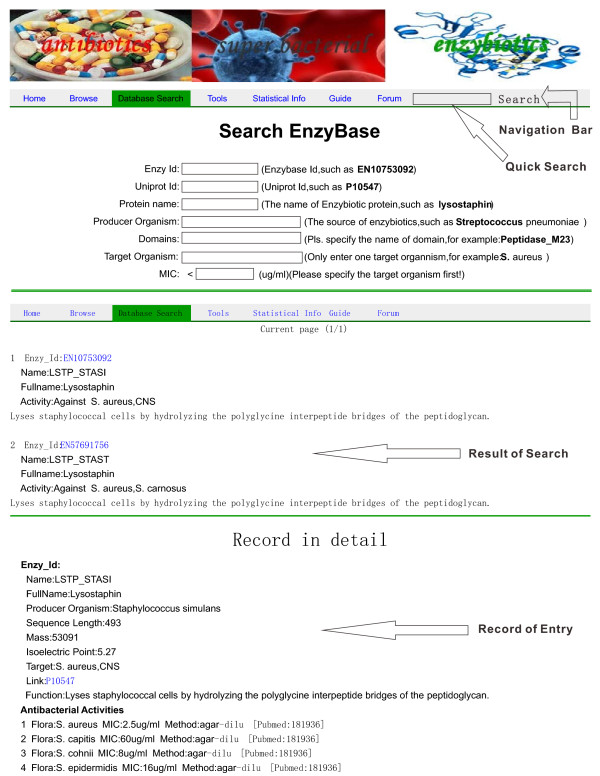
EnzyBase supplies a user-friendly web interface, so that users can easily query and retrieve information on enzybiotics. A concise navigational interface that contains the database browse, search, tools, statistical information, and guide, as well as a forum, were designed to generate a clearly structured database layout that enables fast and easy navigation (Figure 1).
Figure 1.
Screen shots of the EnzyBase search interface. Screen shots of the EnzyBase search interface showing the advanced search and result views. Please note that not all fields are shown.
As a web-based database, all data can be accessed and retrieved directly from the web browser. The database browse interface provides the users with a function of navigating the entire database, whereas the search interface provides the users with the function of retrieving their desired information using either the "quick" or "advanced" options. A "quick" search can be performed using only keywords, while the "advanced" search offers the possibility to specify seven separate fields, namely enzy id, uniprotKB entry number (i.e., uniprot id), protein name, producer organism, domains, target organism, and MIC value. The user can query the database by either one condition (excluding MIC, which requires the type of target organism to be initially stated) or a combination of various conditions. Every enzybiotic has its own results page that contains comprehensive information, including general information, antibacterial activities, sequence, structures, domains, and references. The general information consists of enzy id, protein name, protein full name, producer organism, protein mass, calculated pI, antibacterial activity, and simple function annotations. EnzyBase also provides hyperlinks to other databases, such as UniProt, InterPro, PDB, and PubMed, which allows for easier navigation within the World Wide Web pertaining to additional information on enzybiotics. The tools interface permits the use of BLASTP against EnzyBase, which enables users to search the database for homologous sequences, and then copy obtained results for subsequent research. Owing to limitations of disk space on the host site, we did not implement a local BLASTP against the NCBI database but instead supplied a hyperlink to the BLASTP on the NCBI website. The statistical info interface provides data on sources for enzybiotics, the distribution of sequence length, protein mass, calculated protein pI, and domains (please refer to the 'Statistical description and findings' section below for more information). The guide interface provides simple instructions for potential users on how to use the functions of EnzyBase. Additionally, the forum tools, which are based on UseBB, a free forum software, have been integrated into the database to provide information on updates, bug reports, and user discussions.
Statistical description and findings
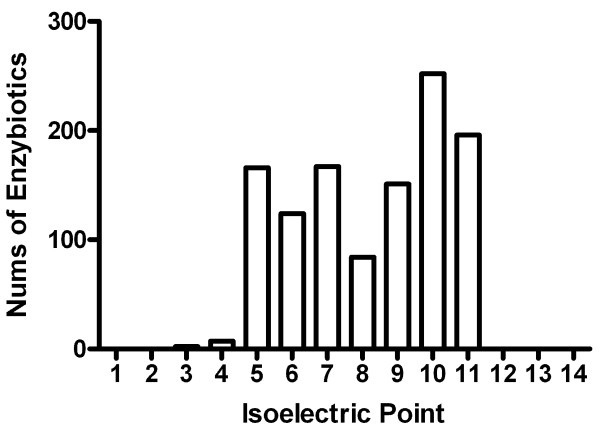
The current version of EnzyBase possesses 1144 enzybiotics from 216 natural sources. The length of the enzybiotic sequences range from 72 to 2337 amino acids. Table 1 presents the top 10 sources for enzybiotics in EnzyBase. The majority (99.2%) of enzybiotics have a calculated pI ranging from 4 to 11 (Figure 2).
Table 1.
