ABSTRACT
Unraveling molecular pathways responsible for regulation of early embryonic development is crucial for our understanding of female infertility. Maternal determinants that control the transition from oocyte to embryo are crucial molecules that govern developmental competence of the newly conceived zygote. We describe a series of defects that are triggered by a disruption of maternal lethal effect gene, Nlrp5. Previous studies have shown that Nlrp5 hypomorph embryos fail to develop beyond the two-cell stage. Despite its importance in preimplantation development, the mechanism by which the embryo arrest occurs remains unclear. We confirmed that Nlrp5 mutant and wild-type females possess comparable ovarian germ pool and follicular recruitment rates. However, ovulated oocytes lacking Nlrp5 have abnormal mitochondrial localization and increased activity in order to sustain physiological ATP content. This results in an accumulation of reactive oxygen species and increased cellular stress causing mitochondrial depletion. Compromised cellular state is also accompanied by increased expression of cell death inducer Bax and depletion of cytochrome c. However, neither genetic deletion (Bax/Nlrp5 double knockout) nor mimetic interference (BH4 domain or Bax inhibitory peptide) were sufficient to alleviate embryo demise caused by depletion of Nlrp5. We therefore conclude that lack of Nlrp5 in oocytes triggers premature activation of the mitochondrial pool, causing mitochondrial damage that cannot be rescued by inactivation of Bax.
Keywords: Bax, cell death, embryo, mitochondria, Nlrp5, two-cell arrest
Lack of Nlrp5 in oocytes triggers premature activation of the mitochondrial pool, causing mitochondrial damage that cannot be rescued by inactivation of Bax.
INTRODUCTION
Maternal effect genes were originally identified in invertebrates such as Drosophila melanogaster and Caenorhabditis elegans [1]. These genes encode mRNAs and proteins that are often restricted to gametes, and actively accumulate during oogenesis to govern the transition from oocyte to embryo. Disruption of these genes almost always results in developmental arrest shortly after fertilization (often during preimplantation stage), causing embryonic lethality and therefore named maternal lethal effect (MLE) genes. In mammals, one of the first identified maternal effect genes resulting in preimplantation lethality in mice was NACHT, leucine-rich repeat (LRR), and Pyrin domain (PYD) containing proteins 5 (NLRP5), a gene originally designated as Mater [2]. Since then, several maternal effect genes, such as Zar1, Dpp3 (Stella), Smarca4 (Brg1), Hsf1, Atg5, 2410004A20Rik (Filia), Padi6, etc., have been identified in mammals [3–9]. Functionally, these genes regulate processes required for the developmental switch from oocyte to embryo, from activation of embryonic genome through degradation of maternal products and regulation of organellar function.
Despite the fact that Nlpr5 was identified as a first member of MLE genes in mammals, very little is known about its mode of action. Nlrp5 belongs to the NLRP family, which contains 14 proteins in humans and approximately 20 orthologs in mice [10]. Members of the NLRP family contain a tripartite structure: PYD-NACHT-LRR domains. The PYD domain at the N-terminal region is mainly involved in inflammation and apoptosis, whereas the LRR domain is known to facilitate protein-protein interactions that regulate different cellular functions. NACHT domains are predicted to bind ATP/guanosine triphosphate [11]. Moreover, these domains are responsible for complex interactions with other proteins, including caspases [12]. Although the majority of NLRP proteins are adaptors involved in the regulation of inflammation [11], it has been suggested that they affect apoptosis and pyroptosis (reviewed in [13]). Overexpression of NLRP1 and NLRP5 has been implicated in regulation of caspase activation and apoptosis in injured neurons [14, 15]. Furthermore, NLRP1 action and caspase 1 activation in macrophages can be inhibited by interaction with antiapoptotic members of BCL2 family: BCL2 and BCL2L1 [16]. However, how NLRP5 regulates embryo development remains unclear. Conserved oocyte-specific expression of NLRP5 is consistent among many mammalian species. Alongside the subcortical enrichment in oocytes and embryos [17], NLRP5 also localizes to the nuclear envelope and mitochondria in germinal vesicle oocytes, suggesting a possible role in mitochondrial function [18, 19]. The only known interacting partners of NLRP5 have been recently identified as members of the subcortical complex—2410004A20RIK (FILIA), OOEP (FLOPED), and TLE6 [17]; however, the precise function of this protein complex remains unknown. Whereas disruption of Ooep (Floped) phenocopies Nlrp5 defects [17], elimination of 2410004A20Rik (Filia) results in subfertility caused by aneuploidies due to disruption of the mitotic spindle [9].
The approach to Nlrp5 disruption has been previously described [2]; since then it has been confirmed that this approach caused severe down-regulation (∼90%–95%) of NLRP5 protein in the oocytes [20], and was termed Nlrp5tm/tm (targeted mutation). Previous studies of Nlrp5tm/tm mice indicate that the ovarian follicle pool in young mice is normal with comparable numbers of ovulated oocytes. Fertilization also appears unaltered, although zygotes conceived from Nlrp5 hypomorph oocytes predominantly arrested at the two-cell stage [2]. Impaired fertility, as a consequence of compromised Nlrp5 function, has also been proposed to exist in other mammalian species such as bovine, pig, and monkey [10, 19–22], and may contribute to a subset of oocyte-based infertility in humans [23, 24]. Interestingly, Nlrp5 expression has also been shown to decrease in oocytes with maternal aging [25, 26], suggesting that a decrease or absence of NLRP5 could result in compromised fertility in women of advanced reproductive age. Although dysfunction of Nlrp5 has not been specifically shown to be responsible for embryonic arrest in humans, its altered expression in abnormal embryos and developmentally arrested two-cell embryos strongly suggests a role in preimplantation embryo development [27, 28].
The aim of this study was to further characterize Nlrp5-mediated phenotype(s) in ovulated oocytes and embryos, with a particular focus on mitochondria. We show that Nlrp5 hypomorph oocytes have elevated ROS and high mitochondrial membrane potential (ΔΨm) accompanied by altered mitochondrial distribution with decreased mitochondrial pool, perhaps because of damage caused by premature recruitment of mitochondria into respiring state. This indicates that Nlrp5 deficiency presents defects prior to fertilization. Few Nlrp5 hypomorph embryos that continue developing beyond the eight-cell stage exhibit elevated apoptosis. In addition, proapoptotic protein BAX is increased at the two-cell stage; however, neutralizing BAX proves unsuccessful in rescuing early embryonic lethality, implying the role of other cell death members.
MATERIALS AND METHODS
Mouse Husbandry and Genotyping
The Nlrp5tm/tm line used in this study was derived from animals that were initially created using stem cell mutagenesis as described previously [29], and maintained on mixed 129Sv x C57BL6 background. Experimental Nlrp5tm/tm females were produced by intercrossing Nlrp5 heterozygous females and males. All animal protocols were approved by the Mount Sinai Hospital Animal Care Committee and met standards for ethical animal treatment. All mice were housed in the Mount Sinai Hospital animal facility with free access to food and water, and kept on a 12L:12D cycle.
