ABSTRACT
Claudins comprise a large family of tight junction (TJ) proteins that are often expressed broadly during development and in adult tissues and constitute the physical barriers that occlude the paracellular space in polarized epithelia. In mouse testis, the integrity of TJs is critical to normal spermatogenesis and is dependent on CLDN11 expression. In the current study, we have generated multiple transgenic mouse lines in which steady-state levels of transgene-derived Cldn11 mRNA are up to fourfold greater than endogenous gene expression. Spermatogenesis in all founder mice harboring two copies of the endogenous Cldn11 gene is normal. These animals breed well, indicating that transgene overexpression, at least at the level of mRNA, is well tolerated by Sertoli cells. In addition, we demonstrate that the promoter/enhancer of the transgene, comprising 5 kb of genomic sequence upstream of exon 1 of the mouse Cldn11 gene, is sufficient to rescue azoospermia in Cldn11-null mice. Finally, using transient transgenic mice, we narrow the location of Sertoli cell-specific cis regulatory elements to a 2-kb region upstream of the Cldn11 transcription start site. Together, these data provide essential information for further investigation of the biological regulation of CLDN11 TJs in the testis.
Keywords: infertility, intercellular junctions, lacZ reporter, OSP, transgenic mice
Transgene-derived expression of claudin 11 in testis rescues azoospermia in claudin 11 knockout mice, and the necessary cis-regulatory elements are within 2 kb upstream of exon 1 of the claudin 11 gene.
INTRODUCTION
Spermatogenesis in the mammalian testis occurs within the seminiferous tubule, which is a polarized epithelium principally comprised of germline lineage cells and somatic Sertoli cells. Germline stem cells reside at the basement membrane of this epithelium, where they undergo intermittent mitosis for self-renewal and the generation of cells for gametogenesis. These differentiating cells complete several mitotic divisions, then migrate centripetally through Sertoli cell tight junctions (TJs) into the central luminal compartment of the seminiferous tubule, undergo meiosis, and subsequently complete their differentiation into mature spermatids [reviewed in 1].
The latter stages of the spermatogenic program beginning at meiosis are critically dependent on the integrity of the Sertoli cell TJs, the major structural component of which is CLDN11 [2]. In the absence of this occluding barrier, Sertoli cells do not generate a luminal compartment that is isolated from interstitial and blood components, and all meiotic germline cells undergo apoptosis during meiosis or shortly thereafter. In addition, the loss of TJ integrity in Cldn11-null mice disrupts Sertoli cell epithelial polarity, and these cells detach from the basement membrane and undergo an epithelial-to-fibroblast transformation [3].
In cultured Sertoli cells, expression of CLDN11 is known to be regulated by multiple growth factors and hormones such as follicle-stimulating hormone, tumor necrosis factor alpha, transforming growth factor beta, and SMADs [4–6]. Further, a GATA/NF-Y motif was reported as a crucial mediator for transactivation of the Cldn11 gene in the TM4 Sertoli cell line [6]. In some cases of maturation-arrested human testis, the absence of the late spermatogenic wave appears to be associated with a significant increase in CLDN11 expression [7], although it is unclear if this is a primary defect that causes disease or a secondary consequence of pathology. Indeed, we have previously shown in mice that testicular pathology secondarily induces CLDN11 expression [2, 3].
Although CLDN11 is crucial for Sertoli cell TJ assembly, function, and germ cell development, these TJs have never been evaluated under conditions when the Cldn11 gene is expressed at supernormal levels in the testis. Previous studies of other claudin family members such as CLDN6 have shown that TJ permeability in epithelial cells, which normally express this protein, is significantly disrupted by CLDN6 overexpression in transgenic mice, resulting in perinatal lethality [8]. In addition, overexpression of CLDN3 and CLDN4 in ovarian cancer cells promotes cell motility, invasion, and survival, although not proliferation [9].
Herein, we report the generation and characterization of transgenic mice overexpressing a Cldn11 transgene in Sertoli cells [Tg(Cldn11)605Gow]. In this genomic transgene, we incorporated all Cldn11 exons (three in total) and their intervening introns as well as 5 kb of upstream sequence. In six independent lines, the transgene expresses onefold to fourfold above endogenous gene expression at the level of transcription, but does not affect Sertoli cell TJ assembly, and these animals breed normally. It is unclear if these increases in mRNA boost protein production; steady-state levels of CLDN11 protein are comparable to wild type, possibly as a result of increased turnover. We also show that the proximal 2 kb of the promoter/enhancer region is sufficient to drive expression of a β-galactosidase reporter gene in Sertoli cells from several additional lines of transgenic mice. Finally, we evaluate the testis phenotype of Cldn11−/−:Tg(Cldn11)605GowTg/+ mice. These double mutant males have normal sperm production and motility and exhibit normal courting behavior, but may sire offspring with lower efficiency as a result of difficulties in copulating with receptive females.
MATERIALS AND METHODS
All experiments involving animals reported in the current study were reviewed and approved by the Institutional Animal Care and Use Committee at Wayne State University. The generation of transgenic mice by pronuclear injection of 0.5-day (C57BL/6 x SJL F1) embryos was performed at the University of Michigan Transgenic Animal Model Core facility (Ann Arbor, MI). All mice used in this study were maintained on mixed 129/C57BL/6tac or 129/C57BL/6tac/SJL genetic backgrounds by sib mating.
Knockout and Transgene Constructs
The entire murine Cldn11 gene cloned from a 129 Sv/Ev library is contained in two overlapping phage clones, and we used an 8-kb fragment of this gene flanking exon 1 to generate Cldn11-null mice [2]. We subsequently removed the floxed PGK-neo selectable marker from the homologously recombined Cldn11-null allele by Cre-mediated recombination to generate Cldn11tm1.1Ral mice (MGI: 5304676). This allele was used exclusively in the current study (designated Cldn11+/+, Cldn11+/−, or Cldn11−/−) and has been maintained on a mixed 129/C57BL/6tac background. To effect this recombination, female heterozygous knockout mice [2] were superovulated and bred with wild-type C57BL/6J mice to harvest 0.5-day embryos. A supercoiled DNA plasmid, pOG231, containing the CMV immediate early promoter driving Cre expression [10] was injected into male pronuclei as described previously [11].
The Osp20 transgene construct (GS605) comprises a 22-kb Sau3A I fragment of genomic sequence from 5 kb upstream of the transcription start site in exon 1 to 1.9 kb downstream of the first polyadenylation site in exon 3. The unique Acc65 I site near the 5′ end of intron 1 was blunted out to distinguish this transgene from the endogenous Cldn11 gene on Southern blots. The genomic fragment was cloned between two Not I sites in a pUC18-based vector [12] and was excised and phenol:chloroform-purified from an agarose gel slice for male pronuclear injection. This allele is referred to as Tg(Cldn11)605Gow (MGI:5306245). Six lines of transgenic mice were generated for this study: Tg(Cldn11)605.5Gow, Tg(Cldn11)605.8Gow, Tg(Cldn11)605.9Gow, Tg(Cldn11)605.10Gow, Tg(Cldn11)605.11Gow, and Tg(Cldn11)605.12Gow. Double mutant mice were generated by breeding males from each of the six Tg(Cldn11)605Gow lines with Cldn11−/− female mice to generate F1 Cldn11+/−:Tg(Cldn11)605GowTg/+ mice. These mice were sib mated to generate F2 (and beyond) Cldn11−/−:Tg(Cldn11)605GowTg/+ mice (as well as sex-matched, age-matched, or littermate controls) for experiments.
The OZG2 transgene includes a 2.1-kb Eco RI–Aat II fragment upstream of the Cldn11 gene transcription start site and the first 108 bp of the 5′ untranslated region (UTR) of exon 1, which replaces the murine myelin basic protein promoter in the MβG transgene construct [12]. This promoter/enhancer region is located immediately upstream of the bacterial lacZ open reading frame (ORF). Splice and polyadenylation signals for this transgene (and those subsequently described) are derived from exon 2 through exon 3 of the human beta-globin gene [12]. The x1x2ZG2 construct includes a 10-kb Nru I–Sau3A I genomic fragment of Cldn11 with 98 bases of 5′ untranslated sequence in exon 1 through 6 kb of intron 2. A 158-bp minimal thymidine kinase (tk) promoter is located immediately downstream of the Cldn11 sequence and immediately upstream of the lacZ/beta-globin cassette. The x3ZG2 construct comprises a 10-kb Fsp I–Sau3A I genomic fragment of Cldn11 with 4 kb of intron 2 through 6 kb downstream of the first polyadenylation signal in exon 3 (fragment). The x1x2 and x3 fragments of Cldn11 overlap by 122 bp. Finally, the tkZG2 construct includes the minimal tk promoter upstream of the lacZ/beta-globin cassette.
