Abstract
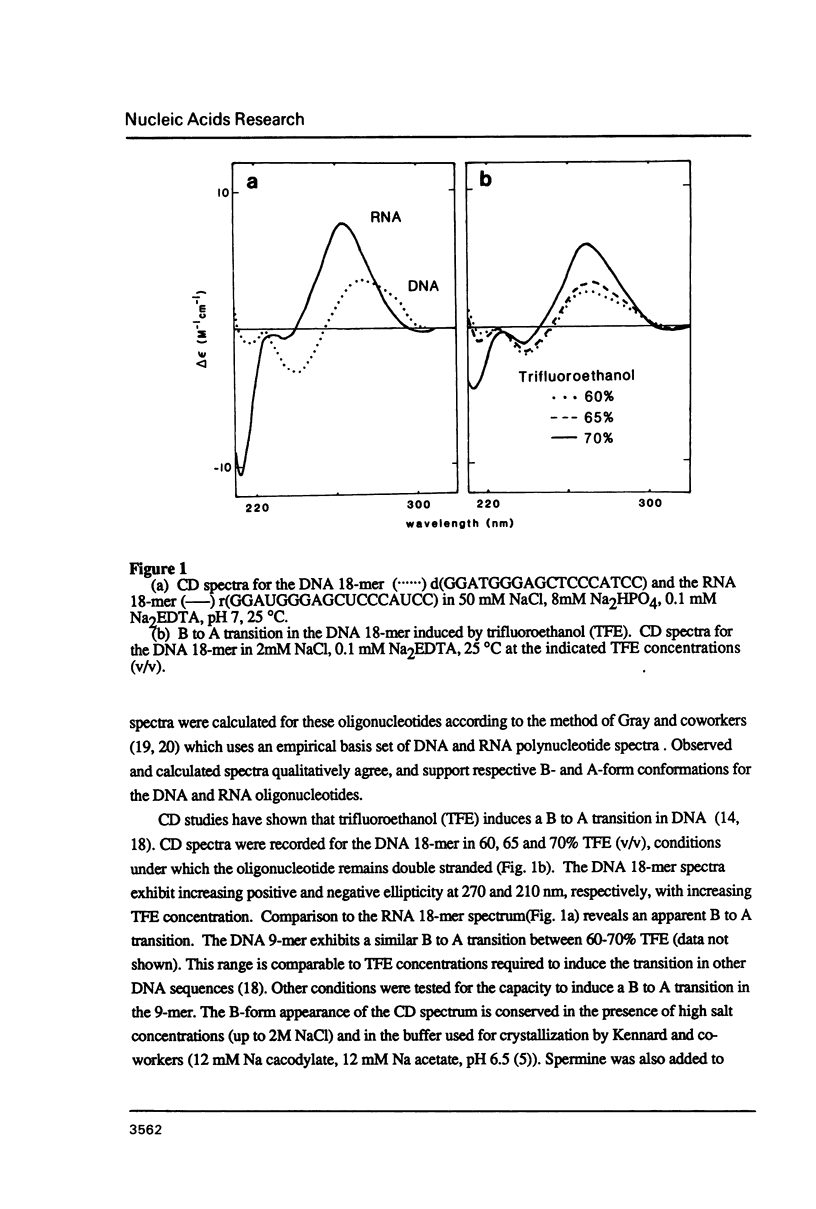
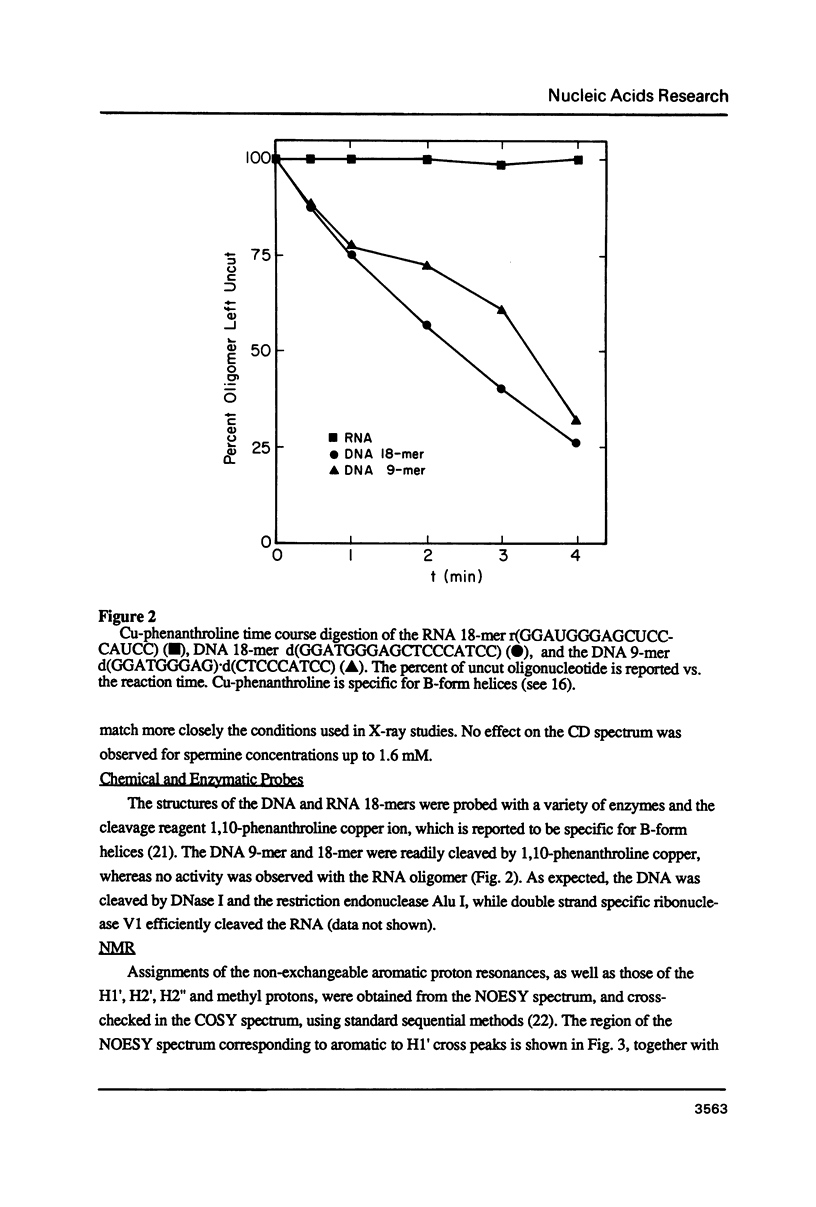
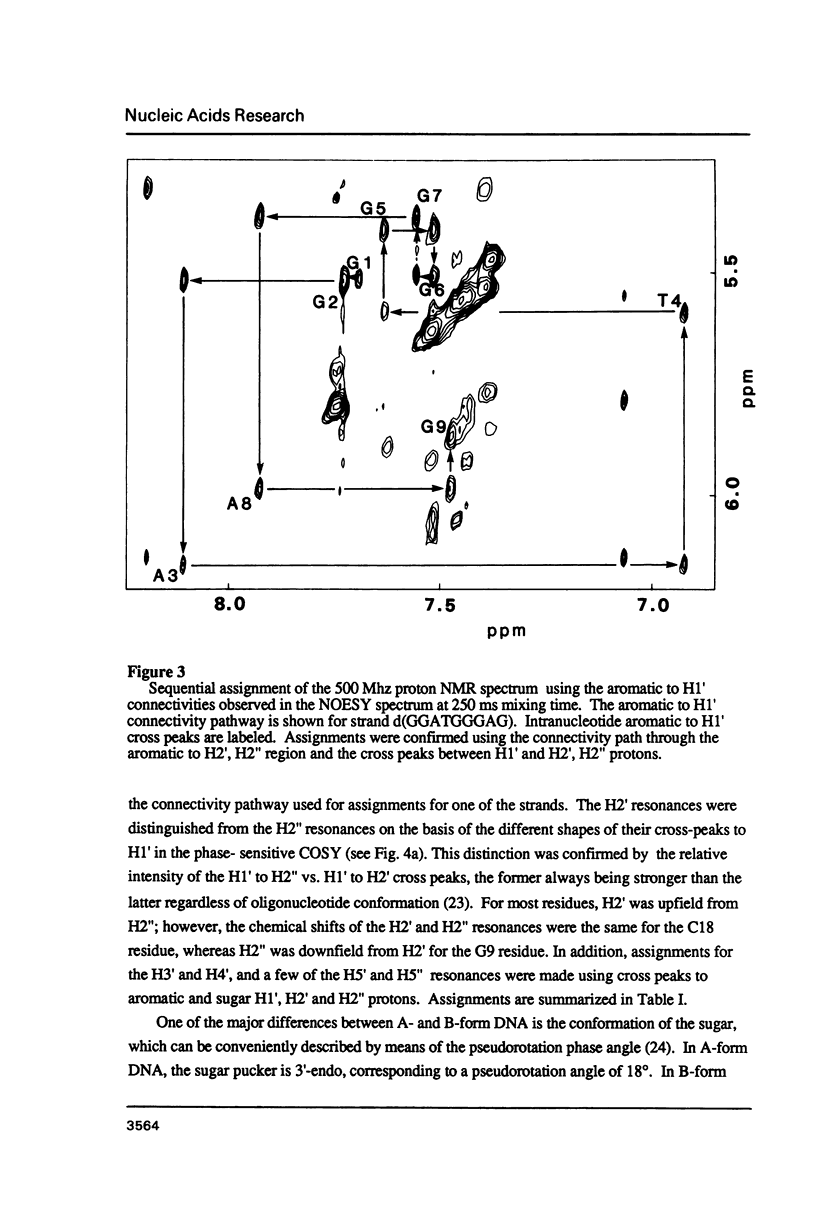
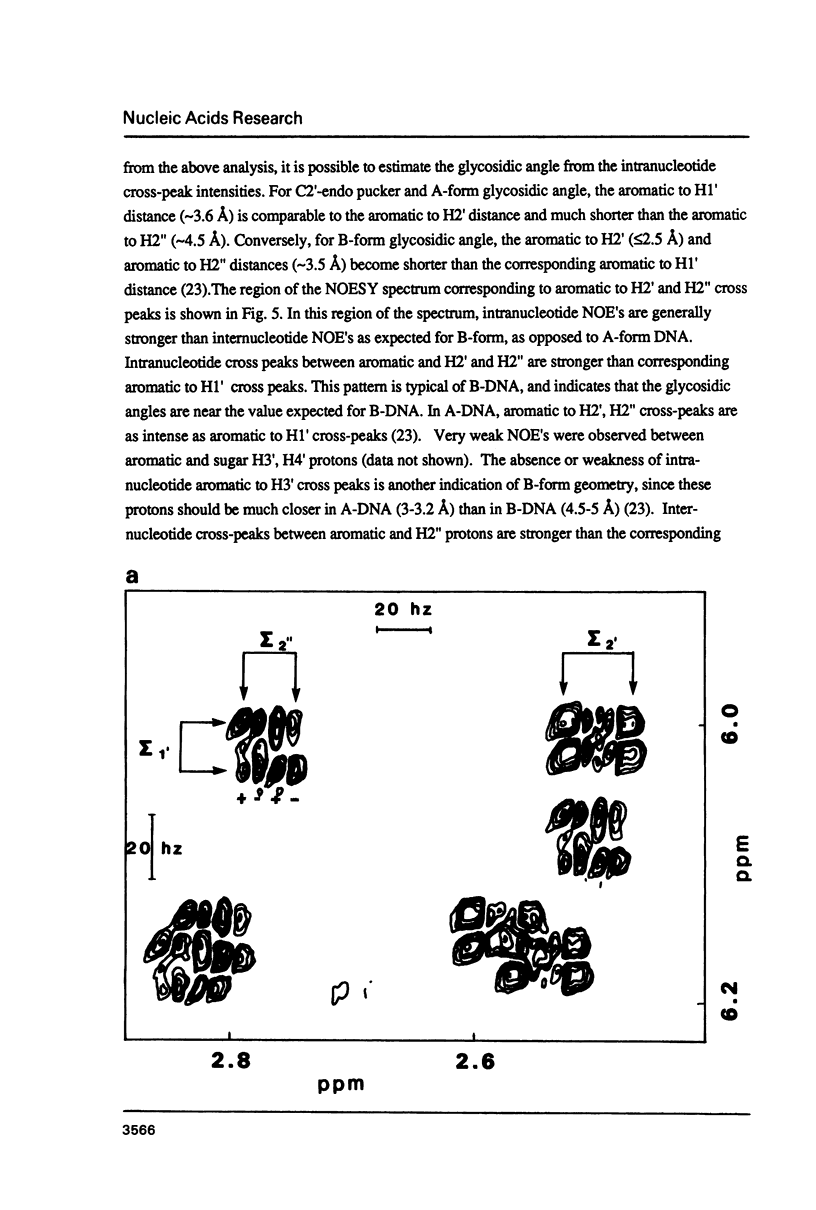
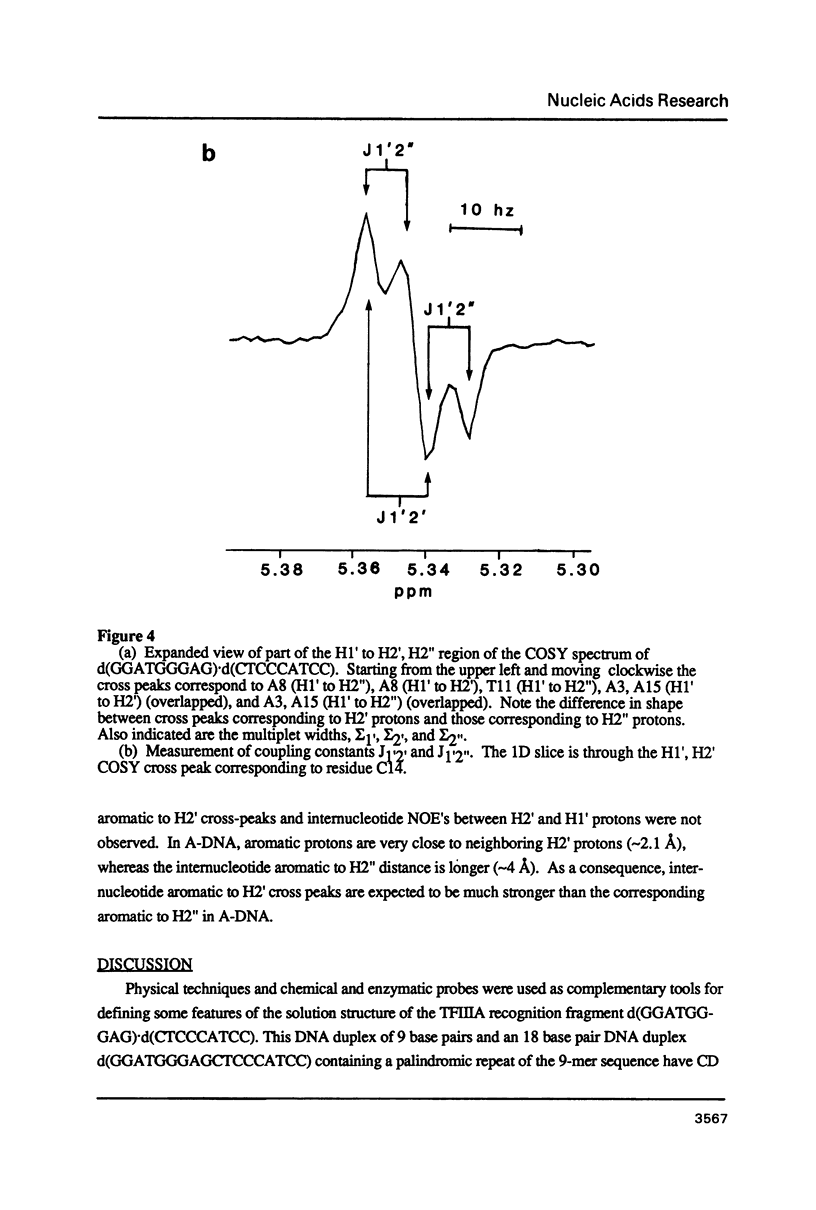
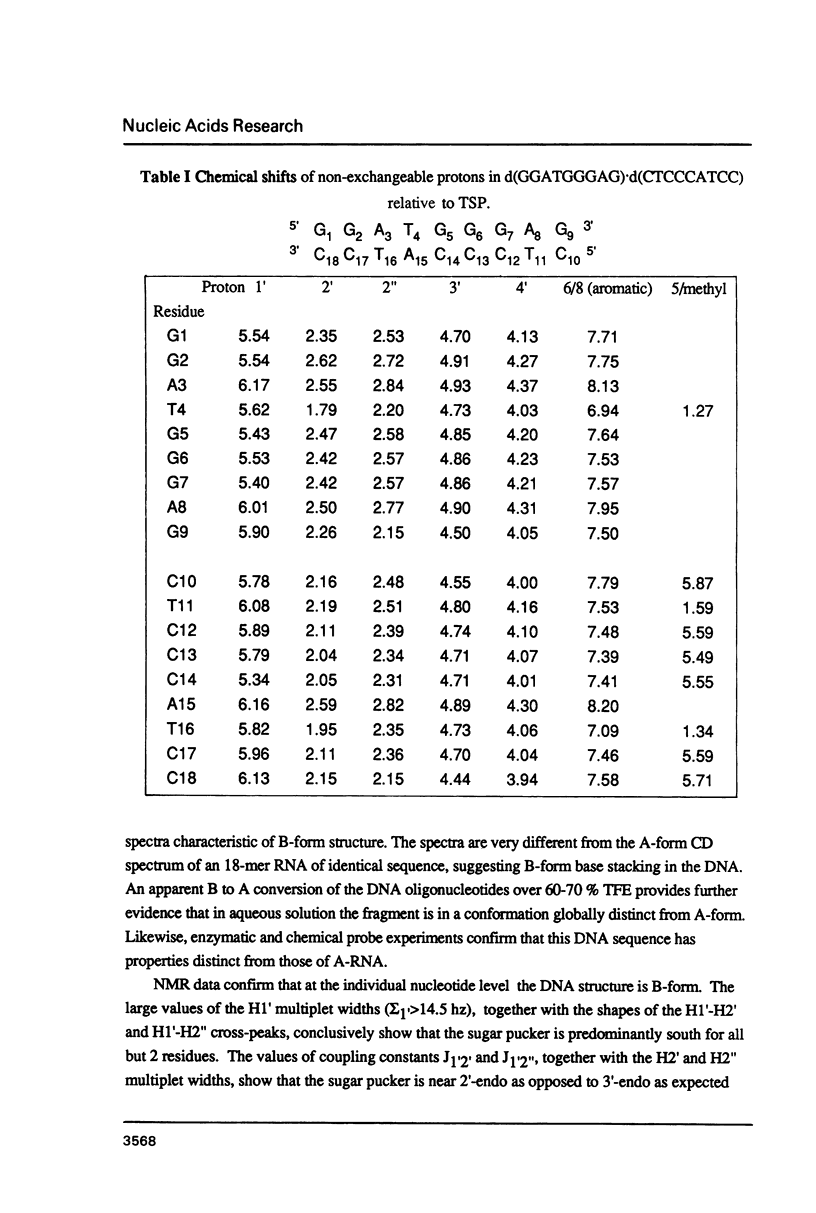
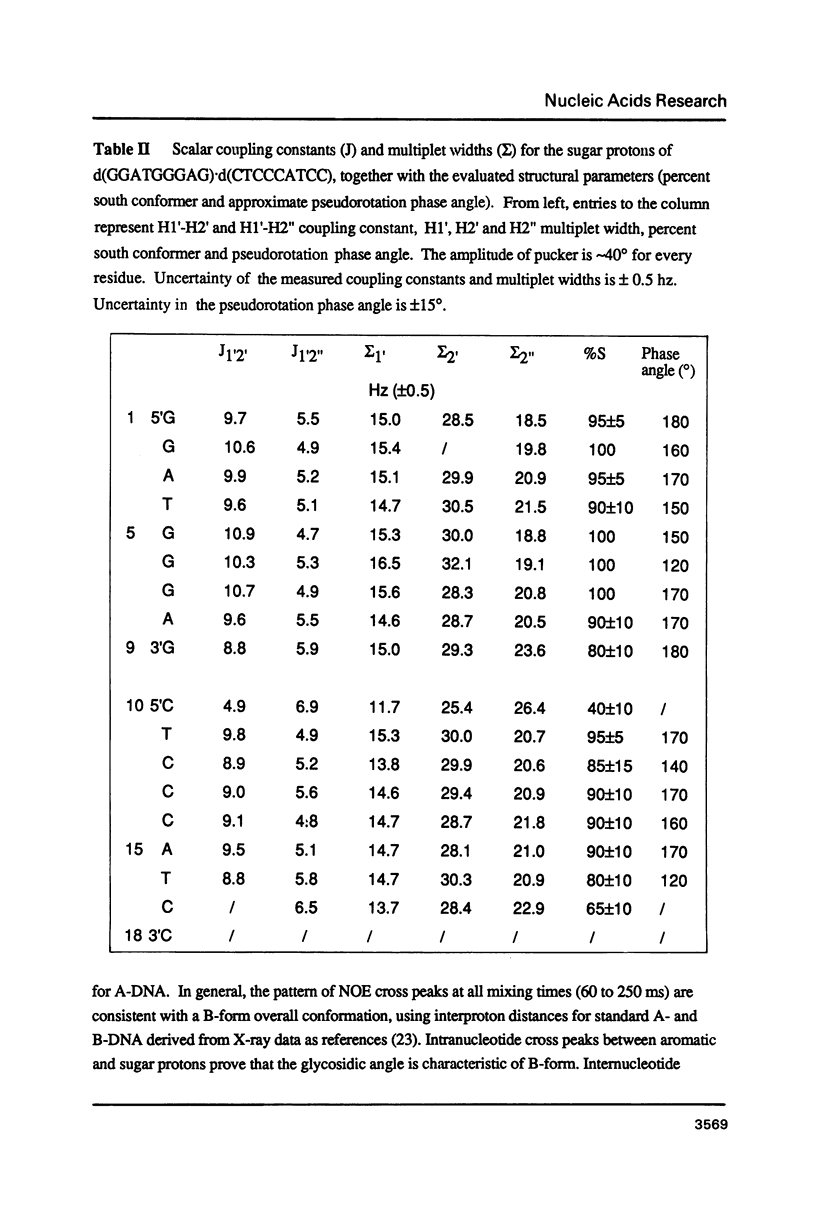
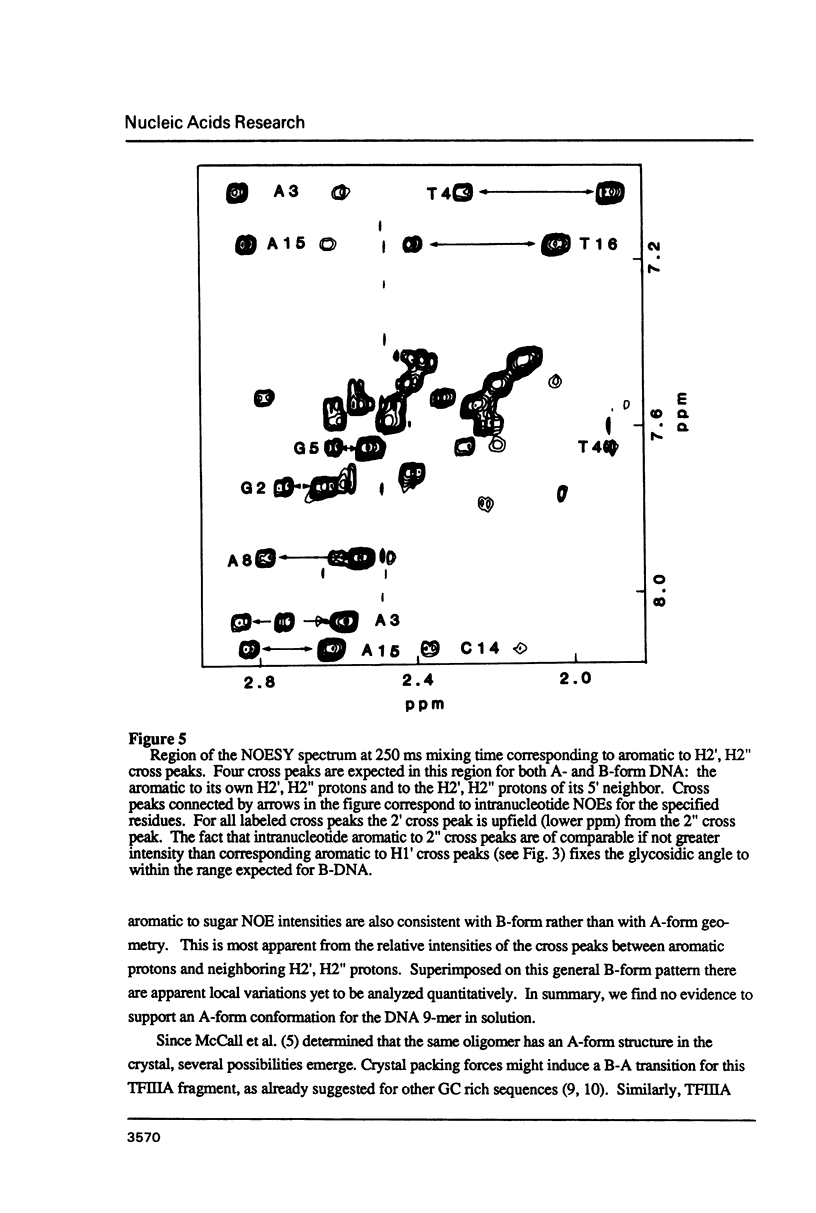
The deoxyoligonucleotide d(GGATGGGAG).d(CTCCCATCC) is a portion of the gene recognition sequence of transcription factor IIIA (TFIIIA). The crystal structure of this oligonucleotide was shown to be A-form (Mc Call, M., Brown, T., Hunter, W.N., and Kennard, O. 1986 Nature 322, 661-664). The present study employs NMR, optical, chemical and