Abstract
Vibrio cholerae cytolysin (VCC) is among the accessory V. cholerae virulence factors that may contribute to disease pathogenesis in humans. VCC, encoded by hlyA gene, belongs to the most common class of bacterial toxins, known as pore-forming toxins (PFTs). V. cholerae infects and kills Caenorhabditis elegans via cholerae toxin independent manner. VCC is required for the lethality, growth retardation and intestinal cell vacuolation during the infection. However, little is known about the host gene expression responses against VCC. To address this question we performed a microarray study in C. elegans exposed to V. cholerae strains with intact and deleted hlyA genes.
Many of the VCC regulated genes identified, including C-type lectins, Prion-like (glutamine [Q]/asparagine [N]-rich)-domain containing genes, genes regulated by insulin/IGF-1-mediated signaling (IIS) pathway, were previously reported as mediators of innate immune response against other bacteria in C. elegans. Protective function of the subset of the genes up-regulated by VCC was confirmed using RNAi. By means of a machine learning algorithm called FastMEDUSA, we identified several putative VCC induced immune regulatory transcriptional factors and transcription factor binding motifs. Our results suggest that VCC is a major virulence factor, which induces a wide variety of immune response- related genes during V. cholerae infection in C. elegans.
Introduction
V. cholerae cytolysin (VCC) is among the accessory V. cholerae virulence factors that may contribute to the sporadic form of diarrheal disease pathogenesis. VCC, encoded by hlyA gene, belongs to the most common class of bacterial toxins, the pore-forming toxins (PFTs), which are important virulence factors. Most of the O1 biotype El Tor, O139, and non-O1/non-O139 V. cholerae isolates, produce a 80-kD water soluble cytolysin (VCC) [1]–[3]. VCC causes tissue and cell damage through apoptosis, autophagy, cellular vacuolization, cell lysis and necrosis. [4]–[9]. Studies using host models such as infant mouse, rabbit ileal loop, streptomycin fed adult C57BL/6 mice models, and nematode C. elegans infection model, suggest that VCC was responsible for the residual toxicity observed with some of the vaccine strains with full or partial coding sequences of hlyA gene [4], [10], [11].
Caenorhabditis elegans has been used as an invertebrate host model to identify and assess virulence factors of several human pathogens including V. cholerae [11]–[14]. V. cholerae causes lethal infection in the nematode Caenorhabditis elegans via a cholera toxin (Ctx) and toxin co-regulated pili (Tcp) independent process, providing a useful host model system to screen for the virulence factors other than Ctx and Tcp. Worm lethality effect inflicted by V. cholerae, is mediated by LuxO-regulated genes in the quorum sensing (QS) pathway, such as hapR, V. cholerae metalloprotease gene PrtV [14], and VCC encoding gene hlyA [11]. hlyA also causes developmental delay and intestinal vacuolation in C. elegans [11].
Host responses to VCC at the molecular level, and the significance of these responses in host organisms' defense during V. cholerae pathogenesis, remain poorly understood. C. elegans provides an excellent model to address these questions. Here we report our findings regarding genome wide host transcriptional response to VCC in C. elegans during V. cholerae infection. We performed a microarray study in C. elegans which was exposed to V. cholerae strains with intact and deleted hlyA genes for 18 hours. Expression profiles of the worms exposed to hlyA(−) V. cholerae strains were compared with the expression profiles of the worms exposed to hlyA(+) V. cholerae strains. Many of the differentially expressed genes previously reported as mediators of innate immune response against other bacteria in C. elegans, suggesting that C. elegans uses common and specific mechanisms against V.cholerae and these defenses are induced by VCC. Among the differentially expressed genes are: C-type lectins, abu (activated in blocked unfolded protein response) genes, which contain Prion-like (glutamine [Q]/asparagine[N]-rich)-domain, and genes regulated by daf-16. Immune response function of the subset of the differentially expressed genes against V. cholerae infection was confirmed using RNAi. Using a machine learning algorithm called FastMEDUSA, we identified putative immune regulatory transcriptional factors, which are regulated by VCC. FastMEDUSA was also used to discover the transcription factor binding motifs, which were later analyzed using the GOMO (Gene Ontology for Motifs) tool to identify the GO-terms associated with these motifs. Go terms related to pathogen recognition and to immune and inflammatory responses, were found to be significantly associated with the motifs identified using FastMEDUSA.
Materials and Methods
Bacterial strains, media and culture conditions
Bacterial strains used in this study include: E7946: V. cholerae Wild-type O1 El Tor, Ogawa strain, HNC45: E7946 ΔhlyA, CVD 109: Δ(ctxAB zot ace) of parental strain E7946, CVD110 Δ(ctxAB zot ace) hlyA::(ctxB mer) Hgr of parental strain E7946.
V. cholerae strains were cultured in tryptic soy broth (TSB, Becton Dicson Microbiology System, BBL, Cockeysville, MD) media supplemented with 1% NaCl at 30°C. E.coli OP 50 was grown in LB culture media.
C. elegans strains, maintenance and microscopy
Strains N2, NL2099 rrf-3 (pk1426), GR1373 eri-1 (mg366), RB711 pqm-1(ok485), VC2169 abu-15(ok2878), VC1806 nhr-234(gk865), RB1590 pax-1 (ok1949), VC1204 nhr-34 (gk556), RB657 nhr-23 (ok407), RB1044 Y47H9C.2 (ok990), and SAL139 pha-1(e2123); denEx17 [dod-22::GFP+pha-1(+)] were acquired from the Caenorhabditis Genetics Center (CGC). All the strains were maintained at 22°C except GR1373 (eri-1), which is maintained at 16°C. The wild type Bristol strain N2, was cultured in C. elegans habitation media (CeHM) in tissue culture flasks on a platform shaker [15]. Nematodes were bleached (0.5 M NaOH, 1% Hypochlorite) to collect eggs which were incubated in M9 media for 24 hours to bring them to synchronized L1 stage and then transferred to CeHM. For microscopy, mixed stage worms grown on OP50 and test bacteria seeded nematode growth media (NGM) plates, were imaged on a Leica MZ16FA stereomicroscope.
RNA Isolation
Synchronized L1 stage N2 animals were transferred to C. elegans habitation media (CeHM) and incubated for 24 hours. Animals were washed with M9 buffer and transferred to NGM plates containing E. coli strain OP50, V. cholerae strains E7946, E7946 Δ hly, CVD109, and CVD110, and incubated at 22°C for 18 hours. Worms were collected and washed in M9 buffer and RNA was extracted using TRIzol reagent (Invitrogen). Residual genomic DNA was removed by DNase treatment (Ambion, Austin, TX). Three independent RNA isolations were performed with each condition for microarray analysis.
Microarray Analysis
For each experimental condition, RNA was isolated from three biological replicate samples. cRNA was synthesized from 10 µg of total RNA, and samples were hybridized to the C. elegans GeneChip (Affymetrix, Santa Clara, CA) by the FDA/CFSAN/DMB Microarray Facility following the manufacturers instruction. The chip represents 22500 transcripts of the expressed C. elegans genome based on the December 2005 genome sequence. The data were processed using Partek Genomics Suite, version 6.4 Partek Inc, St. Louis, MO. The robust multichip averaging algorithm was used to normalize and summarize the probe data into probe set expression values. Analysis of variance, fold-change, and false discovery rate (FDR) calculations were also performed using Partek® Genomics Suite TM version 6.5 (Copyright © 2010 Partek Inc., St. Louis, MO, USA). Transcripts showing a corrected p value of <0.05 and fold change ≤−1.2 or ≥1.2 were considered differentially expressed between experimental treatments groups. The microarray data have been deposited in the Microarray Informatics, EMBL. Accession number is E-TABM-840.