Southern, Northern, and Western Blotting
For Northern blotting, testes from wild-type mice between P1 and P45 were rapidly dissected, pooled (2–10 testes, depending on age), and frozen on dry ice. Partially thawed testes were homogenized in 4 M guanidinium hydrochloride with 0.1 M mercaptoethanol and total RNA was purified by cesium chloride centrifugation [13]. RNAs were electrophoresed on 1% agarose-formaldehyde gels at 125 V for 3 h. For Southern blotting, DNA was purified from tail biopsies using proteinase K digestion and phenol:chloroform extraction, precipitated in ethanol, and dissolved in Tris-EDTA, pH 8.0. DNAs were double digested with Eco R1 and Kpn I for 6 h in the presence of spermidine and electrophoresed on 0.8% agarose gels overnight at 30 V.
Gels for Northern and Southern blots were transferred to Nytran filters overnight (Schleicher & Schuell) using 10× SSPE. Northern probes were at least 400 bp in length and comprised the cDNAs, or parts thereof, from genes of interest. The Tg(Cldn11)605GowTg/+ and Cldn11 alleles were distinguished on Southern blots using a 620 bp Avr II–Acc65 I probe that contained exon 1 and the 5′ end of intron 1. DNA probes were random primed using Klenow for 1 h (32P-dCTP labeling; New England Biolabs) and purified through a G-50 Sephadex column, and blots were hybridized at 42°C in 50% formamide (Fisher Scientific), 2× SSPE, 0.025 M phosphate, 5× Denhardt solution, 0.6% SDS, 0.25 mg/ml sheared salmon sperm DNA with 1–2 × 106 cpm/ml for 24 h as detailed previously [2]. Blots were washed to 0.2× SSPE/0.1% SDS at room temperature and 55°C for 15 min and exposed to Biomax MS film (Kodak) at −80°C.
For Western blotting, adult whole testes were homogenized in cell lysis buffer (Epitomics) containing phosphatase/protease inhibitors (Sigma) and Benzonase (EMD Biosciences) at 4°C, aliquoted, and stored at −80°C. Homogenates were thawed, dissolved in Laemmli sample buffer [14], boiled for 5 min, and centrifuged for 2 min and 100 μg of protein loaded onto 10% SDS-polyacrylamide gels. After electrophoresis, gels were blotted onto nitrocellulose membrane in polyacrylamide gel running buffer containing 20% methanol for 1 h and blocked in 5% Carnation milk powder dissolved in PBS. The blots were washed and incubated in primary antibody overnight at 4°C in 3% milk in PBS, washed, and incubated for 1 h in goat anti-mouse-HRP conjugate (Southern Biotech). After washing, blots were incubated in ECL reagent (Thermo Fisher Scientific) and CL-XPosure film (Thermo Fisher Scientific) used to visualize antibody labeling. Alternatively, blots were incubated with secondary antibody conjugated to alkaline phosphatase and developed with NBT/BCIP (Thermo Fisher Scientific).
ImageJ Quantification of Blots
Quantification of blots that did not exhibit evidence of overexposure was performed by digitizing films using a UMAX 1120 flatbed scanner in transmission mode at a resolution of 300 dpi. Images were processed in NIH ImageJ (v. 1.62) using the gel scan macro set and areas under the curves were calculated after background subtraction.
Antibodies
For immunocytochemistry, we used the following antibodies: mouse anti-CLDN11 [15] and rabbit anti-TJP1 (ZO-1) (Invitrogen). Secondary antibodies were species and isotype specific (where appropriate) and purchased from Southern Biotech. For biotinylated secondary antibodies, we used streptavidin-Alexa488 conjugates for visualization (Invitrogen).
Cytochemistry
For immunocytochemistry, male mice were perfused intracardially using freshly prepared 4% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.2, at room temperature for 15 min. Testes were dissected, infiltrated with 30% sucrose in 0.1 M phosphate buffer, and embedded in Tissue-Tek O.C.T. (Electron Microscopy Sciences) for cryosectioning. Ten-micrometer frozen sections were mounted on Superfrost Plus slides (Thermo Fisher Scientific) and stored at −20°C. Sections were permeabilized in 0.1% Triton X-100 or methanol at room temperature for 10 min, blocked for 30 min with 0.1 M Tris, pH 7.4, containing 1% bovine serum albumin (fraction V; Sigma) and 0.1% gelatin (G-1890; Sigma), and labeled with antibodies as described previously [16]. 4′,6-diamidino-2-phenylindole (DAPI; 2 μg/ml; Sigma) was used to label nuclei in tissue sections.
For histochemistry, male mice were perfused with freshly prepared 2% paraformaldehyde in 0.1 M PIPES buffer, pH 6.9, containing 2 mM MgCl2 at room temperature for 10 min. Testes were dissected, permeabilized overnight with 0.01% deoxycholate (Sigma), 0.02% NP-40 (Sigma), and 2 mM MgCl2 in PBS, then incubated overnight at 37°C in permeabilization solution containing 17.5 mM K3Fe(CN)6, 17.5 mM K4Fe(CN)6, and 1 mg/ml X-gal (5-bromo-4-chloro-3-hydroxyindole; Thermo Fisher Scientific). Finally, testes were fixed overnight in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2.
For histochemistry, testes from 6- to 20-wk-old mice were rapidly dissected, punctured several times at the poles with an 18-gauge hypodermic needle, and immersion fixed in Bouin fixative (Fisher) for 24 h on a rotating shaker. Testes were washed for 36 h in several changes of PBS, then cut in cross section through the equator, dehydrated through an ethanol series into xylene, and embedded in paraffin. Five-micrometer sections were cut, floated on water, mounted on Superfrost slides (Fisher), and incubated overnight at 60°C. Subsequently, sections were dewaxed in xylene and rehydrated through an ethanol series into PBS. Sections were then stained with 0.2% Gill hematoxylin (Fisher) for 20 min followed by washes in tap water for 2 min, 1% HCl in 70% ethanol for 15 sec, 29.7% NH4OH (Sigma) for 1 min, tap water for 5 min, 3 changes of 95% ethanol over 5 min, and dehydration in ethanol. Finally, slides were mounted in Cytoseal 60 (Fisher) and coverslipped for microscopy.
Photography
Fluorescence micrographs were captured using a Leica DMRA2 microscope equipped with a Plan Apo 40×, na 1.25 lens, Yokogawa spinning disk confocal with Argon/He/Ne lasers (QLC100; Visitech International), and Hamamatsu Orca ER digital camera. For histochemistry, sections were photographed using a Leica DFC490 digital color camera. Wholemount tissue specimens were photographed using a Nikon SMZ1500 dissecting microscope equipped with an HR Plan Apo, WD 54 1× lens, and Sony DKC-5000 digital camera. Images were processed in Adobe Photoshop CS2 using whole-field contrast enhancement and background subtraction/elimination (bright-field images).
Analysis of Sperm Number, Morphology, and Motility
Cauda epididymis and testis from P80 mice were rapidly dissected and placed into 2 ml of 0.9% saline at 37°C in 35-mm Petri dishes. The tissue was finely minced with scissors and incubated for 10 min to release sperm into the medium. The number of sperm per milliliter of medium was determined in a hemocytometer [17, 18]. To examine morphology and motility, a drop of medium was placed onto a slide and the proportion of immobile sperm determined by manual counting.
Mating Behavior of Cldn11−/−:Tg(Cldn11)605GowTg/+ Males
For the six independent transgenic lines generated in this study, we mated each of two Cldn11−/−:Tg(Cldn11)605GowTg/+ males (beginning at 7–8 wk of age) with two age-matched wild-type females in standard breeding cages. An equal number of littermate Cldn11+/+:Tg(Cldn11)605GowTg/+ males were mated with wild-type females in the same time frame to serve as mating controls for the colony. The mating behavior of all trios was observed for 30–60 min after the introduction of females into the breeding cages. The breeding pair trios were continuously mated for 4−5 wk to determine if the males could successfully sire young. Testes from all males that failed to generate litters were harvested to determine sperm quality.
To examine specific mating behavior in detail, four Cldn11−/−:Tg(Cldn11)605.12GowTg/+ and four Cldn11+/−:Tg(Cldn11)605.12GowTg/+ male mice were placed individually in 24 × 24-inch clear open-top Perspex cages for 24 h with food and water ad libitum to establish their territory. Day 1 of the study began at 1500 h by placing a wild-type female (2-mo-old C3H purchased from Taconic Farms Inc.) in each cage. The mice were video recorded for four or five nights using a Panasonic CCD color camera, Model WV-CP484 (Panasonic), equipped with a Pentax CCTV 2.8–12 mm 1:1.4 CS zoom lens (Pentax Imaging Company) and an Extreme CCTV ML-2850 Moonlight infrared LED illuminator (Bosch Security Systems Inc.). The camera was controlled by Stellate Harmonie ver. 6.2f software.