Genotypes of animals were determined by isolating DNA from ear tissue using E.Z.N.A Genomic DNA kit (Omega Biotek). The PCR procedure was carried out with 2 μl of DNA in 20 μl total volume using forward primers specific to Nlrp5+/+ 5′-TCA TGT CCT TGG ATG GCA TG-3′ and KO 5′-ACC GGT GGA TGT GGA ATG TG-3′, and a reverse primer 5′-CCA CGT GCT TTC AAG ATT GC-3′. The PCR cycle consisted of denaturation at 95°C for 30 sec, annealing at 59°C for 30 sec, and extension at 72°C for 1 min, with a total of 37 cycles. Amplified products were resolved on agarose gel with bands at 396 bp (+/+) and 242 bp (targeted tm) allele.
Oocyte and Embryo Collection and Culture
Six- to eight-week-old Nlrp5+/+ and Nlrp5tm/tm females were superovulated with 5 IU of equine chorionic gonadotropin (Sigma-Aldrich) and 48 h later with 5 IU of human chorionic gonadotropin (hCG) (Sigma-Aldrich), by intraperitoneal injection. Fifteen hours later the mice were killed by cervical dislocation and oviducts were removed. The oocytes were retrieved in modified human tubal fluid (mHTF; Irvine Scientific) supplemented with 0.1% bovine serum albumin (BSA; Sigma-Aldrich) and denuded of cumulus cells using hyaluronidase (Sigma-Aldrich). For analysis of two-cell embryos, females were mated with males of proven fertility of the same genotype, and embryos were flushed at 1.5 days postcoitum (dpc) from the oviduct. Embryos were cultured in pre-equilibrated 20-μl drops of Global media (Life Global), or Global media containing 3.25 ng/μl BH4 peptide (Calbiochem) supplemented with 0.1% BSA under mineral oil (Sigma-Aldrich) in a humidified incubator at 37°C, 5% CO2, until the time of analysis.
Nlrp5+/+ and Nlrp5tm/tm embryos were also treated with BAX-inhibiting peptide V5 (BIP-V5; Calbiochem) or an inactive form of BAX-inhibiting peptide (BIP-NC; Calbiochem). To determine if the peptide penetrates through the zona, oocytes were exposed to doxorubicin (DXR) as previously described [30]. Zona pellucida from oocytes or embryos was removed by short incubation with acid Tyrode (Sigma-Aldrich), and followed by culture with either BIP-V5, BIP-NC (prepared from a 5 mM stock in dimethyl sulfoxide [DMSO]), or vehicle, at a final concentration of 100 μM. In vitro-cultured embryos were assessed daily from zygote to blastocyst stage using a dissecting microscope. The final values were a mean of the percentages of at least three independent experiments. At Day 4.5, in vitro-cultured embryos were fixed in formalin for 15 min, followed by a 10-min staining with 4′,6-diamidino-2-phenylindole (DAPI), and each embryo was assessed for total cell number and number of mitotic as well as apoptotic cells as previously described [31]. For nocodazole treatment, two-cell embryos were flushed at 1.5 dpc and exposed to either vehicle (DMSO) or nocodazole (10 μM; Sigma) for 30 min. Afterwards, embryos were cultured in the presence of Mitotracker Red and 2′-7′-dichlorodihydrofluorescein diacetate (DCHFDA) as described below. For ovarian reserve and bone mineral density (BMD) studies, please see the Supplemental Materials and Methods (all supplemental data are available online at www.biolreprod.org).
Mitochondrial Distribution and Membrane Potential
JC-1 is a lipophilic cationic dye (5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazolyl carbocyanine iodide; Molecular Probes, Invitrogen), that signals the loss of ΔΨm. JC-1 analysis was performed as previously described [32]. Briefly, JC-1 was prepared at a concentration of 10 μM in HTF medium from 1 M stock solutions directly prior to use. The dye was pre-equilibrated for 30 min prior to the addition of oocytes or embryos, which were stained for 20 min at 37°C, 5% CO2, in a humidified incubator. Following incubation, the embryos were washed in mHTF medium supplemented with 0.1% BSA, and placed in 20 μl of mHTF medium supplemented with 0.1% BSA on a 0.2-mm Hiraki depression slide, covered with a coverslip. Samples were imaged on a deconvolution microscope (Olympus IX70; Applied Precision Inc.) under the fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate filters. Ten 1-μm optical sections for each sample were obtained and analyzed for quantitation of signal intensity as previously described [32]. Live metaphase II oocytes and two-cell embryos were stained with Mitotracker Red or Green (Molecular Probes, Invitrogen) at final concentration of 200 μM and imaged as described above.
Measurements of Oxidized Mitochondrial FAD++Autofluorescence and ATP:ADP Ratio
Following collection of oocytes, FAD++ autofluorescence measurements were carried out as previously described [33]. Ten 1-μm sections were obtained on deconvolution microscope for each oocyte and summed using the softWoRx software (Applied Precision Inc.). Average fluorescence intensities were obtained on a per oocyte basis, and then compiled per group.
The ApoSENSOR ADP/ATP Ratio Assay Kit (BioVision) was used to assess for oocyte viability. Reagents were prepared as per the manufacturer's instructions. Groups of 10 oocytes (triplicate sets for both genotypes) were suspended in 100 μl of nuclear releasing agent and were added to individual wells in a 96-well luminometer plate. Fifty μl of ATP monitoring enzyme was automatically injected to the oocyte-containing solution 60 sec prior to the first reading by the luminometer (MicroLumat Plus; EG&G). The wells were incubated and protected from light for 10 min and were read a second time. Fifty μl of ADP monitoring enzyme was then automatically injected to the oocyte-containing solution 60 sec prior to performing the third luminometer reading. Individual readings for ATP and ADP were recorded and compared.
Quantification of Reduced Glutathione, ROS, and Metabolic Reducing Activity of Oocytes
Quantitation of cellular glutathione (GSH) and ROS in Nlrp5+/+ and Nlrp5tm/tm oocytes was accomplished by the use of monochlorobimane and DCHFDA respectively (Molecular Probes, Invitrogen) and is described in detail elsewhere [34]. Reducing metabolic activity was determined by MTT assay performed in groups of 10 oocytes as previously described [34] with reduction of final DMSO volume to 60 μl.
Mitochondrial DNA Copy Number Using Quantitative PCR
Absolute mitochondrial DNA (mtDNA) copy number for a single oocyte was calculated as described previously in Wai et al. [35] using a standard curve generated from quantitative real-time PCR (qPCR) of 101–106 copies of mouse mtDNA region. The 600-bp mouse mtDNA was amplified using Mito (F): 5′-TCCTATGAATCCGAGCATCC-3′ and Mito (R): 5′-GTACGATGGCCAGGAGGATA-3′ primers and cloned into pTOPO2.1 plasmid (Invitrogen). The standard concentration of the generated pTOPO-Mito was quantitated using nanodrop and utilized in qPCR. Mitochondrial DNA copy number in single oocytes was determined as previously described [36].
Primer sequences used for qPCR for amplifying mtDNA were Mt11 (forward): 5′-CATACCCCGAAAACGTTGGTT-3′ and Mt12 (reverse): 5′-AGGACCTAAGAAGATTGTGAAGTAGATGA-3′. Linear regression analysis for the standard curve generated from 101–106 copies gave a high correlation coefficient of R2 = 0.9975. The absolute mtDNA for a single oocyte was calculated using the standard curve. The oocyte samples and standard reactions were done in triplicate.
