Abstract
Methane-oxidizing bacteria are nature’s primary biological mechanism for suppressing atmospheric levels of the second-most important greenhouse gas via methane monooxygenases (MMOs). The copper-containing particulate enzyme is the most widespread and efficient MMO. Under low-copper conditions methane-oxidizing bacteria secrete the small copper-binding peptide methanobactin (mbtin) to acquire copper, but how variations in the structures of mbtins influence copper metabolism and species selection are unknown. Methanobactins have been isolated from Methylocystis strains M and hirsuta CSC1, organisms that can switch to using an iron-containing soluble MMO when copper is limiting, and the nonswitchover Methylocystis rosea. These mbtins are shorter, and have different amino acid compositions, than the characterized mbtin from Methylosinus trichosporium OB3b. A coordinating pyrazinedione ring in the Methylocystis mbtins has little influence on the Cu(I) site structure. The Methylocystis mbtins have a sulfate group that helps stabilize the Cu(I) forms, resulting in affinities of approximately 1021 M-1. The Cu(II) affinities vary over three orders of magnitude with reduction potentials covering approximately 250 mV, which may dictate the mechanism of intracellular copper release. Copper uptake and the switchover from using the iron-containing soluble MMO to the copper-containing particulate enzyme is faster when mediated by the native mbtin, suggesting that the amino acid sequence is important for the interaction of mbtins with receptors. The differences in structures and properties of mbtins, and their influence on copper utilization by methane-oxidizing bacteria, have important implications for the ecology and global function of these environmentally vital organisms.
Copper is an essential protein cofactor involved in many important cellular processes (1, 2), and copper-trafficking systems have been extensively studied (1, 3–8). Although copper uptake by eukaryotes is well defined (1, 4, 9), acquisition of this metal by prokaryotes remains poorly understood. Methane-oxidizing bacteria secrete the small copper-binding molecule methanobactin (mbtin) when copper is limiting (10–18), presumably for sequestration of this metal. These organisms have conditionally high requirements for copper (19), primarily for the active site (20) of the particulate methane monooxygenase (pMMO). Almost all known methane-oxidizing bacteria use pMMO for the consumption of methane (19), an important greenhouse gas. A subclass of “switchover” organisms exists that can also produce a less efficient iron-containing soluble MMO (sMMO) under copper-deficient conditions, with pMMO expression up-regulated in response to an increase in the copper-to-cell ratio (15, 21).
Methanobactin production has been examined in a number of methane-oxidizing bacteria (22–24), but mbtins from only two organisms have been characterized (13, 18). The mbtin (two forms) from Methylosinus trichosporium OB3b (a switchover organism) is the most extensively studied (13, 15–17, 25–29), and binds a single copper ion coordinated in a distorted tetrahedral arrangement by the nitrogens from two oxazolone rings (29) and the sulfurs from two enethiolate groups. The molecule has a compact arrangement stabilized by a disulfide bridge. The very high affinities for copper that have been determined for the M. trichosporium OB3b molecules are consistent with mbtins playing a role in the acquisition of copper (17). Direct evidence of uptake and cytoplasmic localization has recently been obtained for Cu(I)-mbtin from M. trichosporium OB3b (30). These studies confirm that mbtin is the primary component of an active copper-acquisition system in methane-oxidizing bacteria. Comparisons have been made (13, 15, 16) between mbtins and iron-sequestering siderophores (31, 32), particularly the structurally related pyoverdines. Whereas detailed information is available for siderophore-mediated iron uptake and utilization, almost nothing is known about how mbtins acquire and deliver copper.
In this work mbtins have been isolated and characterized from three Methylocystis strains, including switchover and nonswitchover organisms. All of these mbtins have high Cu(I) affinities that are similar to those of the M. trichosporium OB3b mbtins. The N-terminal group present in the Methylocystis mbtins alters the Cu(II) affinity, which will influence acquisition of the metal, and results in different reduction potentials (Em values) that could dictate the copper release mechanism. Variations in the structures of mbtins affect their ability to provide copper to methane-oxidizing bacteria and to initiate the transition from using sMMO to pMMO in switchover organisms. We have identified features of mbtins that influence how methane-oxidizing bacteria uptake and utilize copper that may influence their capacity to suppress methane in the natural environment.