Mated females were inspected daily for vaginal plugs. Videos were manually inspected first at 8× normal speed to identify individual epochs of courting/copulation behaviors and subsequently at 1× speed to determine the duration of each copulation epoch. For two of the Cldn11−/−:Tg(Cldn11)605.12GowTg/+ and two Cldn11+/−:Tg(Cldn11)605.12GowTg/+ breeding pairs, the video recording failed on the third night. Data presented represent the other four nights of video capture.
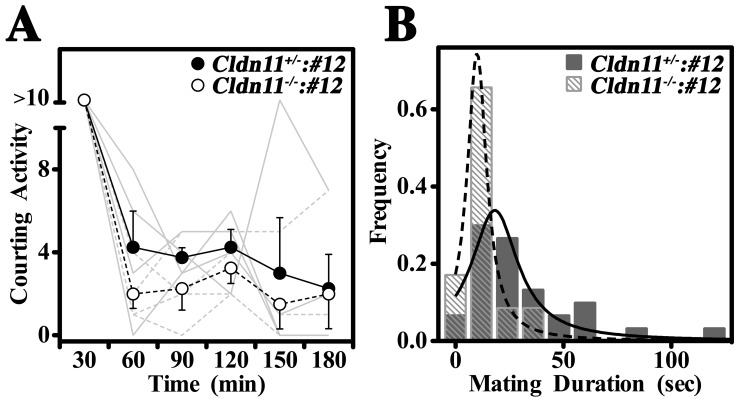
The number of courting/copulation epochs per 30 min for the first 3 h after placing the females in each cage was scored. A courting epoch was defined as the male grabbing or biting the flank/tail of the female, the male sniffing the genitals of the female, or the male attempting to mount the female from behind. In time periods when the level of mating behavior was relatively low, epochs were isolated in time. However, during peak activity, the epochs tended to be more continuous and the number was scored as >10. Copulation epochs were defined as the time elapsed after the male mounted the female from behind and the female was stationary (ceased walking). The results were plotted as a frequency histogram and least squares fitted using a Gaussian (Lorentzian) distribution.
Wholemount X-gal Staining of OZG2 Testis
Male adult mice were anesthetized and perfused with 2% paraformaldehyde in 0.1 M PIPES buffer, pH 6.9, for 15 min as previously described [19]. Briefly, testes were dissected, permeabilized overnight in detergent, and incubated in 1 mg/ml X-gal at 37°C for 24 h, postfixed in 4% paraformaldehyde for 24 h, and washed in PBS. Thereafter, testes were dissected to clean up the surface of the tunica albuginea and photographed using a Nikon SMZ1500 dissecting microscope equipped with a 1.0× Plan Apo lens, Sony DKC-5000 Catseye 3CCD camera, and digital integrator.
Following wholemount photography, testes were cross sectioned and embedded for paraffin or frozen sections to determine the subcellular localization of the X-gal reaction product. Alternately, testes from perfused animals were embedded and labeled with antibodies against β-galactosidase for epifluorescence or 3,3′-diaminobenzidine detection. None of these techniques were successful for characterizing β-galactosidase subcellular localization.
Statistical Analyses
Southern and Northern blots were scanned as indicated above and evaluated using Spearman correlation and F test. A two-way repeated-measures ANOVA was used to analyze courting behavior. Bonferroni multiple comparisons testing was performed between genotypes at each time point. Square-root transformation of the raw data (to correct for unequal variances) did not alter the ANOVA results. The Kolmogorov-Smirnov (KS) test was used to analyze mating duration from the cumulative frequency histograms. All other comparisons were made using ordinary one-way ANOVA by both comparing data from mutant mouse strains with wild-type mice using Dunnett multiple comparison test and comparing all groups against each other using Tukey post hoc t-tests. The P values are indicated in the figure/table legends and figures as appropriate, and asterisks are used to indicate which mutant strains are statistically different from controls.
RESULTS
A 22-kb Genomic Fragment of Cldn11 Rescues Male Sterility in Cldn11−/− Mice
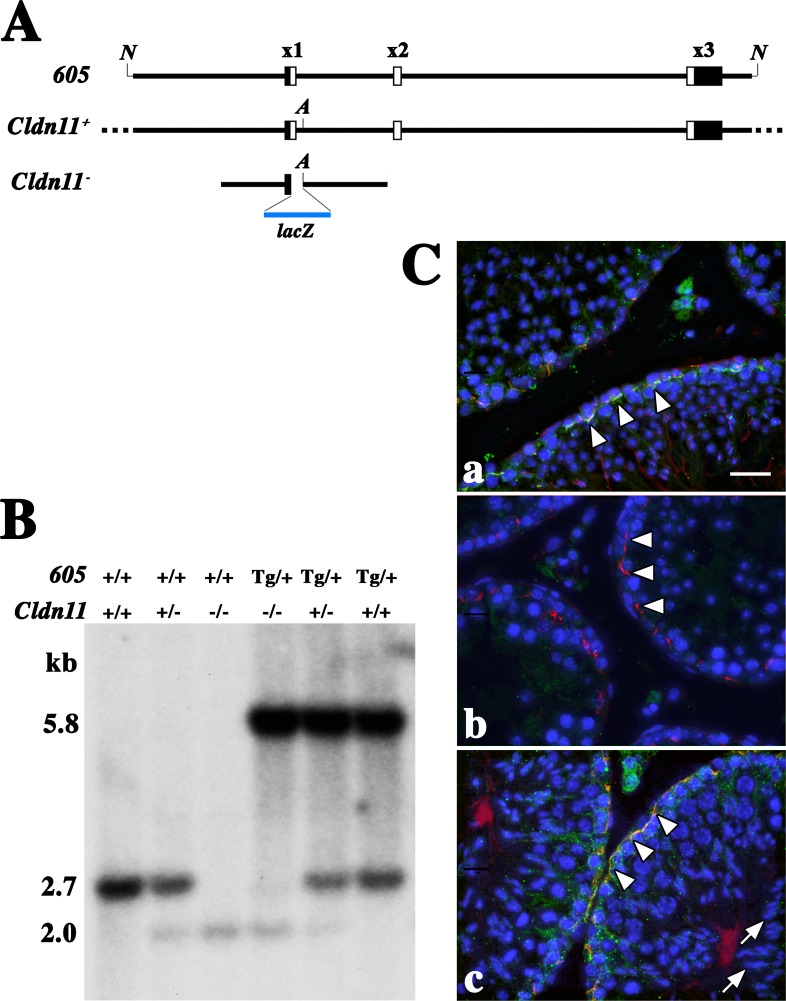
The Cldn11 gene in higher mammals comprised three exons spread over 17 kb, all of which were coding, as shown in Figure 1A. In an earlier study [2], we replaced the coding region in exon 1 of the mouse Cldn11 gene with the bacterial lacZ ORF and characterized the loss-of-function male sterility phenotype in homozygotes as well as the expression pattern of β-galactosidase in these animals.
FIG. 1.
Genotyping the wild-type, Cldn11−/−, and Tg(Cldn11)605Gow transgene alleles. A) The Tg(Cldn11)605Gow transgene construct (605) is aligned to the endogenous Cldn11 gene (Cldn+) and the knockout Cldn11 homologous recombination construct (Cldn−) generated previously [2]. Filled rectangles are the UTRs of exons 1 and 3 and open rectangles represent the CLDN11 coding region. B) Southern blot showing genotyping of the Tg(Cldn11)605GowTg/+ (5.8 kb), Cldn11+ (2.7 kb), and Cldn11− (2.0 kb) alleles. +, wild-type allele; −, knockout allele; Tg, transgene allele. C) Immunofluorescence micrographs from testis of wild-type (a), Cldn11−/− (b), and Cldn11−/−:Tg(Cldn11)605GowTg/+ double mutant mice (c). CLDN11 (green) and TJP1 (ZO-1) (red) are similarly colocalized in TJs near the base of Sertoli cells at the outside edge of seminiferous tubules (arrowheads) in wild type and double mutants. Transgene expression is sufficient to enable secondary spermatocyte development (arrows) in Cldn11−/−:Tg(Cldn11)605GowTg/+ mice. Nuclei are labeled with DAPI (blue) [26]. Bar = 30 μm. N, Not I restriction enzyme sites at the 5′ and 3′ ends of the transgene; A, unique Acc65 I restriction enzyme site in intron 1; lacZ, coding region for bacterial β-galactosidase; x1, exon 1; x2, exon 2; x3, exon 3.