Top 10 sources of enzybiotics in EnzyBase
| Rank | Producer organisms | Numbers of enzybiotics |
|---|---|---|
| 1 | Staphylococcus aureus | 142 |
| 2 | Enterococcus faecalis | 136 |
| 3 | Bacillus cereus | 73 |
| 4 | Streptococcus pneumoniae | 66 |
| 5 | Bacillus thuringiensis | 57 |
| 6 | Staphylococcus phage | 55 |
| 7 | Listeria monocytogenes | 28 |
| 8 | Staphylococcus epidermidis | 27 |
| 9 | Clostridium perfringens | 21 |
| 10 | Enterococcus faecium | 21 |
Figure 2.
Distribution of calculated isoelectric points for the enzybiotics within EnzyBase. Every bar indicates the number of enzybiotics calculated to have their isoelectric point range from pI 1 to 14.
All enzybiotics in EnzyBase contain 55 domains, and only 24 enzybiotics have known 3D structures. The top 10 domains for the enzybiotics within EnzyBase are presented in Table 2. The Amidase_domain is the top domain (till 2012-2-6). In fact, this domain is carried by 392 enzybiotics, representing ca. 34% of the total number of enzybiotics in EnzyBase. Thus, it appears that many of the recorded enzybiotics are amidase like.
Table 2.
Top 10 domains in EnzyBase
| Rank | Interpro Id | Domain Name | Numbers of enzybiotics |
|---|---|---|---|
| 1 | IPR002502 | Amidase_domain | 392 |
| 2 | IPR007921 | CHAP | 224 |
| 3 | IPR017853 | Glycoside_hydrolase_SF | 188 |
| 4 | IPR002053 | Glyco_hydro_25 | 188 |
| 5 | IPR013781 | Glyco_hydro_subgr_catalytic | 187 |
| 6 | IPR002901 | Mano_Glyc_endo_b_GlcNAc | 169 |
| 7 | IPR018392 | Peptidoglycan-bd_lysin | 147 |
| 8 | IPR013667 | SH3_5_bac | 141 |
| 9 | IPR002482 | Peptidoglycan-bd_Lysin_subgr | 141 |
| 10 | IPR003646 | SH3-like_bac | 134 |
Applications
The EnzyBase can be used as a tool to aid researchers in exploring the use of enzybiotics or for designing novel enzybiotics. The most prominent weakness of enzybiotics is their narrow spectrum of antibacterial activity. However, a combination of enzybiotics with different spectra of antibacterial activities and/or different mechanisms of action could be used against a broad spectrum of bacterial infections and/or their resistant strains. Through the use of EnzyBase, users can quickly find a series of enzybiotics with optimum antibacterial activities against specific pathogens, and then combine them as a cocktail to measure their therapeutic effect against bacterial infectious diseases. Similar approaches have been successfully used to design phage cocktail therapies for the treatment of infections [35]. For novel enzybiotics design, users could search for potential domains with high antibacterial activities against specific pathogens on EnzyBase and then combine them to create chimeric enzybiotics. For instance, to search for effective antimicrobial proteins against mastitis-causing pathogens, researchers created a novel chimeric peptidoglycan hydrolase fusion protein between lysostaphin and the endolysin of phage B30, which possesses their respective enzymatic domains, and is capable of degrading both streptococcal and staphylococcal peptidoglycans [36]. Thus, the quantity and quality of the data entered in EnzyBase appears to be very important for successfully applying it in such research applications.
In the future, we plan to implement updates, assess the data quality continuously, and integrate some structural analysis tools, such as RasMol [37], and certain web2.0 functions, such as Wiki, into EnzyBase to improve its interactivity with users and improve research in the field of enzybiotics design and structure function exploration.
Conclusions
In summary, EnzyBase is a comprehensive and web-accessible database of enzybiotics. The current version of EnzyBase has 1144 entries. The database can be queried either by using simply keywords or by combinatorial conditions searches. EnzyBase may aid in enhancing our current understanding of enzybiotics and their mechanisms of action. Its potential applications include the in silico development of combinations of enzybiotics (e.g., cocktails) and the construction of novel enzybiotics against various bacterial infectious diseases. Thus, the database may have implications in the development of new drugs for medical applications.
Availability and requirements
EnzyBase is freely available for academic users at http://biotechlab.fudan.edu.cn/database/EnzyBase/home.php.
Authors' contributions
HW developed the web interface, designed the rational database scheme, and qualified the data. HL and JH primarily contributed to inputting the data into the current database, as well as in writing the manuscript. GL and QH conceived of the initial idea of the database, provided direction for its development, and revised the subsequent drafts of this manuscript. All authors read and approved the final manuscript.
Contributor Information
Hongyu Wu, Email: ak0526@163.com.
Hairong Lu, Email: lhrsh@sohu.com.
Jinjiang Huang, Email: saint7878@hotmail.com.
Guodong Li, Email: leeguod@yahoo.com.cn.
Qingshan Huang, Email: qshuang@fudan.edu.cn.
Acknowledgements
We would like to thank all of our colleagues at the State Key Laboratory of Genetic Engineering at Fudan University and Shanghai High-Tech United Bio-Technological R&D Co., Ltd., of China for their contributions in the literature search and discussions regarding this manuscript. This work was supported in part by the major scientific and technological specialized project of China for 'Significant New Formulation of New Drugs' (grant #: 2008ZX09101-032) and the 'Yangtze River Delta' joint scientific and technological project of China (grant 10495810600).