Results and Discussion
Methanobactin Production by Methylocystis Strains.
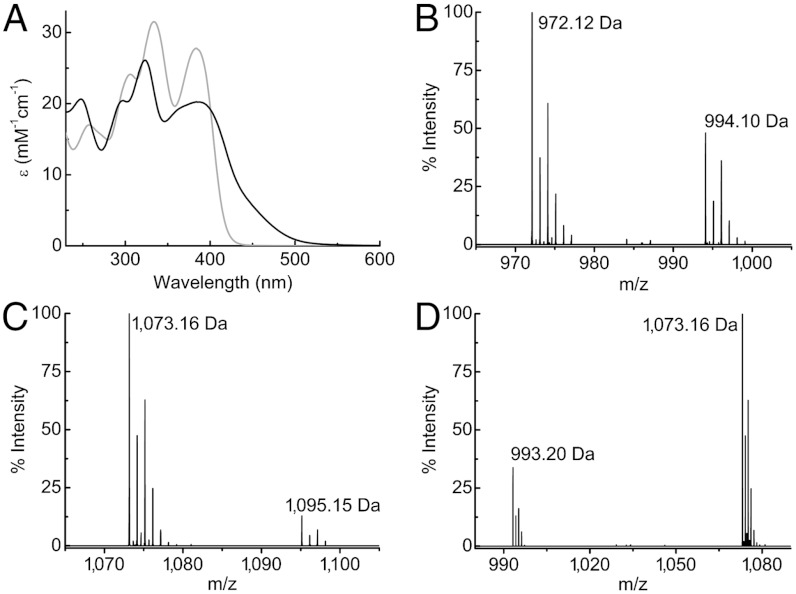
Methanobactins have been isolated (Fig. 1, Fig. S1, Table S1, and SI Text) from Methylocystis strains M (19) and hirsuta CSC1 (33), both switchover organisms, and the nonswitchover Methylocystis rosea (34). Growth under copper-limiting conditions leads to increased amounts of these molecules in the media, consistent with a role in copper acquisition. The two major mbtin forms produced by Methylocystis strains M and hirsuta CSC1 differ by the mass of a Thr residue (101.04 Da, see Fig. 1
B and C and Table S1) and will be referred to as mb (for the full-length molecule) and 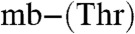 (mb minus a Thr residue) (M. rosea produces more forms, see SI Text). Enzymatic digestion of the mb forms by carboxypeptidase Y shows that this Thr residue is at the C terminus (Fig. S2 A and B). A further modification resulting in a mass difference of 79.96 Da, corresponding to a sulfonate (18) is found by tandem mass spectrometry (MS/MS) (Fig. 1D). Absence of the sulfonate to give the
(mb minus a Thr residue) (M. rosea produces more forms, see SI Text). Enzymatic digestion of the mb forms by carboxypeptidase Y shows that this Thr residue is at the C terminus (Fig. S2 A and B). A further modification resulting in a mass difference of 79.96 Da, corresponding to a sulfonate (18) is found by tandem mass spectrometry (MS/MS) (Fig. 1D). Absence of the sulfonate to give the 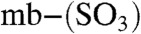 form is also observed in purified extracts of M. hirsuta CSC1 (Fig. S1E and Table S1). We have investigated the stability of the purified Methylocystis mbs under our growth conditions and find no indication that any of these modifications (C-terminal cleavage or loss of the sulfonate) occur in cell-free spent media (Fig. S2 D and E).
form is also observed in purified extracts of M. hirsuta CSC1 (Fig. S1E and Table S1). We have investigated the stability of the purified Methylocystis mbs under our growth conditions and find no indication that any of these modifications (C-terminal cleavage or loss of the sulfonate) occur in cell-free spent media (Fig. S2 D and E).
Fig. 1.
Absorption and mass spectra of the Methylocystis mbtins. (A) UV-visible spectra (25 °C) of M. hirsuta CSC1 apo-mb (gray) and Cu(I)-mb (black) in 20 mM Hepes at pH 7.5. Mass spectra (negative ionization mode) of Methylocystis strain M Cu(I)-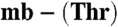 (B), Cu(I)-mb (C), and of the tandem MS analysis of Cu(I)-mb showing the loss of 79.96 Da (sulfonate group) from the main peak (D). In B and C the peaks at 994.10 and 1,095.15 Da are the sodium adducts of the main peaks at 972.12 and 1,073.16 Da (both [M + Cu - 2H]-), respectively.