In the current study, we generated and characterized a Cldn11 genomic transgene construct comprising 5 kb upstream of the transcription start site through 1.9 kb downstream of the polyadenylation site to determine if this construct could rescue any of the phenotypes we previously observed in Cldn11−/− mice [2, 15, 20, 21]. To distinguish the wild-type and knockout Cldn11 alleles from the transgene allele on Southern blots, we deleted the unique Acc65 I restriction site located near the 5′ end of intron 1 in the transgene (Fig. 1A).
Figure 1Ca shows an immunofluorescence micrograph of seminiferous tubules in cross section from a 6-wk-old wild-type mouse. TJs labeled with antibodies against CLDN11 (green) and zonula occludens 1 (TJP1 [ZO-1], red) are visible around the edge of the tubules (arrowheads). CLDN11 is absent in tubules from Cldn11−/− mice (Fig. 1Cb), although TJP1 is apparently localized to cytoplasmic plaques similar to those that normally underlie TJs in Sertoli cells (arrowheads). The diameters of these tubules are clearly reduced compared to wild-type mice because of the absence of secondary spermatocytes and spermatids [2]. Figure 1Cc shows seminiferous tubules from Cldn11−/−:Tg(Cldn11)605.11GowTg/+ mice (see Supplemental Figure S1 [available online at www.biolreprod.org] for CLDN11 and TJP1 labeling of testis from lines 5, 8, and 12). Expression of the transgene restored TJs (arrowheads) and rescued the sterility phenotype in these animals (Fig. 1Cb). Indeed, DAPI labeling reveals the nuclei of maturing spermatids in the central luminal compartment (arrows).
Tg(Cldn11)605GowTg/+ Expression Is Proportional to Transgene Copy Number
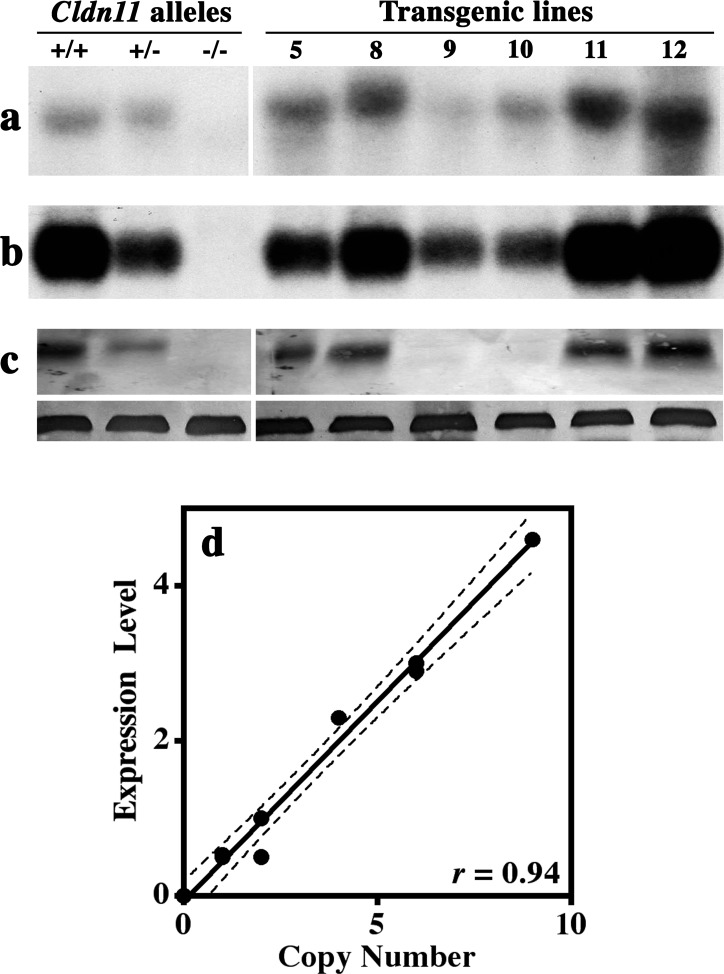
An important measure of transgene function is the correlation between transgene copy number and level of transgene expression in multiple independent lines. To determine transgene copy number in the Tg(Cldn11)605GowTg/+ lines, we performed Southern blotting of tail DNAs from Cldn11−/−:Tg(Cldn11)605GowTg/+ mice, which lack the protein coding region in exon 1 of the endogenous Cldn11 gene and do not synthesize any part of the endogenous CLDN11 protein. DNAs from the three alleles of the Cldn11 gene (wild-type, heterozygous, and homozygous deletion) comprised a standard curve for gene dosage. Of 12 founder Tg(Cldn11)605GowTg/+ transgenic mice that were initially generated, we were able to establish stable lines for 6, and representative DNAs from these lines are shown in Figure 2, a–c. From densitometry scans of the blots after normalization to a single gene dose (lane 2), we found that transgene copy number ranged from 1 to 10.
FIG. 2.
Expression of the Tg(Cldn11)605Gow transgene in testis. Testes from mice harboring 2 (+/+), 1 (+/−), or 0 (−/−) copies of the wild-type Cldn11 gene as well as testes from transgenic mice (lines 5, 8–11, and 12) were dissected at 2 mo of age for Southern blotting with a Cldn11 cDNA probe (a), Northern blotting with a Cldn11 cDNA probe (b), and Western blotting with antibodies against CLDN11 (upper) and β-actin (lower; c). d) We observe a strong and highly statistically significant Spearman correlation coefficient between transgene copy number determined from Southern blots and steady-state expression level from Northern blots (r = 0.94; P = 0.0004). Lines 9 and 10 have the lowest transgene copy numbers, with expression levels similar to Cldn11+/− mice, but synthesize very low levels of CLDN11. Indeed, the apparent level of CLDN11 in lines 5, 8, 11, and 12 is unexpectedly low compared to steady-state mRNA levels, for unknown reasons, and approximates the level of protein in wild-type testis.
Northern blotting of whole testis from P60 Cldn11−/−:Tg(Cldn11)605GowTg/+ mice using a Cldn11 cDNA probe (Fig. 2b) revealed the range of expression levels from the transgene lines. Gene expression from the Cldn11−/− allele was detected by this probe (not shown); however, it included 3 kb of sequence containing the β-galactosidase ORF and most of intron 1, and was much larger than the wild-type or transgene-derived mRNAs. Quantification of the Northern signals by densitometry indicated that transgene expression levels ranged from half to greater than fourfold above wild-type mice (lane 1).
In four of the six transgene lines (5, 8, 11, and 12), the steady-state levels of protein on Western blots were between 80% and 120% of wild-type levels (Fig. 2c, upper panels) when measured from scanned blots using ImageJ. The β-actin loading controls (lower panels) showed even protein loading in each lane. However, two transgenic lines (9 and 10) harboring single copies of the transgene did not synthesize Cldn11 gene products at levels detectable by Western blotting, despite mRNA levels that were comparable to those of Cldn11+/− mice. Currently, we do not understand the reason for the absence of protein in the testis of lines 9 and 10 transgenic mice or for the disproportionately low levels of steady-state protein, compared to mRNA levels, for other Tg(Cldn11)605GowTg/+ lines.
A Cartesian representation of the correlation between transgene copy number and mRNA expression is shown in Figure 2d after normalization. The Spearman coefficient, r = 0.94 (P = 0.0004), indicates a tight correlation between the two parameters and demonstrates that Tg(Cldn11)605GowTg/+ is a stable construct that is not greatly influenced by flanking DNA at the insertion site. The specificity of expression indicated in Figure 1Cc indicated that the major regulatory cis-elements that determine Sertoli cell expression are present within the construct.