Quantitative Real-Time RT-PCR (qRT-PCR)
Following oocyte retrieval, 10 oocytes were transferred into 50 μl of guanidinium isothiocyanate solution and stored at −80°C until further use. Total nucleic acid was precipitated with 100% ethanol, 7.5 mM ammonium acetate, and glycogen. Complementary DNA for real-time PCR analysis was prepared as described [37].
Gene expression levels of the cDNA products were assessed by qRT-PCR using the DNA Engine Opticon 2 thermocycler (MJ Research Inc.) and Opticon Monitor 2 software (MJ Research Inc.). Complementary DNA quality was assessed by analyzing the housekeeping gene Actb (β-actin). Quantitative RT-PCR for proapoptotic genes (Bax, Bok (Mtd), Bak1, Hrk), antiapoptotic genes (Bcl2l10 (Diva) and Bag1), and the housekeeping gene Actb were carried out in 20-μl reactions. The sequences for forward and reverse primers for the selected mRNAs are listed in Supplemental Table S1. With the exception of Hrk, all gene expression experiments utilized the SYBR green PCR mix (Applied Biosystems). All qRT-PCR conditions were as follows: 95°C for 15 sec, 60°C for 1 min. Comparisons of expression levels were determined by normalizing to Actb.
Immunocytochemistry
Nlrp5+/+ and tm/tm oocytes or embryos were flushed at 1.5 dpc (∼42–44 h after hCG), scored, and pretreated with permeabilization buffer (InnoCyte Cytochrome c release kit; Calbiochem) followed by fixation, or just fixed in 10% buffered formalin. Fixed samples were preabsorbed with 10% horse serum + 0.1% Triton-X + phosphate-buffered saline (blocking solution) in a humid chamber for 1 h at room temperature. Following preabsorption, embryos were incubated overnight with an anti-Tom 20 FL-145 (Santa Cruz Biotechnology), anti-cytochrome c (InnoCyte Cytochrome c release kit); anti-total p66 SHC (BD Transduction Laboratories)/anti-Ser36 P66 SHC (Abcam); or anti-total BAX P19 antibody (Santa Cruz Biotechnology). Active BAX was detected on permeabilized samples with antibody recognizing opened confirmation of N terminus (BAX NT; Millipore). After overnight incubation the embryos were incubated for 1 h with appropriate secondary antibody: anti-mouse FITC or Alexa 488 and donkey anti-rabbit FITC or Alexa 594 (Invitrogen) at room temperature. Following incubation, samples were washed and counterstained with the fluorochrome DAPI (Sigma-Aldrich). Embryos were scanned by spinning disk-confocal microscope (Leica) under 20× objective using appropriate filters. Ten optical sections from each sample were taken, deconvolved, and analyzed with Volocity Image Analysis Software (Perkin Elmer). Values obtained from negative control samples without primary antibody were subtracted from experimental signal, and data are shown as sum of fluorescent intensity expressed as random fluorescence units (RFUs).
Statistical Analyses
All results are given as mean ± SEM. All statistics were accomplished with SigmaPlot 11 (Systat Software Inc.). Analysis was performed either with a one-way ANOVA following Tukey posttest, Mann-Whitney rank sum test, or Student t-test, as appropriate. Results were considered statistically significant if P < 0.05.
RESULTS
Nlrp5 Disruption Does Not Affect Oogenesis or Ovulation Rates
As Nlrp5 has been originally identified in a screen for targets of autoimmune oophoritis, we wanted to determine whether the ovarian follicle pool could be affected by Nlrp5 deficiency, and whether this is further impacted by aging. Ovarian histomorphometric analyses revealed the presence of follicles of all developmental stages, and the number of follicles in each stage was comparable between Nlrp5+/+ and Nlrp5tm/tm female mice (Supplemental Fig. S1C). These analyses were performed on both 2- and 6-mo-old mice and neither showed any differences in ovarian reserve. Furthermore, as previously reported [2], ovulation rates were also comparable between both groups. To further confirm that Nlrp5 has no effect on ovarian function, we assessed several physiological changes known to be associated with ovarian failure, such as total BMD, vertebral as well as femoral BMD, and body weight, as well as body fat composition (Supplemental Fig. S1, A and B). Our results did not reveal any alterations, mirroring the comparable follicle pools in Nlrp5+/+ and Nlrp5tm/tm mice. Overall, these data confirm that Nlrp5 does not compromise ovarian follicle pool even with aging.
Characterization of Mitochondrial Parameters in Nlrp5-Deficient Oocytes
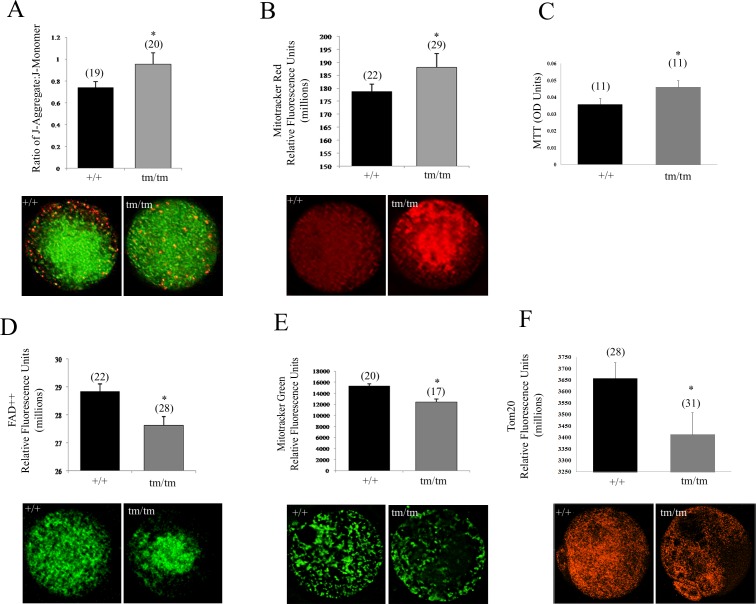
Using live-cell dynamic fluorescence imaging, we assessed the effects of Nlrp5 deficiency on mitochondrial parameters in ovulated oocytes. This technique, in combination with a range of mitochondrial fluorescent probes, enabled evaluation of mitochondrial function within a single oocyte. ΔΨm of Nlrp5+/+ and Nlrp5tm/tm oocytes was measured with the commonly used dye JC1, which stains mitochondria in a membrane potential-dependent manner. Nlrp5tm/tm oocytes exhibit a significantly (P = 0.016) elevated ΔΨm in comparison to +/+ oocytes (Fig. 1A). Furthermore, mitochondrial distribution patterns of active mitochondria were altered in Nlrp5tm/tm oocytes (Fig. 1A). As previously described for the healthy ovulated oocytes, highly polarized (red) mitochondria were enriched in subcortical region in +/+, whereas Nlrp5tm/tm oocytes exhibited a random dispersal of highly polarized organelles throughout the cytoplasm. Mitotracker Red, a dye taken up only by respiring mitochondria, confirmed an increased pool of active mitochondria in the Nlrp5tm/tm oocytes (Fig. 1B).
FIG. 1.