Functional Enrichment Analysis
Genes showing a significant change in expression by microarray analysis (p<0.05) were analyzed using R software (R Development Core Team (2009): A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org). Genes were compared against a 22,500 C. elegans gene data base to identify over-represented Gene Ontology terms (Table S1). Statistical analysis was performed using the chi-square test and the Yates' continuity correction. Significant functional terms were defined as p<0.05.
qRT-PCR
cDNA was synthesized from 5 µg of total RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen). qRT-PCR was performed using SYBR Advantage quantitative PCR premix (Clontech) and gene-specific oligonucleotide primers on the LightCycler (BIO RAD). Primers for qRT-PCR are following: clec-7: (fwd) ttggctgttgtaggcaatca, (rev) tcactgggaatccgttatcc; fmo-2: (fwd) tgctgtcataggagctggtg, (rev) catctgacgcctcaaaacaa; clec-46: (fwd) cttcctcggttcttgcactt, (rev) gcggtttccaacaaaaacac; C23G10.1: (fwd) ccatccactcttggttgctt, (rev) tcacgtgctcctttttcctt; col-41: (fwd) caccaggaactccaggaaac, (rev) gtggggttctgtcgtcttgt; B0024.4: (fwd) caacaacattgagcgcagag, (rev) tgtgtagtcgtctgttggaacc; ttr-21: (fwd) tgtgtcaaggacaaccagcta, (rev) ttccagcaactcgaaaggtt; dct-5: (fwd) gctgcaaaatgtggaaatga, (rev) aagttttgggcacagtccag; pqn-5: (fwd) gctcagccacaacaaactca, (rev) ctggcactgttgctgacatt.
Relative fold-changes for transcripts were calculated using the comparative CT (2−ΔΔCT) method [16]. Cycle thresholds of amplification were determined by Light Cycler software (BIO RAD). All samples were run in triplicates and normalized to internal control.
RNA Interference
E. coli DH5α bacterial strains expressing double-stranded C. elegans RNA [17] were grown in LB broth containing ampicillin (100 µg/ml) at 37°C and plated onto NGM containing 100 µg/ml ampicillin and 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG). RNAi-expressing bacteria were allowed to grow overnight at 37°C. Synchronized L1 stage NL2099 (rrf-3) or GR1373 (eri-1) strains were used for RNAi experiments. NL2099 (rrf-3) has been used for the functional validation of the differentially expressed genes identified through microarray, and GR1373 (eri-1) for the rest of the RNAi experiments regarding prion-like (Q/N rich) domain protein genes, and FastMedusa identified genes. NL2099 (rrf-3) worms were exposed to fresh RNAi expressing bacterial lawn on NGM media for 48 hours, then washed with M9 and plated on NGM plates containing V. cholerae wild type E7946, E7946 Δhly, or E.coli OP50 bacterial lawn, and incubated at 22°C. GR1373 (eri-1) worms were initially treated with V. cholerae wild type E7946, E7946 Δhly or E.coli OP50 bacterial lawn and incubated first at 16°C for 24 hours followed by incubation at 25°C for next 24 hours. Worms were than transferred to the RNAi bacterial lawn and incubated at 25°C for the rest of the experiment. L4440 RNAi which contains the empty vector was included as a control in all experiments.
C. elegans Survival Analysis
Pathogen lawns for survival assays along with food bacteria OP50 were prepared by inoculating NGM (in 6-cm Petri plates) with 50 µl of an overnight bacterial culture. Plates were incubated overnight at room temperature before animals were added. Worms treated with RNAi bacteria, or mutant worms to be tested were transferred to NGM plates containing V. cholerae wild type E7946, E7946 Δhly or E. coli OP 50 bacterial lawns and incubated at 22°C with ∼20–30 L4 stage worms added to each plate. Animals were scored every 24 h for survival and transferred to fresh bacterial lawns everyday to avoid confusion with progeny. Animal survival was plotted using Kaplan-Meier survival curves and analyzed by log rank test using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Survival curves resulting in p values of <0.05 relative to control were considered significantly different.
FastMEDUSA analysis
We used FastMEDUSA software to discover experimental condition-specific transcription factors (TFs) and motifs in C. elegans [18]. FastMEDUSA is a parallelized version of MEDUSA algorithm [19], which trains a model from expression and promoter sequences of genes in a number of experimental conditions. We analyzed the FastMEDUSA model to extract condition-specific significant TFs and motifs in C. elegans.
FastMEDUSA requires discretized gene expression profiles. To this end, we discretized gene expression data by using E7946 samples as reference. We computed differentially expressed genes (DEGs) by using ANOVA (FDR≤0.05) in Partek® Genomics SuiteTM version 6.5 (Copyright © 2010 Partek Inc., St. Louis, MO, USA). For each DEG in a sample, we computed the ratio to its median expression signal across reference samples. A gene in a sample was called upregulated if the ratio ≥1.0 and downregulated otherwise. Genes that had inconsistent expression calls across technical replicates were filtered out. We obtained a list of candidate TFs in C. elegans from EDGEdb [20], and 1,000 bp promoter sequence of all DEGs from BioMart [21].
We ran FastMEDUSA five times on Biowulf cluster at the National Institutes of Health. For each run, we computed significance score of TFs as described in [19] and selected the top 30 significant TFs for each condition. We selected consensus significant TFs that occur in the top list in all runs. We computed significant motifs similarly.
FastMEDUSA applies a machine learning algorithm called boosting [22] to train a predictive model as an alternating decision tree. In order to determine how many boosting iterations are needed to train a model, we ran FastMEDUSA on 90% of the input data and tested the model on the remaining 10%. Running FastMEDUSA with 800 boosting iterations was optimal to learn the model for this data set. When building the model, if the boosting algorithm gives the same score for more than one transcription factor or motif, FastMEDUSA makes a random choice. In other words, FastMEDUSA potentially builds a different model at each run. Thus, we ran FastMEDUSA five times using a different random seed value at each run and selected TFs and motifs that are overrepresented in these models.
Semi-quantitative RT-PCR analysis for V. cholerae virulence genes
The relative transcript abundance and expression of V.cholerae virulence genes at different temperatures was evaluated using semi-quantitative RT-PCR in V. cholerae wild type strain E7946. Cultures of E7946 were grown at 16°C, 22°C, 30°C, and 37°C by shaking at 120 rpm, in TSB with 1% NaCl. Primers for semi-quantitative RT-PCR are following: hlyA: (fwd) TGAGCGTAATGCGAAGAATG, (rev) GCGGGCTAATACGGTTTACA; ace: (fwd) GATGGCTTTACGTGGCTTGT, (rev) AAGCCGCTGTATTGAGGAGA; ctxA: (fwd) TATAGCCACTGCACCCAACA, (rev) CAAGCACCCCAAAATGAACT; zot: (fwd) AGCTTTGAGGTGGCTTTTGA, (rev) GGTAAACTTTGCCCCTAGCC; control gene sanA: (fwd) TTGCTGTGGCTGACTATTGG, (rev) CCAATACCACTGCAACCTGA. First strand cDNA was synthesized by using random hexamers (Invitrogen SuperScriptTM III First-Strand Synthesis System for RT-PCR) according to manufacturer protocol. 1 µl aliquot of resulting cDNA was used in each PCR reaction. Fragments were amplified by 30 cycles of PCR (94°C for 30 sec, 52°C for 30 sec and 72°C for 30 sec. After amplification, 1 µl PCR product was run on a 1.5% agarose gel. The gel image was photographed by video gel documentation system (Universal Hood II-S.N. 76S/04668 by BIO-RAD Laboratories). The semi-quantitative RT-PCR images were evaluated and pixels in respective bands were quantified using Adobe Photoshop and ImageJ (by NIH).