Rescue of Spermatogenesis by Tg(Cldn11)605GowTg/+ Expression
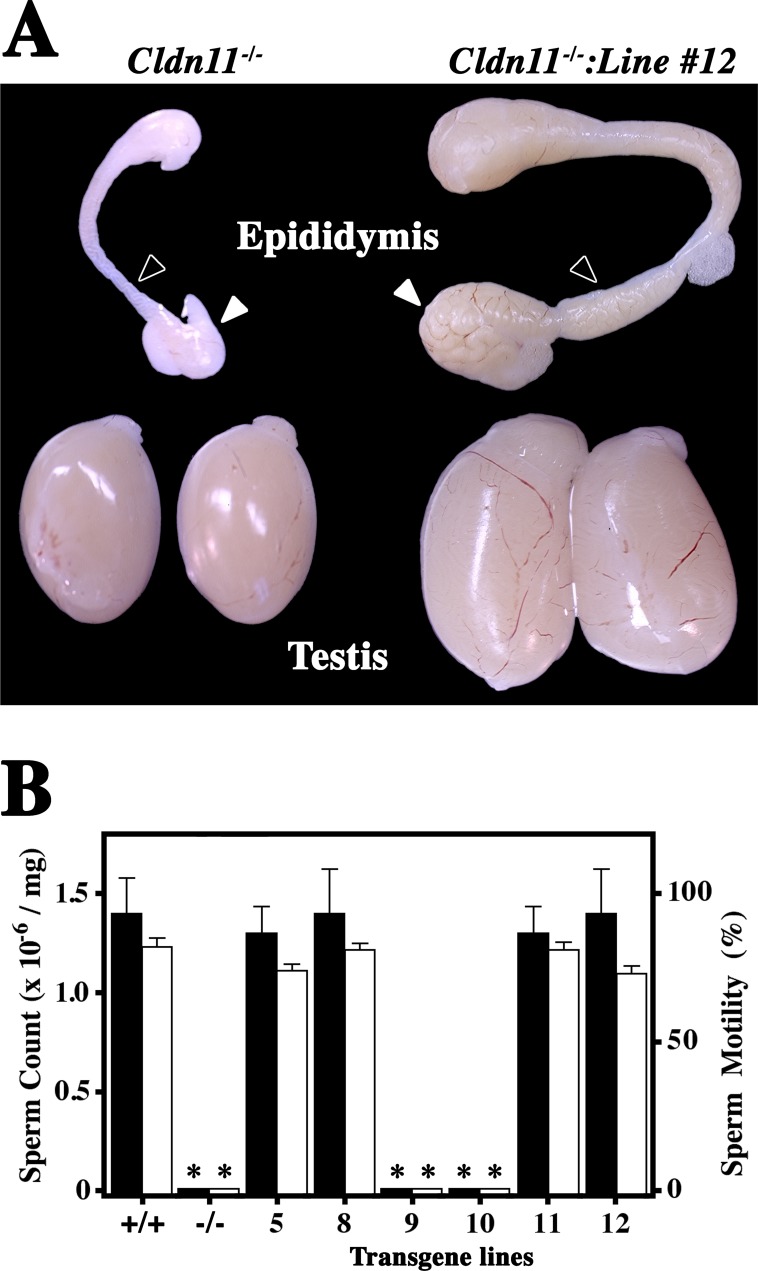
The most important measure of transgene function is the rescue of loss-of-function phenotypes. To evaluate the Tg(Cldn11)605GowTg/+ construct, we examined the morphologies of the epididymis and testes from all lines of Cldn11−/−:Tg(Cldn11)605GowTg/+ mice. Figure 3A shows representative morphologies from Cldn11−/− mice (left). The epididymis has normal form but is very small. Furthermore, the ducts in the head and body of the epididymis are translucent and apparently devoid of their normal content of maturing sperm (black arrowhead). Similar to the epididymis, testes from the knockout are well formed but small, suggesting arrested spermiogenesis.
FIG. 3.
Transgene rescue of the azoospermia phenotype of Cldn11−/− mice. In a previous study [2], we determined that Cldn11−/− mice are sterile. A) Gross dissection of the testes and epididymis from these mice at 2 mo of age (left) demonstrates normal morphology but the size of these organs is small because of the absence of developing sperm. The Tg(Cldn11)605.12Gow transgene (Line 12) rescues the size defects in Cldn11−/− mice (right) and these tissues are indistinguishable from wild type (not shown). Arrowheads show the locations where developing sperm accumulate in the epididymis. Original magnification ×3.5. B) Histograms of sperm counts and sperm motility in wild-type and six lines of transgenic mice. Lines 9 and 10 do not rescue the azoospermia phenotype (Dunnett, P < 0.0001) or motility (Dunnett, P < 0.0001) and are indistinguishable from Cldn11−/− mice in both categories (Tukey, P > 0.05). Means and standard deviations are derived from testes of three to five mice per genotype.
The epididymis and testes from a Cldn11−/−:Tg(Cldn11)605.12GowTg/+ mouse are shown on the right. In addition to the larger size of these tissues, the head and body of the epididymis are clearly swollen in association with the presence of maturing sperm, indicating that the transgene promotes a robust spermatogenic program. Table 1 shows several quantitative weight measures from our analysis and indicates that the double mutants are indistinguishable from controls (Tukey test, P > 0.05). Consistent with the absence of CLDN11 on Western blots from transgenic lines 9 and 10 (Fig. 2c), the epididymis and testis from both of these lines were highly statistically different from the controls (Dunnett test, P < 0.0001) and indistinguishable from each other (Tukey test, P > 0.05). In contrast, the body weights of all mice in this analysis are comparable (Tukey test, P > 0.05).
TABLE 1.
Quantification of sperm counts and motility in Cldn11−/−:Tg(Cldn11)605GowTg/+ double mutant mice.
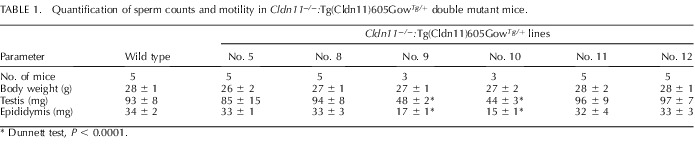
Dunnett test, P < 0.0001.
To determine if restored spermatogenesis in Cldn11−/−:Tg(Cldn11)605GowTg/+ mice also rescues the infertility phenotype, we quantified sperm number and motility in cauda epididymis (Fig. 3B). Sperm counts were approximately 1.5 × 106 per mg of tissue for transgene lines 5, 8, 11, and 12 (n = 5 mice), and were indistinguishable from those of controls (wild type). In addition, sperm motility in these lines was normal at approximately 70%–80% and the majority of sperm had normal head and tail morphologies. These experiments also confirmed the lack of phenotypic rescue for lines 9 and 10 in the absence of CLDN11.
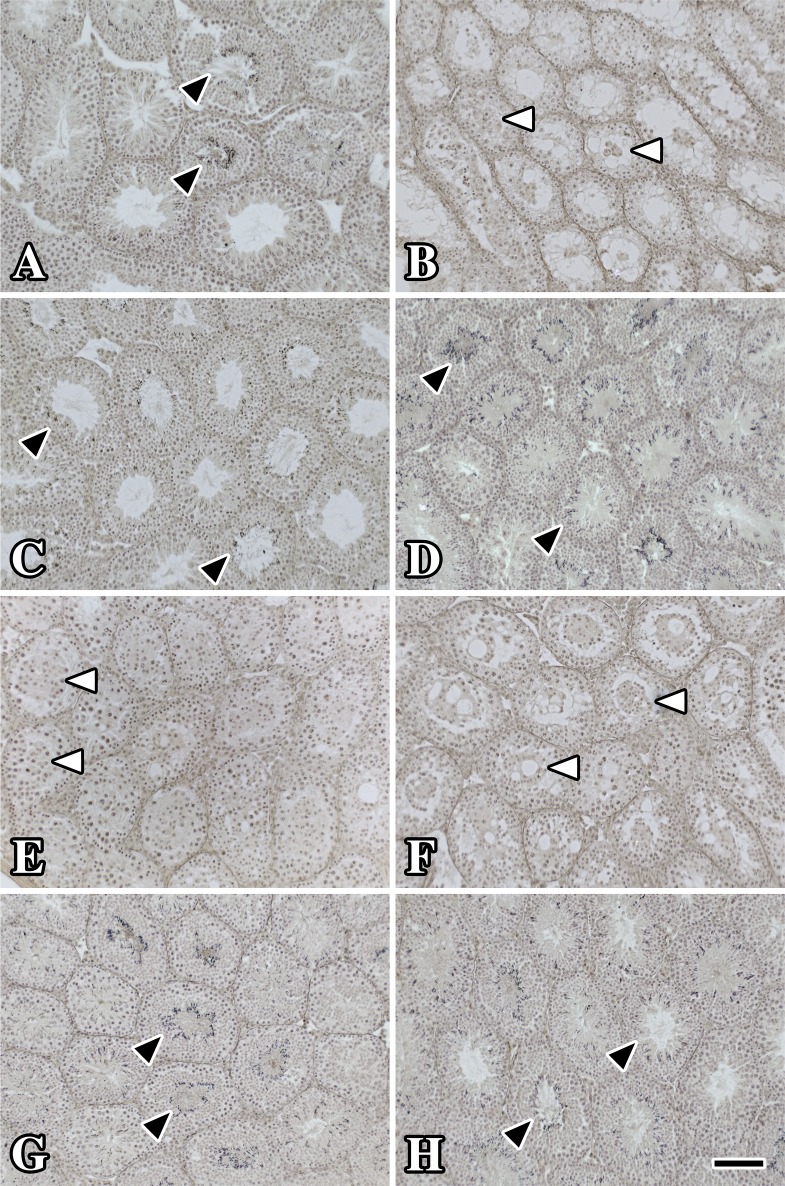
Finally, we examined the morphology of testis from Cldn11−/−:Tg(Cldn11)605GowTg/+ mice. Hematoxylin-stained transverse sections from wild-type mice (Fig. 4A) showed numerous tightly packed seminiferous tubules, each comprising a stratified epithelium bordered by basement membrane. The lumina of several tubules were patent whereas others contained many darkly stained spermatid heads (black arrowheads).