Mitochondrial characterization in Nlrp5-deficient oocytes. A) Mitochondrial membrane potential as measured by JC-1 dye in Nlrp5+/+ and Nlrp5tm/tm oocytes. A significant increase in mitochondrial membrane potential was observed in Nlrp5tm/tm oocytes. Values represent ratio of J-aggregate to J-monomer ± SEM. Asterisk (*) indicates significance (P = 0.016). Representative images are shown below the graph. B) Respiring mitochondrial pool measured by Mitotracker Red in Nlrp5 oocytes. Consistent with JC-1 results, a significant increase in respiring mitochondria was observed in Nlrp5tm/tm oocytes. Values represent mean fluorescence units ± SEM (*P < 0.01). C) MTT assay reflecting reducing activity of mitochondrial enzymes was elevated in mutant oocytes (P = 0.05). Values shown represent colorimetric OD (optical density) reading generated by conversion of dye to colored substrate in pools of 10 oocytes. N indicates number of pools. D) Oxidized FAD++ measurement in Nlrp5 oocytes using an FITC filter. Values shown are relative fluorescence units ± SEM. Numbers in parentheses represent number of oocytes/sample sets used per group; P = 0.008. E) Total mitochondrial pool as measured by Mitotracker Green in Nlrp5 hypomorph oocytes. Representative images are shown below the graph. Values represent mean fluorescence units ± SEM (*P < 0.01). F) Decreased expression of mitochondrial protein Tom20 in mutant oocytes corroborates decreased mitochondrial pool. Values represent mean fluorescence units ± SEM (*P < 0.001). N in each graph (with the exception of MTT assay) indicates number of oocytes used for each assay. Original magnification ×200.
Elevated ΔΨm may culminate from several sources, including increased mitochondrial respiratory activity, higher supply of substrates, or inhibition of ATP synthase activity [38]. Hence, we measured the autofluorescence signal of oxidized mitochondrial flavoproteins (FAD++). Nlrp5tm/tm oocytes exhibited a significantly lower level of FAD++ in comparison to +/+ oocytes (Fig. 1D), as well as increased activity of reducing enzymes in MTT (Fig. 1C). This suggests that the source of elevated ΔΨm may be due to increased mitochondrial respiratory activity. However, assessment of ATP:ADP content in +/+ and mutant oocytes found no significant differences.
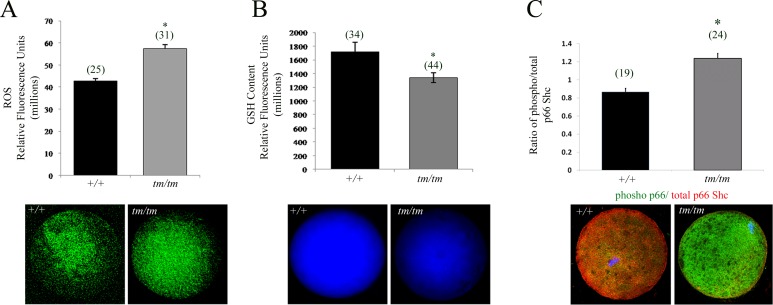
Excessive oxidative stress, another major cause of poor oocyte quality, is an important indicator of early embryo damage [39]. Because ROS are a by-product of mitochondrial respiration, we decided to evaluate accumulation of ROS using H2DCHFDA, a dye that detects H2O2 as well as other ROS species such as NO, lipid peroxides, singlet O2, and superoxide [40]. In line with the elevated ΔΨm and increased respiring mitochondrial pool, we detected increased levels of ROS in Nlrp5tm/tm oocytes (Fig. 2A).
FIG. 2.
Altered GSH levels result in increased ROS production in Nlrp5-deficient oocytes. A) ROS measurement in Nlrp5+/+ and Nlrp5tm/tm oocytes. Values represent relative fluorescence units ± SEM. Increased ROS production was observed in Nlrp5 hypomorph oocytes. Asterisk (*) indicates significance; P < 0.05. Representative images are shown below. B) Baseline GSH content in Nlrp5+/+ and Nlrp5tm/tm oocytes. Values represent mean fluorescence units ± SEM. A significant reduction in the baseline GSH content was observed in tm/tm oocytes (*P < 0.05). C) Expression of p66 SHC in oocytes. Relative fluorescent intensity in RFU generated by phospho p66 (green) to total p66Shc (red) was used to show increased expression of phosphorylated isoform (P < 0.001). There was no change in the signal intensity of total p66SHC, and change in the ratio is due to rise of phospho p66SHC signal (green). Numbers in parentheses represent number of oocytes used in each experiment. Original magnification ×200.
ROS levels have been associated with changes in mitochondrial content [41]. Hence, we hypothesized that the localization of Nlrp5 to mitochondria, together with increased ROS levels, could affect mitochondrial biogenesis. Mitochondrial DNA copy number is an important indicator of organellar status and ranges from 100 000 to 150 000 copies per mature oocyte [42, 43]. Nlrp5+/+ oocytes contained an average of 115 941 ± 37 171 mtDNA copies, in comparison to Nlrp5tm/tm oocytes with an average of 78 225 ± 15 212 copies. However, these results did not reach statistical significance because of variability among individual oocytes (n = 12/genotype). Because a single mitochondrion can contain one to two copies of mtDNA per organelle [44], we decided to use Mitotracker Green, which binds to mitochondrial membranes irrespective of respiratory status. This analysis confirmed that there is a decreased signal in Nlrp5tm/tm oocytes (Fig. 1E). Similarly, expression of Tom 20, a mitochondrial import protein localizing to outer mitochondrial membrane, was also reduced (Fig. 1F; P = 0.008). Taken together, these data indicate that Nlrp5 causes either inadequate mitochondrial biogenesis or depletion of mitochondrial pool.
Finally, we assessed levels of the antioxidant GSH using monochlorobimane. Nlrp5tm/tm oocytes demonstrated a significantly lower baseline GSH level in comparison to +/+ oocytes (Fig. 2B), indicating that the excess ROS levels may in part be a consequence of the lower GSH baseline levels. In order to determine if increased ROS causes compromised cellular state, we next explored if phospho p66SHC, a marker of mitochondrial stress [45] and mediator of embryo arrest [46], becomes elevated. Consistent with increased production of ROS in oocytes, there was an increased accumulation of phosphorylated isoform of p66-SHC in Nlrp5tm/tm in oocytes (Fig. 2C; P < 0.001) as well as two-cell embryos (Supplemental Fig. S2).