enzymatic techniques to investigate the solution structure of this DNA 9-mer. NMR COSY experiments indicate 16 of the 18 residues are predominantly south (C2'-endo) sugar conformation. NMR NOESY indicates glycosidic angles in the range predicted for B-form DNA as opposed to A-form. Related DNA and RNA self-complementary 18-mer sequences, d(GGATGGGAGC-TCCCATCC), with U substituted for T in RNA, were studied by circular dichroism. CD spectra support B-form structures for the DNA 9-mer and the DNA 18-mer, and A-form for the RNA 18-mer. High trifluoroethanol concentrations induce a B- to A-form transition in the DNA oligonucleotides. Enzymatic and chemical probes also illustrate significant differences between the DNA and the RNA oligonucleotides. We find no evidence to support an A-form conformation for the TFIIIA recognition sequence d(GGATGGGAG).d(CTCCCATCC) in solution.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen F. S., Gray D. M., Ratliff R. L. On the first-neighbor analysis of nucleic acid CD spectra: the definitive T matrix and considerations of various methods. Biopolymers. 1984 Nov;23(11 Pt 2):2639–2659. doi: 10.1002/bip.360231133. [DOI] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Benevides J. M., Wang A. H., Rich A., Kyogoku Y., van der Marel G. A., van Boom J. H., Thomas G. J., Jr Raman spectra of single crystals of