FIG. 4.
Histology of testis shows normal spermatogenesis in multiple lines of Cldn11−/−:Tg(Cldn11)605GowTg/+ mice. Testes dissected from 11- to 18-wk-old adult Cldn11−/−:Tg(Cldn11)605GowTg/+ males and controls were immersion fixed in Bouin fixative and embedded in paraffin and sections were stained with Gill hematoxylin. A) Sections from wild-type controls (17 wk old) show dark staining of spermatocyte heads in multiple seminiferous tubule profiles (black arrowheads). B) Seminiferous tubules from age-matched Cldn11−/− mice are significantly smaller than controls and spermatids are absent. However, the lumina of many tubules contain clumped Sertoli cells (white arrowheads) as previously demonstrated [2, 3]. C, D, G, H) The morphology of sections from Cldn11−/−:Tg(Cldn11)605.5GowTg/+ (11 wk old), Cldn11−/−:Tg(Cldn11)605.8GowTg/+ (18 wk old), Cldn11−/−:Tg(Cldn11)605.11GowTg/+ (13 wk old), and Cldn11−/−:Tg(Cldn11)605.12GowTg/+ (12 wk old) are similar to controls and many tubules show the presence of spermatid heads. E, F) Spermatid heads are not observed in Cldn11−/−:Tg(Cldn11)605.9GowTg/+ (17 wk old) and Cldn11−/−:Tg(Cldn11)605.10GowTg/+ (11 wk old) and the pathology is similar to Cldn11−/− mice except that tubule diameters are generally larger. Bar in H = 150 μm.
Seminiferous tubules from Cldn11−/− mice exhibited several pathological features. First, the diameter of the tubules was clearly decreased compared to controls. Second, the seminiferous epithelia were hypocellular, had very few meiotic cells, and completely lacked secondary spermatocytes and spermatids. Third, a number of tubules contained fibroblast-like cell aggregates in their lumina, which we previously identified as Sertoli cells sloughed from the basement membrane [2, 3].
The morphology of the testis from Cldn11−/−:Tg(Cldn11)605GowTg/+ mice was consistent with the level of CLDN11 on Western blots (Fig. 2c), the weight of the testis and epididymis (Table 1), and the sperm count (Fig. 3B). Testes from lines 5, 8, 11, and 12 (Fig. 4C, D, G, and H) had large-diameter tubules, many of which contained hematoxylin-stained spermatids similar to controls. In contrast, tubules from lines 9 and 10 were hypocellular and devoid of spermatids. Nevertheless, the diameters of tubules from these mice were larger than for Cldn11−/− mice, most likely because tubule lumina generally contain larger numbers of cells. Perhaps residual transgene-derived CLDN11 in Sertoli cells from lines 9 and 10 enables meiotic germ cells to survive or proliferate a little longer than in Cldn11−/− mice.
Normal Mating Behavior of Male Cldn11−/−:Tg(Cldn11)605GowTg/+ Mice
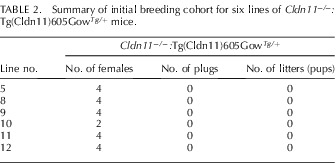
In addition to sperm counts, we mated Cldn11−/−:Tg(Cldn11)605GowTg/+ males from lines 5 and 8–12 with wild-type females (two females per male) for 5 wk in standard breeding cages to determine if they could sire offspring (Table 2). Although all of the double mutant males exhibited courting behavior for at least 1 h after introduction of the wild-type females into the breeding cages, none of the females were observed with vaginal plugs during the first 7 days and none showed signs of pregnancy or delivered pups over the course of 5 wk of continuous mating. Nevertheless, epididymides from all of the males contained normal levels of motile sperm, as determined by tissue harvest at the end of the experiment. During this time period, several other strains of mice in the same room bred normally, indicating that fecundity was not globally affected in our colony.
TABLE 2.
Summary of initial breeding cohort for six lines of Cldn11−/−:Tg(Cldn11)605GowTg/+ mice.
To assess the mating behavior of Cldn11−/−:Tg(Cldn11)605GowTg/+ males in greater detail, we video recorded four Cldn11−/−:Tg(Cldn11)605.12GowTg/+ and four control Cldn11+/−:Tg(Cldn11)605.12GowTg/+ male mice bred with wild-type females (one female per male) for four or five nights. All mice in the study were first-time breeders and exhibited strong mating behaviors in the first few hours of the first night and, in some cages, on the fourth night. Histograms of the average level of courting activity (see Materials and Methods for definitions) for consecutive 30-min intervals during the first 3 h of night 1 (mean ± SEM, n = 4 per genotype) showed that males were most active in the first 30 min and that the Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males pursued the females similarly to controls (Fig. 5A). A two-way ANOVA of this experiment showed that courting activity decreased significantly across the 3-h time frame (P < 0.0001) but that there was no difference in activity between Cldn11−/−:Tg(Cldn11)605.12GowTg/+ mice and controls (P > 0.35).
FIG. 5.
Normal mating behavior in Cldn11−/−:Tg(Cldn11)605GowTg/+ male mice. A detailed examination of the mating behavior of Cldn11−/−:Tg(Cldn11)605GowTg/+ males demonstrates that they exhibit normal courting activity but may find mating a little more difficult than controls. A) Frequency histogram showing the average number of times per 30-min period that Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males or control Cldn11+/−:Tg(Cldn11)605.12GowTg/+ males engaged wild-type females in courting activity (see Materials and Methods for definitions) for the first 3 h after females were placed in the breeding cages. Activity decreased with time (P < 0.0001, two-way ANOVA) but Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males behaved indistinguishably from controls (P > 0.35). Gray traces, courting activity for individual males. B) Frequency histogram showing the copulation times (mating duration) for Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males or control Cldn11+/−:Tg(Cldn11)605.12GowTg/+ males over 4–5 days of the breeding experiment. The median copulation epoch for the Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males was significantly shorter than for controls (10 vs. 22 sec, P < 0.02, KS test) and often ended with knockout males falling over and releasing the female. Controls were rarely observed to lose their balance, even briefly, and did not fall over.
Histograms showing the duration of copulation epochs (mating duration) for the Cldn11−/−:Tg(Cldn11)605.12GowTg/+ males compiled from video recordings (Fig. 5B) suggested that the double mutants may not mate as effectively as the controls. Although the characteristic thrusting movements observed during copulation for Cldn11−/−:Tg(Cldn11)605.12GowTg/+ and Cldn11+/−:Tg(Cldn11)605.12GowTg/+ males appeared comparable, the median duration of the epochs was significantly shorter for the double mutants than the controls (10 vs. 22 sec, P < 0.02). In addition, double mutant mice were prone to losing their balance and falling over during copulation.
Although Cldn11−/−:Tg(Cldn11)605.12GowTg/+ mice appeared less coordinated than the controls, vaginal plugs were observed for two out of four females mated to the double mutants, whereas the control males plugged three out of four females. Of these successful matings, pregnancies were observed for one female mated to a Cldn11−/−:Tg(Cldn11)605.12GowTg/+ male and one mated to a control [Cldn11+/−:Tg(Cldn11)605.12GowTg/+]. The most reasonable explanation for only two pregnancies from five plugged females is the inexperience of the males, which we have observed for a number of mouse strains.
The pregnant female mated to the Cldn11−/−:Tg(Cldn11)605.12GowTg/+ mouse gave birth to seven pups, all of which were determined to be Cldn11+/− by PCR genotyping of DNA from tail biopsies (not shown). These data confirmed the paternal genotype with a high degree of confidence; indeed, Mendelian inheritance theory predicts that the probability of the genotype being correct (i.e., of being Cldn11−/− rather than Cldn11+/−) is >99%. Accordingly, we concluded that the Tg(Cldn11)605Gow transgene fully rescued the azoospermia phenotype of Cldn11−/− male mice, even if these animals cannot mate as effectively as controls.