Nlrp5 and Regulation of Maternal BCL2 Family Endowment
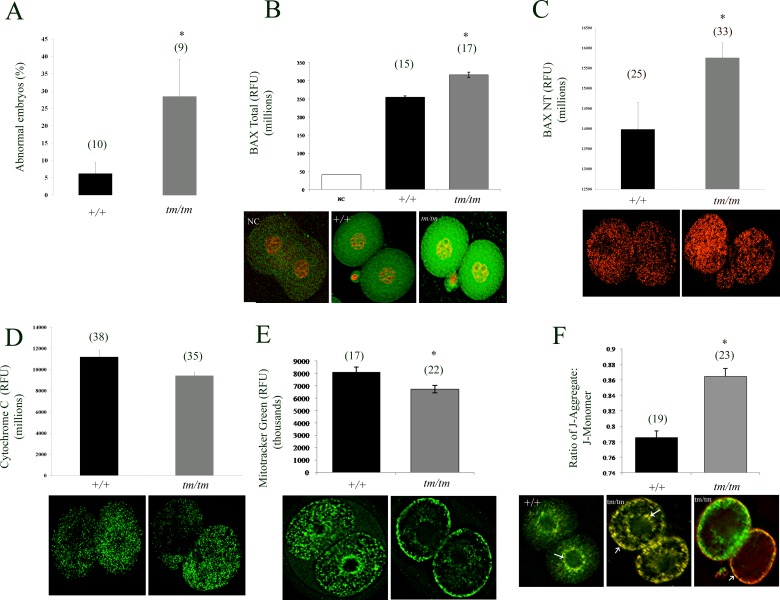
In addition to changes in metabolic mitochondrial status, ΔΨm is also a signal in cell life-death decisions. Nlrp5tm/tm embryos flushed at two-cell stage contained a significantly higher proportion of either fragmented or irregular-sized blastomeres (Fig. 3A) and when maintained in culture failed to progress beyond two-cell stage. The two-cell arrest and fragmentation with hallmarks of oxidative stress in Nlrp5tm/tm embryos further indicates possible dysregulation of cell death/survival molecules. Hence, we profiled mRNA expression of members of the BCL2 family. We detected no change in the expression of the antiapoptotic members, Bcl2l10 (Diva) and Bag1. Of the proapoptotic members (Bax, Bak, Bok (Mtd), and Hrk) assayed, only Bax transcript displayed a significant (P = 0.002) threefold increase in Nlrp5tm/tm oocytes. Because of the increased Bax mRNA expression in Nlrp5tm/tm oocytes, we explored BAX protein expression in Nlrp5tm/tm two-cell embryos by immunocytochemistry. In line with the qRT-PCR data, Nlrp5-deficient embryos express more total as well as active BAX protein (Fig. 3B), suggesting that BAX could be involved in Nlrp5tm/tm embryo demise. Active oligomerized BAX integrates into the outer mitochondrial wall and contributes to release of mitochondrially sequestered apoptosis-inducing factors [47]. Consistent with increased expression of active BAX, we observed reduced levels of cytochrome c retained by mitochondria in Nlrp5tm/tm embryos (Fig. 3C). This indicates either a breach of outer mitochondrial membrane or a decrease in the mitochondrial pool.
FIG. 3.
Mitochondrial and molecular changes in two-cell stage embryos. A) Proportion of abnormal embryos retrieved from WT and Nlrp5tm/tm pregnant females retrieved at Day 1.5 from oviducts. Data are shown per female as percentage of abnormal embryos with fragmented or irregular size blastomeres. B, C) BAX protein (total and activated, NT form respectively) expression in Nlrp5tm/tm two-cell embryos. Consistent with the transcript levels, a significant increase in BAX protein was observed in two-cell embryos. Values represent mean fluorescence units ± SEM. Asterisk indicates significance; P < 0.05. D) Cytochrome c level in mutant embryos was decreased by two-cell stage. Values represent sum of fluorescent signal (in RFUs) ± SEM. Asterisk indicates significance; P = 0.049. E) Total mitochondrial pool as measured by Mitotracker Green in Nlrp5tm/tm two-cell embryos. Representative images are shown below the graph. Values represent sum of fluorescent signal (in RFUs) ± SEM. Asterisk indicates significance; P = 0.007. (F) Mitochondrial membrane potential as measured by JC-1 dye in Nlrp5+/+ and Nlrp5tm/tm two-cell embryos. Values represent ratio of J-aggregate (red) to J-monomer (green) ± SEM. Asterisk means values are significant, P < 0.05. Representative images are shown below the graph. Original magnification ×200. Numbers in parentheses represent number of oocytes per group. For generation of graphs and statistics, negative control (NC) values were subtracted from total RFUs.
BAX protein is known to regulate mitochondrial morphogenesis as well as activity [48]. The elevated ΔΨm, decreased mitochondrial pool, and altered distribution patterns found in the Nlrp5tm/tm oocytes were also persistent in Nlrp5tm/tm two-cell embryos (Fig. 3D). However, the pattern of subcellular distribution of mitochondria was different. Whereas most of the +/+ embryos exhibited a diffuse distribution of active mitochondria throughout the cytoplasm with subtle perinuclear clustering (89%, n = 18), the majority of Nlrp5tm/tm embryos showed an increased subcortical mitochondrial clustering (87%, n = 38). We then compared the total mitochondrial content and distribution using Mitotracker Green, which stains all mitochondria irrespective of their activity. Similar to the profile of JC-1 for active mitochondria, we observed a high enrichment of organelles in the subplasmalemmal cortex of Nlrp5tm/tm embryos (76%; n = 21), whereas all +/+ embryos (n = 18) contained cytoplasmic distribution with subtle perinuclear clustering (Fig. 3C). Merging and analyzing these two sets confirmed significant enrichment of abnormally localized mitochondria in the cortex of Nlrp5tm/tm embryos (chi-square test P < 0.001). Similar to oocytes, fluorescent intensity of Mitotracker Green was significantly lower in Nlrp5tm/tm embryos. NLRP5 has been recently linked to formation of cytoplasmic lattices [20], disruption of which impacts organellar distribution [49]. As altered mitochondrial distribution could be caused by poor tethering of organelles to microtubules, we exposed wild-type two-cell embryos to nocodazole, a microtubule destabilizing agent. Depolymerization of microtubules did not cause redistribution of mitochondria to subcortical region. However, mitochondrial activity, as determined by measurement of Mitotracker Red as well as ROS levels, was significantly decreased (P = 0.001, P < 0.001 respectively; Supplemental Fig. S3). Thus, dissociation of mitochondria from microtubules disrupts mitochondrial function, but this does not mimic changes observed in Nlrp5tm/tm embryos.
Transactivator of Transcription-BH4 and BIP Insufficient to Rescue Nlrp5-Targeted Embryos
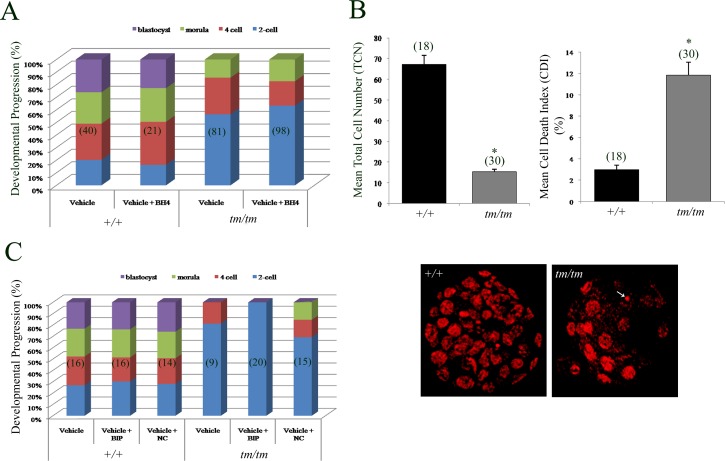
To identify the extent of the role of BAX in the demise of these embryos, we attempted to rescue these embryos by interfering with BAX protein function. We used the cell-permeable antiapoptotic BH4 domain of BCL2L1 (transactivator of transcription [TAT]-BH4) as this peptide can suppress BAX-induced cellular demise [50, 51] and can prevent induction of ROS in embryos [52]. However, we found no impact of TAT-BH4 on progression of Nlrp5tm/tm embryos (Fig. 4A), suggesting that the BH4 domain of BCL2L1 alone is insufficient to improve developmental competence. Few Nlrp5tm/tm embryos progressed beyond the two-cell stage; however, their quality was highly inferior to that of +/+ embryos (Fig. 4B). Those Nlrp5tm/tm embryos that progressed beyond the eight-cell stage in vitro had a significantly lower total cell number, along with a higher percentage of apoptotic cells, evidenced by nuclear condensation and chromatin fragmentation (Fig. 4B).