r(GCG)d(CGC) and d(CCCCGGGG) as models for A DNA, their structure transitions in aqueous solution, and comparison with double-helical poly(dG).poly(dC). Biochemistry. 1986 Jan 14;25(1):41–50. doi: 10.1021/bi00349a007. [DOI] [PubMed] [Google Scholar]
- Brown D. D. How a simple animal gene works. Harvey Lect. 1980;76:27–44. [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Blanco J., Tennant L. L. The 5S gene internal control region is B-form both free in solution and in a complex with TFIIIA. Nature. 1987 Oct 1;329(6138):460–462. doi: 10.1038/329460a0. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Westerink H. P., van der Marel G. A., van Boom J. H. Discrimination between A-type and B-type conformations of double helical nucleic acid fragments in solution by means of two-dimensional nuclear Overhauser experiments. J Biomol Struct Dyn. 1984 Oct;2(2):345–360. doi: 10.1080/07391102.1984.10507572. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Binding of Xenopus transcription factor A to 5S RNA and to single stranded DNA. Nucleic Acids Res. 1984 Mar 26;12(6):2745–2758. doi: 10.1093/nar/12.6.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Marshall L. E., Graham D. R., Reich K. A., Sigman D. S. Cleavage of deoxyribonucleic acid by the 1,10-phenanthroline-cuprous complex. Hydrogen peroxide requirement and primary and secondary structure specificity. Biochemistry. 