Results and Discussion
Transcriptional response to V. cholerae cytolysin (VCC) during infection in C. elegans
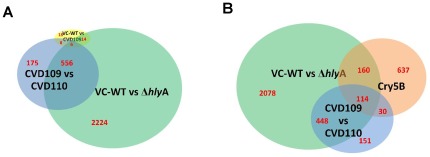
Host responses to VCC at the molecular level, and the significance of these responses in host defense during V. cholerae pathogenesis, remain poorly understood. To address this question, we performed a microarray study of C. elegans, which was exposed to V. cholerae strains with intact and deleted hlyA genes using Affymetrix C. elegans arrays. Gene expression in worms exposed to Wild type O1 el tor V. cholerae strain E7946 was compared with gene expression in worms exposed to the hlyA deletion mutant of E7946. We also compared gene expressions between vaccine strains CVD110 and CVD109 [23], both of which were generated in the E7946 genetic background. In the vaccine strain CVD109 the virulence genes zot, ace, ctxA, ctxB are deleted, but hlyA locus is intact. CVD110 was generated in the CVD109 genetic background by inactivating the hlyA gene via ctxB insertion [23]. We identified 2,800 differentially expressed genes when we compared expression in C. elegans exposed to V. cholerae wild type strain E7946, versus E7946 Δhly, and 743 differentially expressed genes when we compared expression in C. elegans exposed to nearly isogenic V. cholerae vaccine strains CVD109 (hlyA+), versus CVD110 (hlyA−) [considering fold change (+/−) 1.2, FDR = 0.5 and P<0.01]. Microarray data were confirmed using qRT-PCR to measure the expression levels of a set of selected genes (Fig. S1). We found significant overlap between “CVD109 versus CVD110” and “E7946 versus ΔhlyA” comparisons such that there were 562 genes in common between the two (Fig. 1A). A possible explanation for the dissimilarity in the differentially expressed gene number is that presence of the inserted ctxB gene in the CVD110 strain. ctxB, a known immunomodulator, was inserted as an immunologic adjuvant to enhance immune responses against the CVD110 vaccine strain [23]. Hence, over expression of ctxB in CVD110 may cancel out some of the immune response genes induced by hemolysin in CVD109, leading determination of lower number of genes in CVD109 versus CVD110 data set.
Figure 1. Genome-wide expression profile comparisons of the C. elegans genes regulated by pore forming toxins: V. cholerae hlyA, and B. thurigiensis Cry5B.
(A) A Venn diagram illustrating number of genes expressed in CVD109 versus CVD110, V. cholerae wild type versus DhlyA, and V. cholerae wild type versus CVD109 comparisons. (B) A Venn diagram illustrating number of genes expressed in CVD109 versus CVD110, V. cholerae wild type versus DhlyA, and Cry5B induction comparisons.
Induction by wild type V. cholerae strain E7946 versus V. cholerae vaccine strain with a deleted virulence cassette but intact hlyA, CVD109, revealed only 44 differentially expressed genes [considering fold change (+/−) 1.2, FDR = 0.5 and P<0.01]. This suggests that ctxA, ctxB, ace and zot genes of V. cholerae are not involved in immune response induction, and VCC is the major virulence factor inducing immune response during V. cholerae infection in C. elegans (Fig. 1A). Our findings support previous studies that showed cholerae toxin (CTX) may not be involved in the V. cholerae pathogenesis in C. elegans [11], [14,14].
Genome wide transcriptional responses to V. cholerae hemolysin and B. thuringiensis crystal toxin show significant overlap
VCC shows structural similarity to Bacillus thuringiensis crystal toxin, which is also a “small-pore” pore forming toxin [24], [25]. Previous studies revealed that B. thuringiensis crystal toxin causes intestinal damage and lethality in C. elegans [26]. We compared genome wide transcriptional responses to B. thuringiensis crystal toxin [27], with our data regarding transcriptional response to VCC and found a significant overlap between the two genomic responses (Fig. 1B). These results suggest that pore forming toxins induce immune responses in the host organisms through common and specific mechanisms. Since pore forming toxins are the most common bacterial toxins, we speculate that they might contribute to the induction of an immune response during a wide range of bacterial infections.
Previously reported C. elegans innate immune response genes are among the hemolysin responsive genes detected in our microarray analysis
Table 1 shows the list of genes up-regulated ≥2 fold in both ‘E7946 over E7946ΔhlyA’ and ‘CVD109 over CVD110’. About one third of the differentially expressed top ranker genes previously reported to be mediators of innate immune response against other bacteria in C. elegans (Table 1). C-type lectins, such as clec-45, clec-174, clec-209, clec-17, clec-47, are among the differentially expressed top ranker genes. C-type lectins have been proposed to act as pathogen-recognition molecules, and/or as effectors of the antimicrobial response by binding to carbohydrates on the surface of pathogens. The abu (activated in blocked unfolded protein response) genes, which contain a Prion-like (Q/N-rich)-domain, such as pqn-5, abu-6, abu-7, abu-8, were enriched in the Table 1. Lipase related gene lips-6, tollish gene toh-1, genes regulated by daf-16; dod-22 and dod-24, were also among the high rankers listed in this table.
Table 1. Genes induced over twofold following infection of C. elegans with hly(+) V. cholerae strains.