FIG. 4.
Developmental progression rate and rescue using TAT-BH4 and BIP-V5 peptides in Nlrp5 embryos. A) Zygotes cultured either in vehicle or vehicle + BH4 and observed for their progression rate. Significant increases in the formation of blastocysts were seen in Nlrp5+/+ compared to Nlrp5tm/tm embryos. However, presence of BH4 neither rescued the two-cell arrest in tm/tm embryos nor increased the rate of blastocyst formation in +/+ embryos. Values are expressed in percentages. B) Mean cell number and cell death index in Nlrp5 embryos treated with vehicle only. A significant reduction in mean total cell number along with a significant increase in cell death index was observed in Nlrp5tm/tm embryos. Cell death index is shown in percentage and is expressed as mean ± SEM. Representative images of both +/+ and Nlrp5tm/tm blastocysts are shown below. Original magnification ×400. Arrow indicates apoptotic cells. Asterisk indicates values are significant (P < 0.05). Numbers in parentheses represent the number of embryos assessed. C) Nlrp5 zygotes cultured either in vehicle, vehicle + BIP-V5, or vehicle + BIP-NC were assessed for their developmental progression from Day 1.5 to Day 4.5. A significant increase in the formation of blastocysts was seen in Nlrp5+/+ compared to Nlrp5tm/tm embryos. However, presence of BIP neither rescued the two-cell arrest of Nlrp5tm/tm embryos nor increased the rate of blastocyst formation in +/+ embryos. Values are expressed as a percentage of embryos reaching indicated developmental stage.
In order to confirm that BAX is responsible for the developmental defects observed in Nlrp5 embryos, we employed the use of BAX inhibitory peptide BIP-V5, which is known to directly bind and inhibit BAX activation/translocation to the mitochondria. Previous studies had shown that oocytes treated with DXR, a cancer drug, were known to induce death through the BAX pathway [53]. In order to determine the biological activity of BIP-V5, we tested the ability of BIP-V5 to suppress DXR-mediated death in oocytes. These experiments revealed that removal of the zona pellucida was necessary for BIP-V5 to decrease the incidence of oocyte fragmentation triggered by DXR (intact, DXR 30% vs. DXR/BIP-V5, 38%; no zona, DXR 80% vs. DXR/BIP-V5 47%). Thus, for all experiments with embryos, zona pellucida was removed by brief exposure to acid Tyrode. To determine if BIP-V5 is able to rescue the two-cell embryo arrest in Nlrp5 mice, the zona pellucida was removed from zygotes, which were subsequently treated with 100 μM BIP-V5 or BIP-NC (negative control; an inactive form of BIP that does not suppress BAX-mediated apoptosis) in Global medium and cultured until Day 4.5 (Fig. 4C). However, Nlrp5tm/tm embryos showed no improvement in blastocyst formation in the presence of BIP-V5. Finally, we verified this using a genetic approach by utilizing embryos generated from double-knockout mice targeting Bax and Nlrp5. However, double-knockout females (n = 4) did not produce any pups, indicating that BAX is not the sole mediator of the developmental defects observed in Nlrp5tm/tm embryos. Taken together, these data suggest that the BH4 domain or BAX inhibiting peptide are unable to improve embryo developmental potential.
DISCUSSION
Both the underlying cellular function and mode of action of Nlrp5 remain unknown. The goal of this study was to investigate the Nlrp5 mitochondrial phenotype in oocytes, and to determine if there is a defect prior to fertilization. Consistent with previous studies [2], ovarian morphometric analyses along with bone density assays confirmed that Nlrp5tm/tm females have no obvious follicle phenotype even with advanced maternal age. However, oocytes and embryos conceived by hypomorphic females exhibit mitochondrial defects, which could contribute to increased rates of cell death.
It has been previously shown that NLRP5 protein localizes to mitochondria [18], but significance of this observation is currently unknown. Mitochondria, which are maternally inherited, have a well-established role in determining oocyte and embryo quality [54]. ΔΨm, ROS, metabolic outputs, and mitochondrial content are important indicators of cellular health, reflecting overall mitochondrial integrity and metabolic activity [55–57]. Nlrp5tm/tm oocytes exhibit indicators of increased mitochondrial activity, albeit without corresponding change in ATP levels. We have previously observed elevated ΔΨm in oocytes with impaired mitochondrial morphology [34], as well as in arrested mouse embryos in response to culture environment [32]. Whereas mitochondrial depolarization occurs with the onset of death and as a consequence of exposure to mitochondrial poisons, in live cells it is constantly changing. Similar to our findings, increased mitochondrial potential and ROS production with impaired redox state was described in oocytes of females maintained on a high-fat diet [58]. Flickering of mitochondrial membrane potential mirrors metabolic needs and reflects coupling of ΔΨM to ATP synthase [59]. For example, metabolic stress brought on by diabetes is associated with down-regulated mitochondrial biogenesis accompanied by elevated ΔΨm [60]. These results are cell type-specific, as opposite outcomes were also reported in cumulus cells of diabetic mice [61].
Our results imply that Nlrp5tm/tm oocytes have increased mitochondrial respiratory activity, perhaps caused by precocious recruitment of resting mitochondria into respiratory state, likely in the attempt to maintain balanced ATP production. However, this happens at the expense of increased ROS levels, which could cause mitochondrial damage. Indeed, we observed significant decrease in expression of mitochondrial markers, indicating a compromised or diminished mitochondrial pool. Loss of mitochondria appears to be progressive, as no change in mitochondrial fraction was detected by histo-morphometry in germinal vesicle Nlrp5tm/tm oocytes [20], but the change appears severe in two-cell embryos (present study). However, it remains to be determined whether this depletion is a consequence or an initial event contributing to embryo arrest. Alternatively, localization of two mitochondrial proteins, TOM20 and cytochrome c, could be impaired because of Nlrp5 deficiency, and their decreased expression may not necessarily reflect a decreased mitochondrial pool. However, as Mitotracker Green and mtDNA also produced decreased signal, we believe that mitochondrial pool is compromised.