1981 Jan 20;20(2):244–250. doi: 10.1021/bi00505a003. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Kennard O. The crystal structure of d(G-G-G-G-C-C-C-C). A model for poly(dG).poly(dC). J Mol Biol. 1985 Jun 5;183(3):385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- Minchenkova L. E., Schyolkina A. K., Chernov B. K., Ivanov V. I. CC/GG contacts facilitate the B to A transition of DNA in solution. J Biomol Struct Dyn. 1986 Dec;4(3):463–476. doi: 10.1080/07391102.1986.10506362. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Hare D. Nuclear magnetic resonance and distance geometry studies of DNA structures in solution. Annu Rev Biophys Biophys Chem. 1987;16:423–454. doi: 10.1146/annurev.bb.16.060187.002231. [DOI] [PubMed] [Google Scholar]
- Reynolds W. F., Gottesfeld J. M. 5S rRNA gene transcription factor IIIA alters the helical configuration of DNA. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1862–1866. doi: 10.1073/pnas.80.7.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Riazance J. H., Baase W. A., Johnson W. C., Jr, Hall K., Cruz P., Tinoco I., Jr Evidence for Z-form RNA by vacuum UV circular dichroism. Nucleic Acids Res. 1985 Jul 11;13(13):4983–4989. doi: 10.1093/nar/13.13.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel L. J., Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J Biomol Struct Dyn. 1987 Feb;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., Sanderson M. R., van der Marel G. A., van Boom J. H., Altona C. Conformational analysis of the octamer d(G-G-C-C-G-G-C-C) in aqueous solution. A one-dimensional and two-dimensional proton NMR study at 300 MHz and 500 MHz. Eur J Biochem. 1986 Aug 15;159(1):85–93. doi: 10.1111/j.1432-1033.1986.tb09836.x. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of the base sequence on the conformational behaviour of DNA polynucleotides in solution. Eur J Biochem. 1987 Jul 1;166(1):87–101. doi: 10.1111/j.1432-1033.1987.tb13487.x. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]