| Gene Name | Description | CVD109/CVD110 fold change | DhlyA/E7946 fold change | Reported Immune response function |
| clec-45 | C-type Lectin | 14.4 | 17.8 | Schulenburg et al, 2007 |
| clec-174 | C-type Lectin | 9.9 | 10.1 | Schulenburg et al, 2007 |
| C23G10.11 | hypothetical protein/Confirmed | 6.0 | 9.7 | Shapira et al, 2006 |
| fmo-2 | Flavin-containing MonoOxygenase family | 2.3 | 8.9 | Irazoqui et al, 2008 |
| dct-5 | DAF-16/FOXO Controlled, germline Tumor affecting | 4.7 | 7.9 | |
| col-41 | Collagen | 5.7 | 6.2 | |
| col-90 | Collagen | 6.5 | 5.8 | |
| grd-6 | Groundhog (hedgehog-like family) | 3.6 | 5.6 | |
| B0024.4 | hypothetical protein/Confirmed | 3.0 | 5.2 | |
| F35B3.4 | hypothetical protein/Confirmed | 3.9 | 4.9 | |
| F55G11.4 | hypothetical protein/Confirmed | 3.7 | 4.5 | Alper et al, 2007 |
| Y47D7A.13 | hypothetical protein/Partially confirmed | 3.2 | 4.2 | |
| col-54 | Collagen | 3.6 | 4.1 | |
| C25H3.10 | hypothetical protein | 2.6 | 4.1 | |
| pqn-5 | Prion-like-(Q/N-rich)-domain-bearing protein | 3.3 | 3.9 | Russell et al, 2008 |
| C42D4.3 | hypothetical protein/Confirmed | 2.5 | 3.7 | |
| abu-6 | Activated in Blocked Unfolded protein response | 3.2 | 3.6 | Russell et al, 2008 |
| C50F7.5 | hypothetical protein/Partially confirmed | 2.8 | 3.6 | |
| F53A9.2 | hypothetical protein/Confirmed | 2.3 | 3.5 | |
| F49H6.13 | hypothetical protein/Partially confirmed | 3.2 | 3.4 | |
| abu-8 | Activated in Blocked Unfolded protein response | 2.5 | 3.2 | Russell et al, 2008 |
| toh-1 | Tollish (Tolloid and BMP-1 family) | 2.1 | 3.1 | |
| abu-7 | Activated in Blocked Unfolded protein response | 2.9 | 3.1 | Russell et al, 2008 |
| lips-6 | Lipase related | 5.1 | 3.1 | |
| F22H10.2 | hypothetical protein/Partially confirmed | 2.2 | 3.0 | |
| C35C5.8 | hypothetical protein | 2.5 | 3.0 | |
| F44G3.10 | hypothetical protein/Confirmed | 2.2 | 2.9 | |
| col-156 | Collagen | 3.0 | 2.8 | |
| dod-24 | Downstream Of DAF-16 (regulated by DAF-16) | 3.0 | 2.8 | Troemel et al, 2006Styer et al, 2008 |
| toh-1 | Tollish (Tolloid and BMP-1 family) | 2.0 | 2.8 | |
| clec-209 | C-type Lectin | 3.7 | 2.8 | Schulenburg et al, 2007 |
| T22F3.11 | hypothetical protein | 2.1 | 2.8 | |
| glc-1 | Glutamate-gated ChLoride channel | 2.1 | 2.7 | |
| ZK180.5 | hypothetical protein | 4.0 | 2.7 | |
| Y54G2A.11 | hypothetical protein | 2.4 | 2.7 | |
| dod-22 | Downstream Of DAF-16 (regulated by DAF-16) | 2.1 | 2.5 | Shapira et al, 2006Alper et al, 2007 |
| clec-17 | C-type Lectin | 2.8 | 2.5 | O'Rourke et al, 2006Schulenburg et al, 2007 |
| Y95B8A.2 | hypothetical protein/Confirmed | 2.4 | 2.5 | |
| grd-14 | Groundhog (hedgehog-like family) | 3.6 | 2.4 | |
| nspb-12 | Nematode Specific Peptide family, group B | 2.5 | 2.4 | |
| F54B8.4 | hypothetical protein/Partially_confirmed | 2.1 | 2.4 | |
| clec-47 | C-type Lectin | 2.3 | 2.4 | Schulenburg et al, 2007 |
| lgc-21 | Ligand-Gated ion Channel | 3.2 | 2.4 | |
| F53A9.8 | hypothetical protein/Confirmed | 2.5 | 2.3 | O'Rourke et al, 2006 |
| C05E7.2 | hypothetical protein/Partially_confirmed | 2.6 | 2.3 | |
| D2096.6 | hypothetical protein/Partially_confirmed | 2.7 | 2.3 | |
| Y42A5A.3 | hypothetical protein/Confirmed | 2.0 | 2.3 | Troemel et al, 2006 |
| C06E8.5 | bacterial permeability-increasing protein | 2.0 | 2.2 | |
| C28H8.5 | hypothetical protein | 4.3 | 2.2 | |
| F41E6.11 | hypothetical protein/Partially_confirmed | 2.2 | 2.1 | |
| nspb-11 | Nematode Specific Peptide family, group B | 2.3 | 2.1 | |
| T12D8.5 | hypothetical protein/Confirmed | 2.0 | 2.1 | |
| ZK1307.2 | hypothetical protein/Confirmed | 2.3 | 2.1 | |
| T22B2.6 | hypothetical protein/Confirmed | 2.5 | 2.1 | |
| mup-4 | Muscle Positioning | 2.7 | 2.0 |
Only, genes induced over twofold in both CVD109/CVD110 and ΔhlyA/E7946 comparisons are listed.
The immune response function of the subset of the genes up regulated against V. cholerae hemolysin was confirmed using RNAi
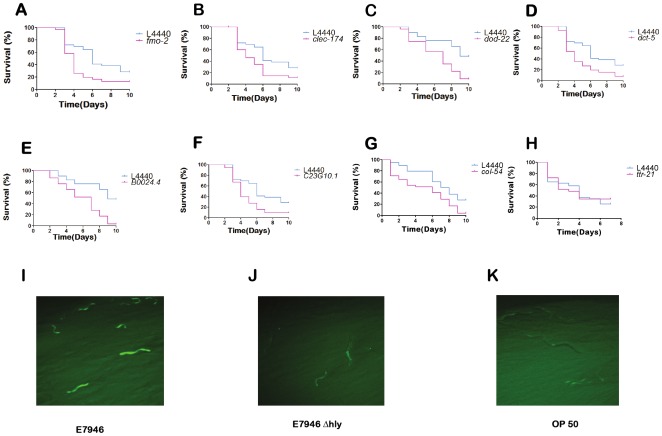
Seven, out of the nine genes tested, caused increased lethality when knocked down using RNAi (Fig. 2). These data suggest that these genes may have immune response functions. Some of these genes such as fmo-2, clec-174, and dod-22 were previously reported as immune response genes in C. elegans against other bacterial species. The flavin-containing MonoOxigenase family gene fmo-2 expression is up-regulated against Staphylococcus aureus through the β-catenin pathway [28]. The C-type lectin family gene clec-174 was shown to be up-regulated during Pseudomonas aureginosa and Photorhabdus luminescens infections in C. elegans [29]. dod-22, a CUB domain containing gene, was previously shown to be involved in the immune response against gram negative organisms S. Marcescens and P. aeruginosa via nsy-1 MAP kinase and daf-16 insulin signaling pathways [30]. Expression of the dod-22 gene is regulated by the insulin signaling pathway gene daf-16 [31]. We found that dod-22::GFP expression is induced in the C. elegans intestine during V. cholerae infection, and this induction is hlyA dependent (Fig. 2I, 2J, 2K). dct-5 was identified as a direct DAF-16 target [32], and found to regulate tumor growth in C. elegans [33]. B0024.4 and C23G10.1 genes were not previously characterized. B0024.4 gene encodes a putative glycoprotein, and C23G10.1 gene encodes a serine/threonine specific protein phosphatase PP1, with 64.9% similarity to human serine/threonine-protein phosphatase PP1-alpha catalytic subunit. col-54 encodes a protein similar to type IV and type XIII collagens which are located in basement membranes. We found that col-54 RNAi causes increased lethality in C. elegans exposed to V. cholerae wild type strain E7946 (Fig. 2G). Recent studies reported a dose-dependent antimicrobial activity of extracellular matrix collagens against group A, C, and G streptococci [34], [35]. Our data suggest a previously unrecognized innate immune response function for col-54, against V. cholerae.
Figure 2. VCC- induced C. elegans genes mediate immune response.
(A) fmo-2, p = 0.0063 (B) clec-174, p = 0.0135 (C) dod-22, p = 0.0007 (D) dct-5, p = 0.0045 (E) B0024.4, p = 0.0001(F) C23G10.1, p = 0.0038 (G) col-54, p = 0.0001 (H) ttr-21, p = 0.7436, RNAi result in lethality. Expression of dod-22::GFP in worms fed on (I) V. cholerae wild type strain E7946, (J) DhlyA, and (K) OP50.
Altogether our microarray and RNAi experiments indicate that V. cholerae hemolysin induces a variety of immune response genes in C. elegans during V. cholerae infection.