Proper mitochondrial localization in oocytes and embryos is important for signaling events associated with fertilization and developmental competence [62]. Altered mitochondrial aggregation patterns were previously identified in preimplantation embryos undergoing arrest [63]. Studies in hamster two-cell embryos suggested that the translocation of mitochondria to the subcortical regions of the cells, and especially to areas of cell-to-cell contact, were mediated by microfilaments, whereas translocation of mitochondria to the perinuclear region was mediated by microtubules [64]. It is plausible that NLRP5, which localizes to mitochondria, may be involved in translocating/tethering a subset of the active organelles to either the subcortical or the peri-nuclear region at appropriate times during embryo development. We have not been able to replicate abnormal mitochondrial distribution seen in Nlrp5tm/tm embryos by disruption of microtubules. Similarly, nocodazole and vinblastine were not able to affect mitochondrial distribution in oocytes [49]. Perhaps mitochondrial reorganization could be mediated by cytoplasmic lattices rather than by microtubules. Recently NLRP5 was shown to localize to a cytoplasmic lattice network together with PADI6, an essential component of this structure [20]. Cytoplasmic lattices are poorly formed and labile in the Nlrp5tm/tm hypomorph oocytes [20]. Lattices serve as sites of ribosomal storage and transport [65] but likely have additional roles in regulating organellar distribution. These results are consistent with the recent observation of altered mitochondrial and ER localization in Padi6-deficient oocytes [49]. In addition, Nlrp5tm/tm oocytes also exhibit a lower baseline level of the antioxidant GSH, suggesting that the elevated ROS could be in part also a consequence of insufficient clearance. Excess ROS levels have negative impact on embryo development and could also affect mitochondrial biogenesis [66]. As a marker of mitochondrial stress, we detected an increase in phosphorylated p66Shc. This TP53-inducible alternatively spliced isoform of a growth factor adapter becomes phosphorylated in response to oxidative stress [45]. A fraction of phosphorylated p66Shc localizes to mitochondria, where it binds to cytochrome c and acts as an oxidoreductase, further promoting generation of ROS and causing organelle malfunction and eventually cell death [45]. Previous work in bovine embryos identified this adaptor protein as a mediator of permanent embryo arrest [46, 67]. In addition, we have recently shown that injection of recBCL2L1 stimulates mitochondrial function in stressed embryos, prevents excessive generation of ROS, and also maintains low levels of phosphorylated p66Shc [52], implicating BCL2 family members as regulators of developmental competence.
Consequently, further expression analysis revealed increased expression of BAX in Nlrp5tm/tm embryos. This was accompanied by decreased mitochondrial retention of cytochrome C and increased predisposition to cell death. We attempted to improve embryo developmental competence with the use of two peptides that directly or indirectly inhibit BAX activity: BIP-V5 and TAT-BH4 respectively. However, neither supplementation of TAT-BH4 nor suppression of BAX (chemically or genetically) was sufficient for improving developmental potential in Nlrp5 mutant embryos. It is likely that impaired mitochondrial function could lead to a myriad of downstream effects too vast to rescue. It is also possible that additional proapoptotic members not efficiently mitigated by these peptides could be activated in this model. Alternatively, the BH4 domain alone may not be sufficient to alleviate the developmental block, as this approach also failed to rescue medium induced embryo arrest [52]. However, as Bax/Nlrp5 double knockout animals produced no live births, we conclude that BAX is likely only one piece of a bigger puzzle involved in the death of these embryos.
Supplementary Material
Footnotes
Supported by the Grants from Canadian Institutes of Health Research (CIHR MOP 14058, 84328), Canadian Fund for Innovation (CFI#203771), and Canada Research Chairs Program. This work was also supported in part by the Intramural Research Program on Reproductive and Adult Endocrinology, National Institute of Child Health and Human Development, National Institutes of Health.
REFERENCES
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 1989; 121: 101 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 2000; 26: 267 268 [DOI] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature 2000; 407: 693 694 [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy 2008; 4: 1076 1078 [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 2006; 20: 1744 1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. Maternal control of early mouse development. Development 2010; 137: 859 870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 2003; 13: 2110 2117 [DOI] [PubMed] [Google Scholar]
- Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet 2003; 33: 187 191 [DOI] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A 2009; 106: 7473 7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta). Hum Reprod 2009; 24: 415 424 [DOI] [PubMed] [Google Scholar]
- Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol 2003; 4: 95 104 [DOI] [PubMed] [Google Scholar]
- Dade S, Callebaut I, Paillisson A, Bontoux M, Dalbies-Tran R, Monget P. In silico identification and structural features of six new genes similar to MATER specifically expressed in the oocyte. Biochem Biophys Res Commun 2004; 324: 547 553 [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol 2011; 12: 121 128 [DOI] [PubMed] [Google Scholar]
- Frederick Lo C, Ning X, Gonzales C, Ozenberger BA. Induced expression of death domain genes NALP1 and NALP5 following neuronal injury. Biochem Biophys Res Commun 2008; 366: 664 669 [DOI] [PubMed] [Google Scholar]
- Liu F, Lo CF, Ning X, Kajkowski EM, Jin M, Chiriac C, Gonzales C, Naureckiene S, Lock YW, Pong K, Zaleska MM, Jacobsen JS. et al. Expression of NALP1 in cerebellar granule neurons stimulates apoptosis. Cell Signal 2004; 16: 1013 1021 [DOI] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, Matsuzawa S, Terskikh AV. et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell 2007; 129: 45 56 [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 2008; 15: 416 425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, De Pol A, Vanevski K, Dorward H, Sena P, Palumbo C, Bondy CA, Nelson LM. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology 2004; 145: 1427 1434 [DOI] [PubMed] [Google Scholar]
- Pennetier S, Perreau C, Uzbekova S, Thelie A, Delaleu B, Mermillod P, Dalbies-Tran R. MATER protein expression and intracellular localization throughout folliculogenesis and preimplantation embryo development in the bovine. BMC Dev Biol 2006; 6: 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kan R, Anguish L, Nelson LM, Coonrod SA. Potential role for MATER in cytoplasmic lattice formation in murine oocytes. PLoS One 2010; 5: e12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development 2008; 135: 259 269 [DOI] [PubMed] [Google Scholar]
- Ma J, Milan D, Rocha D. Chromosomal assignment of the porcine NALP5 gene, a candidate gene for female reproductive traits. Anim Reprod Sci 2009; 112: 397 401 [DOI] [PubMed] [Google Scholar]
- Tong ZB, Bondy CA, Zhou J, Nelson LM. A human homologue of mouse Mater, a maternal effect gene essential for early embryonic development. Hum Reprod 2002; 17: 903 911 [DOI] [PubMed] [Google Scholar]