Sertoli Cell-Specific Expression of Cldn11 Resides Within 2 kb Upstream of Exon 1
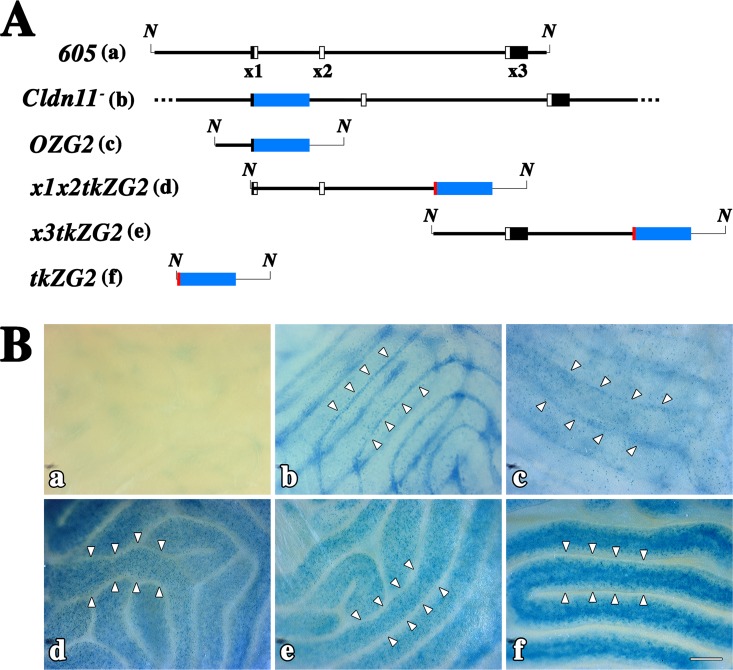
In light of our success in rescuing the infertility phenotype in Cldn11−/−:Tg(Cldn11)605GowTg/+ mice, we sought to locate the regulatory cis-elements responsible for Sertoli cell-specific expression using transient transgenic mice. Details of our original Cldn11 allele [2] and four additional lacZ reporter transgene constructs are shown in Figure 6A. Each of the four reporter constructs harbored a portion of the initial Tg(Cldn11)605Gow transgene, beginning 2 kb upstream of the transcription start site and covering the downstream region to 5 kb downstream of exon 3. The OZG2 transgene utilized the Cldn11 gene proximal promoter region, whereas the exon 1/2- and exon 3-containing constructs harbored a thymidine kinase gene (tk) minimal promoter to ensure that unknown Cldn11 proximal promoter elements could not complicate interpretation of the analysis. This minimal promoter was also included in a transgene in the absence of Cldn11 genomic sequence.
FIG. 6.
Testis-specific enhancer located within 2 kb upstream of the transcription-initiation site. A) Alignment of the Tg(Cldn11)605Gow transgene (605) with five reporter transgene constructs. The reporters include the original knockout allele (Cldn11−) used to characterize the knockout phenotype [2], which expresses the bacterial β-galactosidase protein (blue box) instead of CLDN11, and different regions of the Tg(Cldn11)605Gow transgene (thick line) inserted upstream of a thymidine kinase (tk) minimal promoter (red)/lacZ reporter (blue)/splice-polyadenylation (thin line) cassette. Each reporter was used to generate 5–20 lines of transient transgenic mice. B) Micrographs of seminiferous tubules from wild-type (a) and Cldn11+/− (b) mice for comparison with representative examples from each of the transient transgenic lines from A (c–f). For Cldn11+/− and OZG2 mice (c), β-galactosidase labeling is observed at the lateral edges of the seminiferous tubules (arrowheads) and represents expression in Sertoli cells. In x1x2tkZG2 and x3tkZG2 mice, the reporter activity is strong but is localized to cells in the germ cell lineage in the central region of the tubules. Labeling for these two constructs is indistinguishable from negative control tkZG2 mice and not likely to arise from enhancers in the Cldn11 gene. Indeed, the minimal tk promoter can drive expression in germ cells under at least some conditions [26]. N, Not I restriction enzyme sites; x1, exon 1; x2, exon 2; x3, exon 3. Bar = 200 μm.
Figure 6B demonstrates representative X-gal histochemical labeling of wholemount testes from wild type, Cldn11+/−, and each transgene reporter construct. In wild-type testis, faint nondescript staining was observed (Fig. 6Ba), indicating there was little endogenous β-galactosidase activity in testis. In contrast, epididymis was intensely stained (not shown), which precluded histochemical analysis of this tissue. Staining of testis from Cldn11+/− mice, in which β-galactosidase is expressed by Sertoli cells, showed that these cells were located at the outside edge of the seminiferous tubules (arrowheads in Fig. 6Bb). Blue puncta across the surface of the tubules highlighted the relatively low density of Sertoli cells in the seminiferous epithelium, which contrasts with the large number of cells in the germ cell lineage.
Expression of β-galactosidase from the OZG2 transgene (Fig. 6Ac/6Bc) was reminiscent of that in Cldn11+/− mice and demonstrated that Sertoli cell-specific cis regulatory elements were located within 2 kb upstream of the Cldn11 transcription start site. A summary of the expression analysis from multiple independent lines of OZG2 is presented in Table 3. Four out of five lines of male founder mice expressed β-galactosidase in Sertoli cells. It is likely that the transgene in the nonexpressing founder had inserted into a region of the genome that is transcriptionally silent (heterochromatin).
TABLE 3.
Summary of β-galactosidase expression in testis of transient transgenic mice.a

ND, not determined.
Surprisingly, we observed β-galactosidase expression in testis from tkZG2 transgenic mice (Table 3); however, staining in the seminiferous epithelium was distinct from that in Cldn11+/− mice. Indeed, cells at the perimeter of seminiferous tubules from seven of nine tkZG2 lines did not express the transgene, as indicated by the X-gal negative regions in Figure 6Af/6Bf (arrowheads). Rather, the X-gal staining was present in a large number of cells toward the center of the tubules, suggesting that secondary spermatocytes expressed the transgene and that the minimal tk promoter contained promiscuous regulatory cis-elements for this cell lineage. The β-galactosidase staining for one of the nine tkZG2 founder mice resembled that of Cldn11+/− mice (not shown), which is indicative of Sertoli cell expression; however, we regarded this to be fortuitous expression associated with DNA sequences flanking the transgene insertion site because the preponderance of founders exhibited germ cell lineage-specific staining (Table 3).
Staining patterns generated from the remaining transgene constructs (Fig. 6Ad/6Bd and 6Ae/6Be) were indistinguishable from tkZG2 mice and likely arose because of the tk minimal promoter. A single mouse from each of these x1x2ZG2 and x3ZG2 founders expressed β-galactosidase in Sertoli cells (data not shown), which we regard as fortuitous expression (Table 3).
DISCUSSION
In the current study, we generated and characterized a genomic transgene comprising all coding exons of the mouse Cldn11 gene as well as 5 kb of upstream flanking sequence. Of 12 founder mice harboring this construct, colonies were established from six founders for breeding with Cldn11−/− females (F0 generation) to generate F2 Cldn11−/−:Tg(Cldn11)605GowTg/+ double mutant mice for experiments. All six lines express transgene-derived mRNA in the testis at levels proportional to transgene copy number, thereby demonstrating that the major transcription regulatory elements contained within are stable and operate independently of flanking genomic sequences. Four of these lines also synthesize the protein at approximately wild-type levels, and rescue the male sterility phenotype of Cldn11−/− mice as indicated by normal sperm counts, morphology, and motility as well as breeding success (line 12). Several aspects of this rescue study are noteworthy.
The Tg(Cldn11)605Gow Transgene Rescues Spermatogenesis
In addition to the normal fecundity of six of the Tg(Cldn11)605Gow transgenic founder mice (lines 5, 8–12) generated for this study, four lines of Cldn11−/−:Tg(Cldn11)605GowTg/+ mice (lines 5, 8, 11, and 12) have normal sperm count, morphology, and motility and testes are of normal size and morphology. Accordingly, these double mutants would be expected to exhibit normal fecundity. However, the double mutants appear to be less effective at siring pups compared to littermate controls. We do not regard this deficit as being associated with a developmental defect in spermatogenesis but, rather, a loss-of-function phenotype in additional tissue(s) not rescued by the Tg(Cldn11)605Gow transgene.
For example, in earlier studies we demonstrated that CLDN11 is broadly expressed in a number of tissues and different cell types during embryogenesis and that congenital loss of both alleles causes pleiotropic symptoms in Cldn11−/− mice including azoospermia, late-onset deafness, reduced central nervous system (CNS) conduction velocity, and hind limb weakness [2, 15, 20, 21]. In this light, we speculate that the lower effectiveness of the Cldn11−/−:Tg(Cldn11)605GowTg/+ males in siring young is likely related to their diminished balance and effectiveness during copulation, which may stem from complications associated with slower CNS conduction [20, 21] and/or hind limb weakness [2].