PQN/ABU Unfolded Protein response (UPR) genes are regulated through VCC
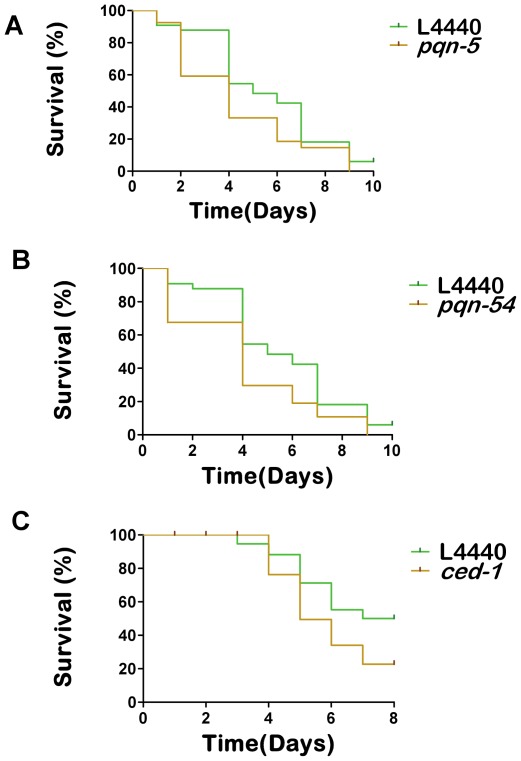
In C. elegans the pqn (prion-like glutamine [Q]/asparagines [N]) genes are identified as part of an alternative UPR pathway involved in regulating immune response against bacteria [36], [37]. A subgroup of pqn genes named as abu (activated in blocked UPR) genes, were induced to higher levels in endoplasmic reticulum (ER) stressed canonical UPR pathway mutants than in ER stressed wild type animals [38]. We found that many of pqn/abu genes are regulated through VCC in C. elegans. Twenty nine of seventy one pqn genes, and nine of eleven abu genes present in the C. elegans genome, are found to be differentially expressed in worms fed with hlyA deletion strains (Table 2). We knocked down seven of these genes using RNAi and found that two out of seven, namely pqn-5 and pqn-54, showed increased lethality during V. cholerae infection in C. elegans (Fig. 3A, 3B). It was previously reported that PQN/ABU proteins have a distant similarity to the C. elegans cell corpse engulfment protein CED-1, and to a mammalian scavenger receptor of endothelial cells (hSREC), which are transmembrane cell surface proteins [39]. ced-1 mutants are immuno-compromised, and are rapidly killed by live Salmonella enterica serovar Typhimurium and E. coli. Full-genome microarray analyses in C. elegans demonstrated that CED-1 upregulates expression of proteins with prion-like glutamine/asparagine (Q/N)-rich domains, which are known to be activated by ER stress and believed to aid in the unfolded protein response in Salmonella Typhimurium fed C. elegans [36]. A cluster of ced-1 regulated genes identified by Haskins et al., found to be down-regulated in worms exposed to hemolysin deletion strains of V. cholerae (Table S2). We found that ced-1 RNAi shows increased lethality when fed with V. cholerae wild type strain E7946 (Fig. 3C). Altogether our data suggest that PQN/ABU Unfolded Protein response genes and ced-1 are regulated through VCC.
Table 2. Prion-like (Q/N rich) domain protein genes regulated by hlyA.
| ORF NAME | GENE NAME | E7946/E7946ΔhlyA | CVD109/CVD110 |
| C03A7.4 | pqn-5 | +3.9 | +3.35 |
| C03A7.7 | abu-6/pqn-6 | +3.64 | +3.21 |
| ZC15.8 | pqn-94 | +3.27 | ND |
| C03A7.14 | abu-8 | +3.26 | +2.83 |
| F21C10.8 | pqn-31 | +3.2 | ND |
| C03A7.8 | abu-7/pqn-7 | +3.11 | +2.99 |
| ZK1067.7 | pqn-95 | +1.98 | +3.46 |
| R09B5.5 | pqn-54 | +1.9 | +2.12 |
| T01D1.6 | abu-11/pqn-61 | +1.87 | +1.85 |
| W01B11.5 | pqn-72 | +1.81 | ND |
| W02A2.3 | pqn-74 | +1.78 | +1.85 |
| AC3.3/AC3.4 | abu-1/pqn-2 | +1.7 | +1.98 |
| F35A5.3 | abu-10/pqn-33 | +1.67 | +1.68 |
| R09F10.7 | pqn-57 | +1.64 | ND |
| F39D8.1 | pqn-36 | +1.54 | ND |
| D1044.3 | pqn-25 | +1.45 | ND |
| T06E4.11 | pqn-63 | +1.4 | +1.73 |
| T23F1.6 | pqn-71 | +1.38 | ND |
| Y105C5A.4 | abu-5/pqn-77 | +1.35 | ND |
| Y5H2A.3 | abu-4 | +1.34 | ND |
| T16G1.1 | pqn-67 | +1.34 | ND |
| F31A3.1 | abu-3 | +1.31 | ND |
| W03D2.1 | pqn-75 | +1.31 | ND |
| C03A7.14 | abu-8/pqn-4 | +1.28 | +2.82 |
| M01E11.4 | pqn-52 | −1.2 | ND |
| R09E10.7 | pqn-55 | −1.23 | −1.52 |
| F35B3.5 | pqn-34 | −1.25 | ND |
| Y73B6BR.1 | pqn-89 | −1.33 | ND |
| F52D1.3 | pqn-40 | −1.79 | ND |
| F57B9.9 | pqn-46 | −2.46 | ND |
| F29C12.1 | pqn-32 | −3.01 | ND |
ND: No Difference,(+) up-regulated, (−) down-regulated.
Figure 3. RNAi of prion-like (Q/N rich) domain protein genes pqn-5 and pqn-54 causes increased lethality in C. elegans during V. cholerae infection.
(A) pqn-5 RNAi survival plot, p = 0.04 (B) pqn-54 RNAi survival plot, p = 0.02 (C) ced-1 RNAi survival plot p<0.0001.
Determination of the regulatory genes involved in the transcriptional response against VCC
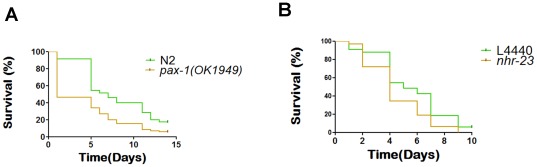
High throughput expression data collected from C. elegans in response to V. cholerae strains with intact and deleted hlyA loci provides us with a platform to search for regulatory genes involved in innate immune response, particularly the ones responsive to VCC. We used FastMEDUSA [18], a machine learning algorithm which integrates promoter sequence data, and microarray expression data, to determine the regulatory genes involve in the transcriptional response against VCC. FastMEDUSA is an open source implementation of MEDUSA [40] in C++ that uses parallel computing to decrease the execution time of MEDUSA. Using genome-wide expression changes in response to V. cholerae strains with intact and deleted hemolysin locus, 11 transcription factors were identified as immune regulatory genes during V. cholerae infection (Table 3). Five of these eleven genes are known to be expressed in C. elegans intestines (wormbase). We tested whether or not these transcriptional factors are required for the C. elegans' defense against V. cholerae using mutants and RNAi of these genes in lethality assay. Three genes, pax-1, nhr-23 and nhr-234, were tested for their contribution to the organisms response to infection. We found that the nhr-23 RNAi and the pax-1(ok1949) mutants exhibited increased lethality, suggesting that these genes induce the immune response in C. elegans (Fig. 4). nhr-23 gene encodes a conserved nuclear hormone receptor (NHR) in C. elegans [41]. Human homolog of NHR-23, RAR-related orphan receptor gamma (RORγ) involves in thymopoesis [42], which is the maturation process of immune T-cells. This is the first report demonstrating a possible immune function for nhr-23 in C. elegans. nhr-234(gk865) worms did not exhibit a significant decrease in C. elegans life span in lethality assay [p = 0.7011]. C. elegans has a large family of NHRs with 284 genes [43]. The C. elegans NHR family is a lot larger than its drosophila (18 genes), mouse (49 genes), and human (48 genes) counterparts [44]. Despite the large NHR component, C. elegans genome encodes only 15 conserved NHRs that belong to five of the six NHR subfamilies [41], [43], [45]. pax-1 gene encodes a paired box transcription factor, which is involved in skeletal system development [46], and has oncogenic potential in tissue cultures and in mice [47]. This is the first report indicating a possible immune function for pax-1. Molecular mechanisms underlying NHR-23 and PAX-1 function in innate immune response remains to be understood.