- Sena P, Riccio M, Marzona L, Nicoli A, Marsella T, Marmiroli S, Bertacchini J, Fano RA, La Sala GB, De Pol A. Human MATER localization in specific cell domains of oocytes and follicular cells. Reprod Biomed Online 2009; 18: 226 234 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Falco G, Piao Y, Poosala S, Becker KG, Zonderman AB, Longo DL, Schlessinger D, Ko M. Effects of aging and calorie restriction on the global gene expression profiles of mouse testis and ovary. BMC Biol 2008; 6: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet 2004; 13: 2263 2278 [DOI] [PubMed] [Google Scholar]
- Zhang P, Dixon M, Zucchelli M, Hambiliki F, Levkov L, Hovatta O, Kere J. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLoS One 2008; 3: e2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM. Reijo Pera RA. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010; 28: 1115 1121 [DOI] [PubMed] [Google Scholar]
- Tong ZB, Nelson LM, Dean J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm Genome 2000; 11: 281 287 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Lee HJ, D'Estaing SG, Tilly J, Perez GI. Molecular requirements for doxorubicin-mediated death in murine oocytes. Cell Death Differ 2006; 13: 1466 1474 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Latham KE, Casper RF, Casper RF, Varmuza SL. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol Reprod Dev 1998; 51: 243 253 [DOI] [PubMed] [Google Scholar]
- Acton BM, Jurisicova A, Jurisica I, Casper RF. Alterations in mitochondrial membrane potential during preimplantation stages of mouse and human embryo development. Mol Hum Reprod 2004; 10: 23 32 [DOI] [PubMed] [Google Scholar]
- Dumollard R, Marangos P, Fitzharris G, Swann K, Duchen M, Carroll J. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 2004; 131: 3057 3067 [DOI] [PubMed] [Google Scholar]
- Perez GI, Acton BM, Jurisicova A, Perkins GA, White A, Brown J, Trbovich AM, Kim MR, Fissore R, Xu J, Ahmady A, D'Estaing SG. et al. Genetic variance modifies apoptosis susceptibility in mature oocytes via alterations in DNA repair capacity and mitochondrial ultrastructure. Cell Death Differ 2007; 14: 524 533 [DOI] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet 2008; 40: 1484 1488 [DOI] [PubMed] [Google Scholar]
- Perumalsamy A, Fernandes R, Lai I, Detmar J, Varmuza S, Casper RF, Jurisicova A. Developmental consequences of alternative Bcl-x splicing during preimplantation embryo development. FEBS J 2010; 277: 1219 1233 [DOI] [PubMed] [Google Scholar]
- Rambhatla L, Patel B, Dhanasekaran N, Latham KE. Analysis of G protein alpha subunit mRNA abundance in preimplantation mouse embryos using a rapid, quantitative RT-PCR approach. Mol Reprod Dev 1995; 41: 314 324 [DOI] [PubMed] [Google Scholar]
- Davidson SM, Yellon D, Duchen MR. Assessing mitochondrial potential, calcium, and redox state in isolated mammalian cells using confocal microscopy. Methods Mol Biol 2007; 372: 421 430 [DOI] [PubMed] [Google Scholar]
- Guerin P, El Mouatassim S, Menezo Y. Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum Reprod Update 2001; 7: 175 189 [DOI] [PubMed] [Google Scholar]
- Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol 2010; 594: 57 72 [DOI] [PubMed] [Google Scholar]
- Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res 2009; 37: 749 761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaratti MR, Meirelles FV. Mitochondrial DNA copy number, a marker of viability for oocytes. Biol Reprod 2010; 83: 1 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piko L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 1987; 123: 364 374 [DOI] [PubMed] [Google Scholar]
- Shoubridge EA, Wai T, Mitochondrial DNA. and the mammalian oocyte. Curr Top Dev Biol 2007; 77: 87 111 [DOI] [PubMed] [Google Scholar]
- Gertz M, Steegborn C. The Lifespan-regulator p66Shc in mitochondria: redox enzyme or redox sensor? Antioxid Redox Signal 2010; 13: 1417 1428 [DOI] [PubMed] [Google Scholar]
- Favetta LA, Madan P, Mastromonaco GF, St John EJ, King WA, Betts DH. The oxidative stress adaptor p66Shc is required for permanent embryo arrest in vitro. BMC Dev Biol 2007; 7: 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Valentijn AJ, Zhang L, Gilmore AP. The N-terminal conformation of Bax regulates cell commitment to apoptosis. Cell Death Differ 2007; 14: 932 942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland MM, Norris KL, Karbowski M, Wang C, Suen DF, Jiao S, George NM, Luo X, Li Z, Youle RJ. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ 2011; 18: 235 247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol 2011; 350: 311 322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto P, Smith LC. BH4 peptide derived from Bcl-xL and Bax-inhibitor peptide suppresses apoptotic mitochondrial changes in heat stressed bovine oocytes. Mol Reprod Dev 2009; 76: 637 646 [DOI] [PubMed] [Google Scholar]
- Perez GI, Jurisicova A, Matikainen T, Moriyama T, Kim MR, Takai Y, Pru JK, Kolesnick RN, Tilly JL. A central role for ceramide in the age-related acceleration of apoptosis in the female germline. FASEB J 2005; 19: 860 862 [DOI] [PubMed] [Google Scholar]
- Liu X, Fernandes R, Gertsenstein M, Perumalsamy A, Lai I, Chi M, Moley KH, Greenblatt E, Jurisica I, Casper RF, Sun Y, Jurisicova A. Automated microinjection of recombinant BCL-X into mouse zygotes enhances embryo development. PLoS One 2011; 6: e21687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez GI, Knudson CM, Leykin L, Korsmeyer SJ, Tilly JL. Apoptosis-associated signaling pathways are required for chemotherapy-mediated female germ cell destruction. Nat Med 1997; 3: 1228 1232 [DOI] [PubMed] [Google Scholar]
- Thouas GA, Trounson AO, Wolvetang EJ, Jones GM. Mitochondrial dysfunction in mouse oocytes results in preimplantation embryo arrest in vitro. Biol Reprod 2004; 71: 1936 1942 [DOI] [PubMed] [Google Scholar]
- Reers M, Smiley ST, Mottola-Hartshorn C, Chen A, Lin M, Chen LB. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol 1995; 260: 406 417 [DOI] [PubMed] [Google Scholar]
- Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006; 85: 584 591 [DOI] [PubMed] [Google Scholar]
- May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod 2005; 20: 593 597 [DOI] [PubMed] [Google Scholar]
- Igosheva N, Abramov AY, Poston L, Eckert JJ, Fleming TP, Duchen MR, McConnell J. Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLoS One 2010; 5: e10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiffault C, Bennett JP., Jr Cyclical mitochondrial deltapsiM fluctuations linked to electron transport, F0F1 ATP-synthase and mitochondrial Na+/Ca+2 exchange are reduced in Alzheimer's disease cybrids. Mitochondrion 2005; 5: 109 119 [DOI] [PubMed] [Google Scholar]
- Widlansky ME, Wang J, Shenouda SM, Hagen TM, Smith AR, Kizhakekuttu TJ, Kluge MA, Weihrauch D, Gutterman DD, Vita JA. Altered mitochondrial membrane potential, mass, and morphology in the mononuclear cells of humans with type 2 diabetes. Transl Res 2010; 156: 15 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Frolova AI, Purcell S, Adastra K, Schoeller E, Chi MM, Schedl T, Moley KH. Mitochondrial dysfunction and apoptosis in cumulus cells of type I diabetic mice. PLoS One 2011; 5: e15901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol 2007; 77: 21 49 [DOI] [PubMed] [Google Scholar]
- Muggleton-Harris AL, Brown JJ. Cytoplasmic factors influence mitochondrial reorganization and resumption of cleavage during culture of early mouse embryos. Hum Reprod 1988; 3: 1020 1028 [DOI] [PubMed] [Google Scholar]
- Kabashima K, Matsuzaki M, Suzuki H. Both microtubules and microfilaments mutually control the distribution of mitochondria in two-cell embryos of golden hamsters. J Mamm Ova Res 2007; 24: 120 125 [Google Scholar]
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 2008; 135: 2627 2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol 2005; 37: 822 834 [DOI] [PubMed] [Google Scholar]
- Favetta LA, St John EJ, King WA, Betts DH. High levels of p66shc and intracellular ROS in permanently arrested early embryos. Free Radic Biol Med 2007; 42: 1201 1210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.