(B), Cu(I)-mb (C), and of the tandem MS analysis of Cu(I)-mb showing the loss of 79.96 Da (sulfonate group) from the main peak (D). In B and C the peaks at 994.10 and 1,095.15 Da are the sodium adducts of the main peaks at 972.12 and 1,073.16 Da (both [M + Cu - 2H]-), respectively.
A High Affinity for Cu(I) Is a Common Feature of Methanobactins.
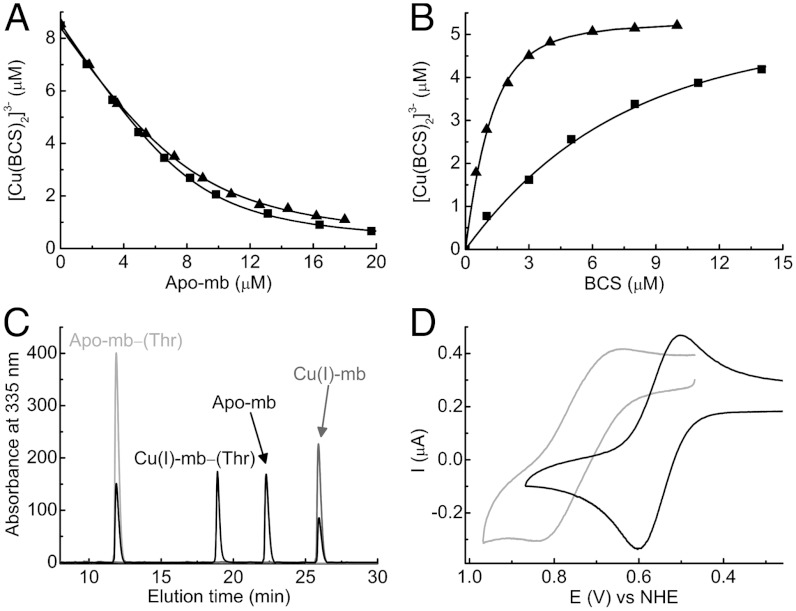
The Cu(I) affinities (Kb values) of the Methylocystis mbtins have been determined from competition titrations with the chromophoric copper chelator bathocuproine disulfonate (BCS) (Fig. 2 A and B) and also using an mbtin with a Cu(I) affinity that already has been determined (Fig. 2C). The composition of mbtins differ (see below), but this has a limited effect on the Cu(I) affinities, which are typically in the 1021 M-1 range (Table S2). The absence of the sulfonate group in the M. hirsuta CSC1 mbtins decreases the Cu(I) affinity by more than an order of magnitude (Fig. 2B and Table S2), similar to the effect of reduction of the disulfide bond in M. trichosporium OB3b mbtins (17). A very high affinity for Cu(I) is a distinctive feature of this class of copper-binding molecule.
Fig. 2.