Table 3. Putative regulatory transcription factors identified using the Medusa program.
| Gene name | Description | GFP expression* | ||
| CVD110 | ΔhlyA | |||
| nhr-23 | ✓ | nuclear hormone receptor | Not known | |
| F22D6.2 | ✓ | ✓ | predicted Zn-finger protein | intestinal |
| egl-44 | ✓ | similar to vertebrate TEF proteins | Intestinal | |
| pqm-1 | ✓ | ✓ | C2H2-type zn-finger and leucine zipper containing protein | Intestinal |
| nhr-34 | ✓ | divergent nuclear receptor | Not known | |
| pax-1 | ✓ | ✓ | paired box transcription factor | Not known |
| mep-1/gei-2 | ✓ | Zn-finger protein | Intestinal | |
| peb-1 | ✓ | DNA binding protein containing FLYWCH type Zn-finger domain | Not known | |
| nhr-234 | ✓ | ✓ | nuclear hormone receptor | Not known |
| dhhc-2 | ✓ | Zn-finger protein, DHHC type | Not known | |
| ZK1320.3 | ✓ | ✓ | Unnamed protein | Intestine only |
GFP expression data retrieved from WormBase.
Figure 4. Lethality assays of the knock-downs of the FastMedusa identified immune response regulator transcription factors.
(A) pax-1(ok1949), p = 0.0013 and, (B) nhr-23, p = 0.0308 RNAi survival plots.
Binding motifs identified via FastMEDUSA are associated with GO terms related to pathogen recognition, innate immune response, and inflammatory response
FastMEDUSA analysis identified DNA motifs which may constitute putative transcription factor binding sites. Top 30 motifs were identified for each; ‘CVD110 versus V. cholerae wild type strain E7946’ and ‘ΔhlyA versus E7946’ comparisons. Eighteen of these motifs were common in the two comparisons. We ran 18 common motifs against human genome using GOMO (Gene Ontology for Motifs) tool to identify the GO-terms significantly associated with the identified motifs. Using the motifs GOMO scored the promoter region of each gene in the selected organism according to its binding affinity for the motif. Using these scores and the GO annotations of the organism's genes, GOMO determined the GO terms associated with the putative target genes of the binding motif [48]. Top five GO predictions considered for each motif identified (Table 4). We found that GO terms related to pathogen recognition, innate immune response, and inflammatory response predicted as high rankers. Eleven in eighteen common motifs were found to be associated with “olfactory receptor activity” and “sensory perception of smell” functions. C. elegans protects itself from pathogens not only through innate immunity pathways but also through behavioral strategies such as leaving the lawn of pathogenic bacteria ([49]–[54] and our unpublished data). C. elegans modifies its olfactory preferences after exposure to pathogenic bacteria, avoiding odors from the pathogen, becoming more attracted to odors from familiar nonpathogenic bacteria [55]. Recently, Sun et al. showed that, in C. elegans ASH and ASI sensory neurons involve in the regulation of immune responses via pqn/abu UPR pathway [56]. Olfactory nervous system functions are known to be important in the murine nervous system which has the ability to detect molecules related disease or inflammation, through the vemoronasal organ [57]. Five out of eighteen common motifs were found to be associated with immune and inflammatory responses related GO terms (Table 4). One in eighteen common motifs was found to be associated with unfolded protein response, a function shown to be important in immune response against pore forming toxins [58] and to VCC (this work). Interestingly, FastMEDUSA determined C. elegans putative binding sites found to be enriched in the promotors of the genes belong to these categories in human genome, suggesting functional homology between C. elegans and human genomes.
Table 4. GO terms associated with binding motifs identified via FastMEDUSA.
| Motif Logo | Predictions | Top 5 specific predictions |
| AATCGCTT | 36 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| MF RNA binding | ||
| CC spliceosomal complex | ||
| BP chromosome segregation | ||
| AATGGAC | 21 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| BP innate immune response | ||
| MF taste receptor activity | ||
| ACAGAGG | 146 | CC extracellular space |
| CC integral to plasma membrane | ||
| MF calcium ion binding | ||
| MF hormone activity | ||
| BP cell adhesion | ||
| ACCAGAGCT | 52 | CC integral to plasma membrane |
| CC extracellular space | ||
| MF heme binding | ||
| BP excretion | ||
| CC keratin filament | ||
| ACGTGAT | 57 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP intracellular protein transport | ||
| MF RNA binding | ||
| BP DNA repair | ||
| ACGTTCG | 408 | CC nucleolus |
| CC spliceosomal complex | ||
| BP rRNA processing | ||
| MF structural constituent of ribosome | ||
| MF translation regulator activity | ||
| AGATTTC | 28 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| BP innate immune response | ||
| BP inflammatory response | ||
| ATCGCTA | 114 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| CC mitochondrial matrix | ||
| MF RNA binding | ||
| BP ncRNA metabolic process | ||
| ATGCCCC | 112 | BP regulation of striated muscle contraction |
| BP regulation of signal transduction | ||
| CC terminal button | ||
| BP cardiac muscle tissue morphogenesis | ||
| CC keratin filament | ||
| ATTGTTCCA | 64 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| BP inflammatory response | ||
| BP regulation of lymphocyte mediated immunity | ||
| CGCTAGA | 158 | BP nuclear mRNA splicing, via spliceosome |
| CC spliceosomal complex | ||
| CC nucleolus | ||
| BP rRNA processing | ||
| MF structural constituent of ribosome | ||
| CTGGAACT | 0 | |
| GAAATCT | 34 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| BP innate immune response | ||
| BP inflammatory response | ||
| TAATCGCTG | 5 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| TACGTTCT | 37 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| BP response to stimulus | ||
| MF unfolded protein binding | ||
| TAGAACG | 71 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| MF RNA binding | ||
| BP nuclear mRNA splicing, via spliceosome | ||
| MF structural constituent of ribosome | ||
| TCAGAGAA | 94 | MF olfactory receptor activity |
| BP sensory perception of smell | ||
| BP G-protein coupled receptor protein signaling pathway | ||
| CC extracellular space | ||
| BP immune response | ||
| TCCAGAGG | 70 | CC integral to plasma membrane |
| CC extracellular space | ||
| CC proteinaceous extracellular matrix | ||
| MF calcium ion binding | ||
| BP regulation of monooxygenase activity |
The effect of temperature on expression of V. cholerae virulence factors
Many bacterial pathogens regulate the expression of virulence factors in response to changes in the environment. For example the levels of Listeria monocytogenes virulence gene expression depend on the amounts of the PrfA protein, which is expressed at high levels at 37°C, the temperature of the warm-blooded animal host [59]. C. elegans is a soil nematode with an optimal culture temperature range between 16°C and 25°C, therefore this host system is not amenable to conduct experiments at 37°C. In spite of this limitation, C. elegans host-pathogen interaction studies regarding human pathogens such as Salmonella species, Staphylococcus aureus, E. coli have yielded a body of C. elegans host response data showing high correlation to human immune response against these pathogens [60]–[64]. V. cholerae virulence genes are coordinately regulated by external stimuli, such as temperature, pH and osmolarity [65]. The expression of toxR and toxR regulated virulence genes including ctxA, was reduced between 12- and 32-fold by growth at 37°C in comparison with 30°C growth [66]. It is not known whether the decreased levels of virulence gene expression at 37°C in vitro, correlates with the intraintestinal expression. [66]. Effects of incubation temperature and time to the hemolytic activity of El Tor V. cholerae was reported by Feeley and Pittman. They found that at 35°C maximum hemolytic activity were observed at 24 hours, followed by a decline. At 30°C, maximum titres were also observed in 24 hours but the rate of decrease was less pronounced. With cultures incubated at 22°C, they measured comparable levels of maximum hemolytic activity in 48 hours, and rate of decay of activity was greatly retarded [67].