References
- English BK, Gaur AH. The use and abuse of antibiotics and the development of antibiotic resistance. Adv Exp Med Biol. 2010;659:73–82. doi: 10.1007/978-1-4419-0981-7_6. [DOI] [PubMed] [Google Scholar]
- Heddini A, Cars O, Qiang S, Tomson G. Antibiotic resistance in China-a major future challenge. Lancet. 2009;373:30. doi: 10.1016/S0140-6736(08)61956-X. [DOI] [PubMed] [Google Scholar]
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- Nelson D, Loomis L, Fischetti VA. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci USA. 2001;98:4107–4112. doi: 10.1073/pnas.061038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Crespo P, Ageitos JM, Poza M, Villa TG. Enzybiotics: a look to the future, recalling the past. J Pharm Sci. 2007;96:1917–1924. doi: 10.1002/jps.20853. [DOI] [PubMed] [Google Scholar]
- Hermoso JA, Garcia JL, Garcia P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Loessner MJ. Bacteriophage endolysins-current state of research and applications. Curr Opin Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Gordon YJ, Romanowski EG, McDermott AM. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr Eye Res. 2005;30:505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysowski J, Weber-Dabrowska B, Gorski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med (Maywood) 2006;231:366–377. doi: 10.1177/153537020623100402. [DOI] [PubMed] [Google Scholar]
- Kusuma C, Jadanova A, Chanturiya T, Kokai-Kun JF. Lysostaphin-resistant variants of Staphylococcus aureus demonstrate reduced fitness in vitro and in vivo. Antimicrob Agents Chemother. 2007;51:475–482. doi: 10.1128/AAC.00786-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos MdCdF, Coutinho BG, Coelho MLV. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. pharmaceuticals. 2010;3:1139–1161. doi: 10.3390/ph3041139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gao Y, Feng J, Huang Y, Li S, Liu Y, Liu C, Fan M, Shen B, Shao N. C-terminus of TRAP in Staphylococcus can enhance the activity of lysozyme and lysostaphin. Acta Biochim Biophys Sin (Shanghai) 2008;40:452–458. doi: 10.1111/j.1745-7270.2008.00415.x. [DOI] [PubMed] [Google Scholar]
- Kumar JK. Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol. 2008;80:555–561. doi: 10.1007/s00253-008-1579-y. [DOI] [PubMed] [Google Scholar]
- Rainard P. Tackling mastitis in dairy cows. Nat Biotechnol. 2005;23:430–432. doi: 10.1038/nbt0405-430. [DOI] [PubMed] [Google Scholar]
- Tenovuo J. Clinical applications of antimicrobial host proteins lactoperoxidase, lysozyme and lactoferrin in xerostomia: efficacy and safety. Oral Dis. 2002;8:23–29. doi: 10.1034/j.1601-0825.2002.1o781.x. [DOI] [PubMed] [Google Scholar]
- Donovan DM. Bacteriophage and peptidoglycan degrading enzymes with antimicrobial applications. Recent Pat Biotechnol. 2007;1:113–122. doi: 10.2174/187220807780809463. [DOI] [PubMed] [Google Scholar]
- Gil-Montoya JA, Guardia-Lopez I, Gonzalez-Moles MA. Evaluation of the clinical efficacy of a mouthwash and oral gel containing the antimicrobial proteins lactoperoxidase, lysozyme and lactoferrin in elderly patients with dry mouth-a pilot study. Gerodontology. 2008;25:3–9. doi: 10.1111/j.1741-2358.2007.00197.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:D590–592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachary M, Krishnan SP, Koh JL, Khan AM, Seah SH, Tan TW, Brusic V, Bajic VB. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Karnik S, Barai RS, Jayaraman VK, Idicula-Thomas S. CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Zouhir A, Ben Hamida J, Fliss I. BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007;7:89. doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 2010;10:22. doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Ben Hamida J, Vergoten G, Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009;37:963–968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen Y, Garnier J, Robert L, Lefranc MP, Mougenot I, de Lorgeril J, Janech M, Gross PS, Warr GW, Cuthbertson B. et al. PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev Comp Immunol. 2006;30:283–288. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Seebah S, Suresh A, Zhuo S, Choong YH, Chua H, Chuon D, Beuerman R, Verma C. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2007;35:D265–268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CK, Kaas Q, Chiche L, Craik DJ. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. The Peptaibol Database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R. et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L. et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge LD. In: Enzybiotics: antibiotic enzymes as drugs and therapeutics. 1. Villa TG, Veiga-crespo P, editor. New Jersey: John Wiley &Sons, Inc.,Publication; 2010. Design of phage cocktails for therapy from a host range point of view; pp. 199–218. [Google Scholar]
- Donovan DM, Dong S, Garrett W, Rousseau GM, Moineau S, Pritchard DG. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl Environ Microbiol. 2006;72:2988–2996. doi: 10.1128/AEM.72.4.2988-2996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/S0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]