The Tg(Cldn11)605Gow Transgene Fails to Yield Endogenous-Level Expression of the Encoded Protein
Two of the transgenic lines, 9 and 10, harbor single copies of the transgene but do not rescue azoospermia in Cldn11−/− mice, and Western blots from these lines (Figure 2b) demonstrate that the encoded protein is essentially undetectable. This is unexpected because CLDN11 is detectable in testis of Cldn11+/− mice, with only one functional allele of the endogenous gene, and these mutants exhibit normal fecundity. Thus, although the Tg(Cldn11)605Gow transgene contains the cis-regulatory elements necessary for cell-type specific expression of CLDN11 in Sertoli cells (Fig. 1C), it may lack another cis-element(s) that renders lines 9 and 10 hypomorphic alleles at the protein level (i.e., reduced protein per gene copy compared to the endogenous gene). It is unclear how such a mechanism might operate at the posttranslational level, but the influence of this element to gene expression seems relatively small because steady-state mRNA levels for Tg(Cldn11)605.9GowTg/+ and Tg(Cldn11)605.10GowTg/+ are comparable to Cldn11+/− controls (Fig. 2b).
We regard as unlikely the possibility that a missing cis-element is the binding site for a translation-promoting protein(s) in the UTRs of transgene-derived mature mRNA [22] for two reasons. First, we utilize the canonical 5′ UTR of the Cldn11 gene. Second, the Cldn11 gene in mice contains at least two polyadenylation signals separated by more than 1 kb, but the 5′-most signal is the one used overwhelmingly in testis and is present in the transgene. This is demonstrated by the correspondence of the mRNA sizes on Northern blots for the endogenous gene and the transgene (Fig. 2b). Thus, the simplest explanation for lower than expected CLDN11 steady-state levels is that the protein turns over more rapidly in Tg(Cldn11)605GowTg/+ testis compared to controls.
Overexpression of the Tg(Cldn11)605Gow Transgene Is Well Tolerated in Sertoli Cells
Cldn11 expression at the level of mRNA in the Tg(Cldn11)605GowTg/+ founder mice and their progeny is substantially greater than in mice harboring two copies of the endogenous Cldn11 gene, including two lines (11 and 12) with more than fourfold overexpression. We do not observe a gain-of-function phenotype stemming from this overexpression in any of the lines, irrespective of the number of endogenous gene copies. These mice breed successfully and transmit the transgene at Mendelian ratios, suggesting that supernormal levels of Cldn11 expression in Sertoli cells are well tolerated. However, a mitigating aspect of this observation is that CLDN11 expression at the level of protein may not coincide with the increased mRNA levels. For example, we cannot rule out the possibility that CLDN11 turnover rates are increased in transgenic mice, which would cause steady-state protein levels to approach those in wild-type mice. Indeed, we do not observe substantially increased CLDN11 levels on Western blots from Tg(Cldn11)605GowTg/+ testis on Western blots (Fig. 2c). Protein turnover could increase if the available target sites at Sertoli TJs are limiting and excess protein simply overflows into the endocytic compartment and is degraded in lysosomes.
Other studies involving the overexpression of claudin family members, such as CLDN6 in the epidermis of transgenic mice (Inv-Cldn6 mice), report strong phenotypes amounting to dissolution of the epidermal permeability barrier and leading to perinatal death in multiple lines [23]. These data are similar to the phenotype associated with ablation of the Cldn1 gene [24] and suggest that the gain-of-function phenotype in Inv-Cldn6 mice at least functionally disrupts TJs or epithelial polarization. In view of the molecular heterogeneity of TJs in epidermal cells, comprised of six or more claudin family members, as well as the possibility that epidermal cells segregate these claudins into different domains like other cell types [25], changes in the relative abundance of component claudins may underlie the observed disruption to the permeability barrier.
An alternative explanation to account for the supernormal expression phenotype in Inv-Cldn6 mice involves consideration of the relatively unique biological properties of this claudin family member. Thus, TJs in most tissues are identified at the ultrastructural level in terms of the characteristic close apposition of the plasma membranes between adjacent cells, the pentalaminar organization of the membrane at cell-cell contact points, and the paucity of cytoskeletal elements recruited to the cytoplasmic surface of the TJs. However, CLDN6-containing TJs differ in this last regard because the protein is able to recruit cytoplasmic adaptor proteins that assemble a cytoskeletal plaque at cell-cell contact points in vitro and in vivo [25]. Accordingly, the phenotype of Inv-Cldn6 mice may stem from the excessive adhesiveness of TJs in the epidermis. In this light, the overexpression of CLDN11 may not significantly perturb Sertoli cell biology because its biological activities are similar to the canonical claudins in most TJs.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Jeff Loeb, Dept of Neurology, WSU, for access to mouse open field digital cameras and monitoring equipment.
Footnotes
Supported by grants to A.G. from NIDCD, NIH (DC006262), and the William and Marie Carls Foundation, Detroit, Michigan.
REFERENCES
- Hess R. Spermatogenesis, overview. : Knobil E, Neill JD. (eds.), Encyclopedia of Reproduction, vol. 4. San Diego: Academic Press; 1999: 539 545 [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and Sertoli cell tight junction strands are absent In Osp/Claudin 11-null mice. Cell 1999; 99: 649 659 [DOI] [PubMed] [Google Scholar]
- Mazaud-Guittot S, Meugnier E, Pesenti S, Wu X, Vidal H, Gow A, Le Magueresse-Battistoni B. Claudin 11 deficiency in mice results in loss of the Sertoli cell epithelial phenotype in the testis. Biol Reprod 2010; 82: 202 213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WY, Lee WM, Cheng CY. Transforming growth factor-beta3 perturbs the inter-Sertoli tight junction permeability barrier in vitro possibly mediated via its effects on occludin, zonula occludens-1, and claudin-11. Endocrinology 2001; 142: 1865 1877 [DOI] [PubMed] [Google Scholar]
- Florin A, Maire M, Bozec A, Hellani A, Chater S, Bars R, Chuzel F, Benahmed M. Androgens and postmeiotic germ cells regulate claudin-11 expression in rat Sertoli cells. Endocrinology 2005; 146: 1532 1540 [DOI] [PubMed] [Google Scholar]
- Lui WY, Wong EW, Guan Y, Lee WM. Dual transcriptional control of claudin-11 via an overlapping GATA/NF-Y motif: positive regulation through the interaction of GATA, NF-YA, and CREB and negative regulation through the interaction of Smad, HDAC1, and mSin3A. J Cell Physiol 2007; 211: 638 648 [DOI] [PubMed] [Google Scholar]
- Nah WH, Lee JE, Park HJ, Park NC, Gye MC. Claudin-11 expression increased in spermatogenic defect in human testes. Fertil Steril 2011; 95: 385 388 [DOI] [PubMed] [Google Scholar]
- Arabzadeh A, Troy TC, Turksen K. Role of the Cldn6 cytoplasmic tail domain in membrane targeting and epidermal differentiation in vivo. Mol Cell Biol 2006; 26: 5876 5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005; 65: 7378 7385 [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 1991; 251: 1351 1355 [DOI] [PubMed] [Google Scholar]
- Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VLJ, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM-20 cannot replace PLP in CNS myelin. J Neurosci 2000; 20: 4002 4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Friedrich VL, Lazzarini RA. Myelin basic protein gene contains separate enhancers for oligodendrocytes and Schwann cell expression. J Cell Biol 1992; 119: 605 616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 1979; 18: 5294 5299 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680 685 [DOI] [PubMed] [Google Scholar]
- Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DM, Kachar B. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci 2004; 24: 7051 7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Lazzarini RA. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J Cell Biol 1998; 140: 925 934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Mumford D, Lactate Gordon H. and malate dehydrogenase and alpha-esterases in oligospermia. Fertil Steril 1976; 27: 832 835 [DOI] [PubMed] [Google Scholar]
- Wyrobek AJ, Bruce WR. Chemical induction of sperm abnormalities in mice. Proc Natl Acad Sci U S A 1975; 72: 4425 4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood C, He C, Garbern J, Kamholz J, Arroyo E, Gow A. CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure. J Neurosci 2004; 24: 11215 11225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux JJ, Gow A. Tight junctions potentiate the insulative properties of small CNS myelinated axons. J Cell Biol 2008; 183: 909 921 PMC2592840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Devaux JJ. A model of tight junction function In CNS myelinated axons. Neuron Glia Biol 2008; 4: 307 317 PMC2957896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 2003; 28: 182 188 [DOI] [PubMed] [Google Scholar]
- Turksen K, Troy TC. Permeability barrier dysfunction in transgenic mice overexpressing claudin 6. Development 2002; 129: 1775 1784 [DOI] [PubMed] [Google Scholar]
- Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol 2002; 156: 1099 1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, Kachar B. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci 2006; 119: 4819 4827 [DOI] [PubMed] [Google Scholar]
- Widlak W, Scieglinska D, Vydra N, Malusecka E, Krawczyk Z. In vivo electroporation of the testis versus transgenic mice model in functional studies of spermatocyte-specific hst70 gene promoter: a comparative study. Mol Reprod Dev 2003; 65: 382 388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.