We wanted to explore the effect of temperature on the expression of hlyA and other virulence genes deleted in vaccine strains CVD109 and CVD110. The relative transcript abundance and expression of V. cholerae virulence genes hlyA, ace, zot, and ctxA at different temperatures was evaluated using semi-quantitative RT-PCR in V. cholerae wild type strain E7946. We found that all four of the genes tested were expressed at comparable levels at all the temperatures tested (Fig. S2). Our data suggest that, at our experimental conditions [22°C], V. cholerae virulence gene expression levels are comparable to the expression levels at the human body temperature 37°C.
In summary, we report a genome scale study regarding host responses against V. cholerae hemolysin. V. cholerae hemolysin induces expression of wide variety of immune response genes, some of which known to be responsive to other pathogenic bacteria. We found that PQN/ABU Unfolded Protein response (UPR) pathway involves in immune response against V. cholerae hemolysin. Using bioinformatics tools together with experimental validation, we identified transcriptional factors and transcriptional factor binding motifs involved in the immune response against VCC.
Supporting Information
qRT-PCR results for selected high ranker genes.
(TIF)
Semi-quantitative RT-PCR results showing expression levels of V. cholerae virulence genes at different temperatures. Corresponding gel images are shown at the top of each column.
(TIF)
GO terms enriched in CVD109/CVD110 and/or E7946/Δ hlyA comparisons.
(DOC)
Cluster of C. elegans CED-1 regulated genes#, and fold differences in expressional response to hly (+) versus hly (−) V. cholerae .
(DOC)
Acknowledgments
C. elegans strains used in this paper were provided by the Caenorhabditis Genetics Center. We are grateful to Nick Olejnick, Thomas Black, and Oluwakemi Odusami for technical help, to Christopher Grim and Augusto Franco-Mora for their help on semi-quantitative RT-PCR analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Ichinose Y, Yamamoto K, Nakasone N, Tanabe MJ, Takeda T, et al. Enterotoxicity of El Tor-like hemolysin of non-O1 Vibrio cholerae. Infect Immun. 1987;55:1090–1093. doi: 10.1128/iai.55.5.1090-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson R, Gouaux E. Vibrio cholerae cytolysin is composed of an alpha-hemolysin-like core. Protein Sci. 2003;12:379–383. doi: 10.1110/ps.0231703. 10.1110/ps.0231703 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikigai H, Akatsuka A, Tsujiyama H, Nakae T, Shimamura T. Mechanism of membrane damage by El Tor hemolysin of Vibrio cholerae O1. Infect Immun. 1996;64:2968–2973. doi: 10.1128/iai.64.8.2968-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm RA, Mayrhofer G, Kotlarski I, Manning PA. Amino-terminal domain of the El Tor haemolysin of Vibrio cholerae O1 is expressed in classical strains and is cytotoxic. Vaccine. 1991;9:588–594. doi: 10.1016/0264-410x(91)90247-4. [DOI] [PubMed] [Google Scholar]
- 5.Zitzer A, Palmer M, Weller U, Wassenaar T, Biermann C, et al. Mode of primary binding to target membranes and pore formation induced by Vibrio cholerae cytolysin (hemolysin). Eur J Biochem. 1997;247:209–216. doi: 10.1111/j.1432-1033.1997.00209.x. [DOI] [PubMed] [Google Scholar]
- 6.Coelho A, Andrade JR, Vicente AC, Dirita VJ. Cytotoxic cell vacuolating activity from Vibrio cholerae hemolysin. Infect Immun. 2000;68:1700–1705. doi: 10.1128/iai.68.3.1700-1705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra R, Figueroa P, Mukhopadhyay AK, Shimada T, Takeda Y, et al. Cell vacuolation, a manifestation of the El tor hemolysin of Vibrio cholerae. Infect Immun. 2000;68:1928–1933. doi: 10.1128/iai.68.4.1928-1933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa-Arredondo P, Heuser JE, Akopyants NS, Morisaki JH, Giono-Cerezo S, et al. Cell vacuolation caused by Vibrio cholerae hemolysin. Infect Immun. 2001;69:1613–1624. doi: 10.1128/IAI.69.3.1613-1624.2001. 10.1128/IAI.69.3.1613-1624.2001 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saka HA, Gutierrez MG, Bocco JL, Colombo MI. The autophagic pathway: a cell survival strategy against the bacterial pore-forming toxin Vibrio cholerae cytolysin. Autophagy. 2007;3:363–365. doi: 10.4161/auto.4159. 4159 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Olivier V, Haines GK, III, Tan Y, Satchell KJ. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cinar HN, Kothary M, Datta AR, Tall BD, Sprando R, et al. Vibrio cholerae hemolysin is required for lethality, developmental delay, and intestinal vacuolation in Caenorhabditis elegans. PLoS One. 2010;5:e11558. doi: 10.1371/journal.pone.0011558. 10.1371/journal.pone.0011558 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–390. doi: 10.1038/nrg1067. 10.1038/nrg1067 [doi];nrg1067 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, et al. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc Natl Acad Sci U S A. 2006;103:9280–9285. doi: 10.1073/pnas.0601754103. 0601754103 [pii];10.1073/pnas.0601754103 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprando RL, Olejnik N, Cinar HN, Ferguson M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a Complex Object Parametric Analyzer and Sorter, and axenic liquid media. Food Chem Toxicol. 2009;47:722–728. doi: 10.1016/j.fct.2009.01.007. S0278-6915(09)00008-8 [pii];10.1016/j.fct.2009.01.007 [doi] [DOI] [PubMed] [Google Scholar]
- 16.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. 10.1038/nature01278 [doi];nature01278 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Bozdag S, Li A, Wuchty S, Fine HA. FastMEDUSA: a parallelized tool to infer gene regulatory networks. Bioinformatics. 2010;26:1792–1793. doi: 10.1093/bioinformatics/btq275. btq275 [pii];10.1093/bioinformatics/btq275 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundaje A, Xin X, Lan C, Lianoglou S, Zhou M, et al. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput Biol. 2008;4:e1000224. doi: 10.1371/journal.pcbi.1000224. 10.1371/journal.pcbi.1000224 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrasa MI, Vaglio P, Cavasino F, Jacotot L, Walhout AJ. EDGEdb: a transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. 1471-2164-8-21 [pii];10.1186/1471-2164-8-21 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider S, Ballester B, Smedley D, Zhang J, Rice P, et al. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. gkp265 [pii];10.1093/nar/gkp265 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freund Y, Schapire R. A decision-theoretic generalization of on-line learning and an application to boosting. Journal of Computer and System Sciences. 1997;55:119–139. [Google Scholar]
- 23.Michalski J, Galen JE, Fasano A, Kaper JB. CVD110, an attenuated Vibrio cholerae O1 El Tor live oral vaccine strain. Infect Immun. 1993;61:4462–4468. doi: 10.1128/iai.61.10.4462-4468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huffman DL, Bischof LJ, Griffitts JS, Aroian RV. Pore worms: using Caenorhabditis elegans to study how bacterial toxins interact with their target host. Int J Med Microbiol. 2004;293:599–607. doi: 10.1078/1438-4221-00303. [DOI] [PubMed] [Google Scholar]
- 25.Bellier A, Chen CS, Kao CY, Cinar HN, Aroian RV. Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans. PLoS Pathog. 2009;5:e1000689. doi: 10.1371/journal.ppat.1000689. 10.1371/journal.ppat.1000689 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marroquin LD, Elyassnia D, Griffitts JS, Feitelson JS, Aroian RV. Bacillus thuringiensis (Bt) toxin susceptibility and isolation of resistance mutants in the nematode Caenorhabditis elegans. Genetics. 2000;155:1693–1699. doi: 10.1093/genetics/155.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, et al. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. 10.1073/pnas.0404073101 [doi];0404073101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc Natl Acad Sci U S A. 2008;105:17469–17474. doi: 10.1073/pnas.0809527105. 0809527105 [pii];10.1073/pnas.0809527105 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulenburg H, Hoeppner MP, Weiner J, III, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213:237–250. doi: 10.1016/j.imbio.2007.12.004. S0171-2985(08)00002-8 [pii];10.1016/j.imbio.2007.12.004 [doi] [DOI] [PubMed] [Google Scholar]
- 30.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. MCB.02070-06 [pii];10.1128/MCB.02070-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. 10.1038/nature01789 [doi];nature01789 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. ng1723 [pii];10.1038/ng1723 [doi] [DOI] [PubMed] [Google Scholar]
- 33.Pinkston-Gosse J, Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 2007;39:1403–1409. doi: 10.1038/ng.2007.1. ng.2007.1 [pii];10.1038/ng.2007.1 [doi] [DOI] [PubMed] [Google Scholar]
- 34.Bober M, Enochsson C, Collin M, Morgelin M. Collagen VI is a subepithelial adhesive target for human respiratory tract pathogens. J Innate Immun. 2010;2:160–166. doi: 10.1159/000232587. 000232587 [pii];10.1159/000232587 [doi] [DOI] [PubMed] [Google Scholar]
- 35.Abdillahi SM, Balvanovic S, Baumgarten M, Morgelin M. Collagen VI Encodes Antimicrobial Activity: Novel Innate Host Defense Properties of the Extracellular Matrix. J Innate Immun. 2012 doi: 10.1159/000335239. 000335239 [pii];10.1159/000335239 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. S1534-5807(08)00206-2 [pii];10.1016/j.devcel.2008.05.006 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–732. doi: 10.1126/science.1203411. science.1203411 [pii];10.1126/science.1203411 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. 10.1083/jcb.200203086 [doi];200203086 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. S0092-8674(01)00190-8 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Kundaje A, Lianoglou S, Li X, Quigley D, Arias M, et al. Learning regulatory programs that accurately predict differential expression with MEDUSA. Ann N Y Acad Sci. 2007;1115:178–202. doi: 10.1196/annals.1407.020. annals.1407.020 [pii];10.1196/annals.1407.020 [doi] [DOI] [PubMed] [Google Scholar]
- 41.Antebi A. Nuclear hormone receptors in C. elegans. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.64.1. 10.1895/wormbook.1.64.1 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. 10.1038/ni1022 [doi];ni1022 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Sluder AE, Mathews SW, Hough D, Yin VP, Maina CV. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- 44.Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, et al. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2:RESEARCH0029. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van GM, Gissendanner CR, Sluder AE. Diversity and function of orphan nuclear receptors in nematodes. Crit Rev Eukaryot Gene Expr. 2002;12:65–88. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.40. [DOI] [PubMed] [Google Scholar]
- 46.Deutsch U, Dressler GR, Gruss P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell. 1988;53:617–625. doi: 10.1016/0092-8674(88)90577-6. 0092-8674(88)90577-6 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Maulbecker CC, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12:2361–2367. doi: 10.1002/j.1460-2075.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buske FA, Boden M, Bauer DC, Bailey TL. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 2010;26:860–866. doi: 10.1093/bioinformatics/btq049. btq049 [pii];10.1093/bioinformatics/btq049 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. S0960-9822(01)00241-X [pii] [DOI] [PubMed] [Google Scholar]
- 50.Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H. The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda: Rhabditidae). Environ Microbiol. 2007;9:12–25. doi: 10.1111/j.1462-2920.2006.01099.x. EMI1099 [pii];10.1111/j.1462-2920.2006.01099.x [doi] [DOI] [PubMed] [Google Scholar]
- 51.Beale E, Li G, Tan MW, Rumbaugh KP. Caenorhabditis elegans senses bacterial autoinducers. Appl Environ Microbiol. 2006;72:5135–5137. doi: 10.1128/AEM.00611-06. 72/7/5135 [pii];10.1128/AEM.00611-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, et al. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. 0610281104 [pii];10.1073/pnas.0610281104 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. 323/5912/382 [pii];10.1126/science.1166527 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy KC, Hunter RC, Bhatla N, Newman DK, Kim DH. Caenorhabditis elegans NPR-1-mediated behaviors are suppressed in the presence of mucoid bacteria. Proc Natl Acad Sci U S A. 2011;108:12887–12892. doi: 10.1073/pnas.1108265108. 1108265108 [pii];10.1073/pnas.1108265108 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. nature04216 [pii];10.1038/nature04216 [doi] [DOI] [PubMed] [Google Scholar]
- 56.Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–732. doi: 10.1126/science.1203411. science.1203411 [pii];10.1126/science.1203411 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. nature08029 [pii];10.1038/nature08029 [doi] [DOI] [PubMed] [Google Scholar]
- 58.Bischof LJ, Kao CY, Los FC, Gonzalez MR, Shen Z, et al. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog. 2008;4:e1000176. doi: 10.1371/journal.ppat.1000176. 10.1371/journal.ppat.1000176 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. S0092867402009054 [pii] [DOI] [PubMed] [Google Scholar]
- 60.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. S0952-7915(04)00189-X [pii];10.1016/j.coi.2004.11.007 [doi] [DOI] [PubMed] [Google Scholar]
- 61.Kurz CL, Ewbank JJ. Infection in a dish: high-throughput analyses of bacterial pathogenesis. Curr Opin Microbiol. 2007;10:10–16. doi: 10.1016/j.mib.2006.12.001. S1369-5274(06)00186-X [pii];10.1016/j.mib.2006.12.001 [doi] [DOI] [PubMed] [Google Scholar]
- 62.Shivers RP, Youngman MJ, Kim DH. Transcriptional responses to pathogens in Caenorhabditis elegans. Curr Opin Microbiol. 2008;11:251–256. doi: 10.1016/j.mib.2008.05. S1369-5274(08)00071-4 [pii];10.1016/j.mib.2008.05.014 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2010;708:105–121. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 64.Glavis-Bloom J, Muhammed M, Mylonakis E. Of model hosts and man: using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv Exp Med Biol. 2012;710:11–17. doi: 10.1007/978-1-4419-5638-5_2. 10.1007/978-1-4419-5638-5_2 [doi] [DOI] [PubMed] [Google Scholar]
- 65.Skorupski K, Taylor RK. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 66.Parsot C, Mekalanos JJ. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc Natl Acad Sci U S A. 1990;87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feeley JC, Pittman M. Studies on the haemolytic activity of El Tor vibrios. Bull World Health Organ. 1963;28:347–356. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR results for selected high ranker genes.
(TIF)
Semi-quantitative RT-PCR results showing expression levels of V. cholerae virulence genes at different temperatures. Corresponding gel images are shown at the top of each column.
(TIF)
GO terms enriched in CVD109/CVD110 and/or E7946/Δ hlyA comparisons.
(DOC)
Cluster of C. elegans CED-1 regulated genes#, and fold differences in expressional response to hly (+) versus hly (−) V. cholerae .
(DOC)