Determination of Cu(I) affinities and Em values of the Methylocystis mbtins. (A) Titrations of apo-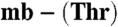 from Methylocystis strains M (squares) and hirsuta CSC1 (triangles) into a mixture of BCS (5 mM) and Cu(I) (8.6 and 8.4 μM respectively). In (B) titrations of BCS into Cu(I)-mb (5.3 μM, squares) and Cu(I)-
from Methylocystis strains M (squares) and hirsuta CSC1 (triangles) into a mixture of BCS (5 mM) and Cu(I) (8.6 and 8.4 μM respectively). In (B) titrations of BCS into Cu(I)-mb (5.3 μM, squares) and Cu(I)-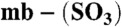 (5.3 μM, triangles), both from M. hirsuta CSC1, are shown. Titrations were performed at 25 °C in 20 mM Hepes pH 7.5 plus 200 mM NaCl. The lines are fits of the data to equation 2 in ref. 17 and give Kb values respectively of (1.5 ± 0.1) × 1021 and (1.0 ± 0.1) × 1021 M-1 (A) and (7.4 ± 0.2) × 1020 and (2.2 ± 0.1) × 1019 M-1 (B). (C) An overlay of analytical HPLC profiles obtained from the injection of a mixture of Methylocystis strain M apo-
(5.3 μM, triangles), both from M. hirsuta CSC1, are shown. Titrations were performed at 25 °C in 20 mM Hepes pH 7.5 plus 200 mM NaCl. The lines are fits of the data to equation 2 in ref. 17 and give Kb values respectively of (1.5 ± 0.1) × 1021 and (1.0 ± 0.1) × 1021 M-1 (A) and (7.4 ± 0.2) × 1020 and (2.2 ± 0.1) × 1019 M-1 (B). (C) An overlay of analytical HPLC profiles obtained from the injection of a mixture of Methylocystis strain M apo-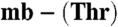 (40 μM, light gray), M. trichosporium OB3b Cu(I)-mb (40 μM, dark gray), and a mixture of the two mbtins (both 40 μM, black) incubated for 6 h at 25 °C in 20 mM Hepes pH 7.5. Copper [(24 ± 1) μM] has transferred between the two mbtins allowing a Kb value of (1.6 ± 0.3) × 1021 M-1 to be obtained for Methylocystis strain M
(40 μM, light gray), M. trichosporium OB3b Cu(I)-mb (40 μM, dark gray), and a mixture of the two mbtins (both 40 μM, black) incubated for 6 h at 25 °C in 20 mM Hepes pH 7.5. Copper [(24 ± 1) μM] has transferred between the two mbtins allowing a Kb value of (1.6 ± 0.3) × 1021 M-1 to be obtained for Methylocystis strain M 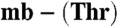 using Eq. 1. (D) CVs (24 ± 2 °C) of the copper-loaded
using Eq. 1. (D) CVs (24 ± 2 °C) of the copper-loaded 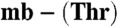 from Methylocystis strains M (gray) and hirsuta CSC1 (black) at a scan rate of 20 mV/s in 20 mM Hepes pH 7.5 plus 90 mM NaCl.
from Methylocystis strains M (gray) and hirsuta CSC1 (black) at a scan rate of 20 mV/s in 20 mM Hepes pH 7.5 plus 90 mM NaCl.
Methanobactins Exhibit a Range of Reduction Potentials.
The Em values of Methylocystis (copper-loaded) mbtins have been measured by cyclic voltammetry (CV) (Fig. 2D and Fig. S3 A and B) and range from 483 mV for M. hirsuta CSC1 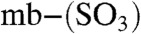 to 745 mV for M. rosea
to 745 mV for M. rosea
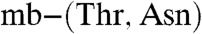 (Table S2) [Methylocystis strain M
(Table S2) [Methylocystis strain M 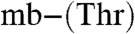 has an Em of 740 mV]. The mbtins from M. trichosporium OB3b have Em values around 600 mV (Table S2). These Em values, and particularly those of the Methylocystis strains M and rosea mbtins, are consistent with a strong preference for Cu(I) over Cu(II) (Table S2). The Em differences are primarily due to Cu(II) affinities that are approximately three orders of magnitude higher in the M. hirsuta CSC1 mbtins, compared to the Methylocystis strains M and rosea molecules (Table S2). Such variations in copper affinity can have significant physiological consequences (6–8). The Em value [and Cu(II) affinity] of an mbtin does not appear to correlate with the ability of that organism to switch over, as Methylocystis strain M (switchover) and M. rosea (nonswitchover) both produce mbtins with similarly high Em values.
has an Em of 740 mV]. The mbtins from M. trichosporium OB3b have Em values around 600 mV (Table S2). These Em values, and particularly those of the Methylocystis strains M and rosea mbtins, are consistent with a strong preference for Cu(I) over Cu(II) (Table S2). The Em differences are primarily due to Cu(II) affinities that are approximately three orders of magnitude higher in the M. hirsuta CSC1 mbtins, compared to the Methylocystis strains M and rosea molecules (Table S2). Such variations in copper affinity can have significant physiological consequences (6–8). The Em value [and Cu(II) affinity] of an mbtin does not appear to correlate with the ability of that organism to switch over, as Methylocystis strain M (switchover) and M. rosea (nonswitchover) both produce mbtins with similarly high Em values.
The Crystal Structures of the Methylocystis Methanobactins.
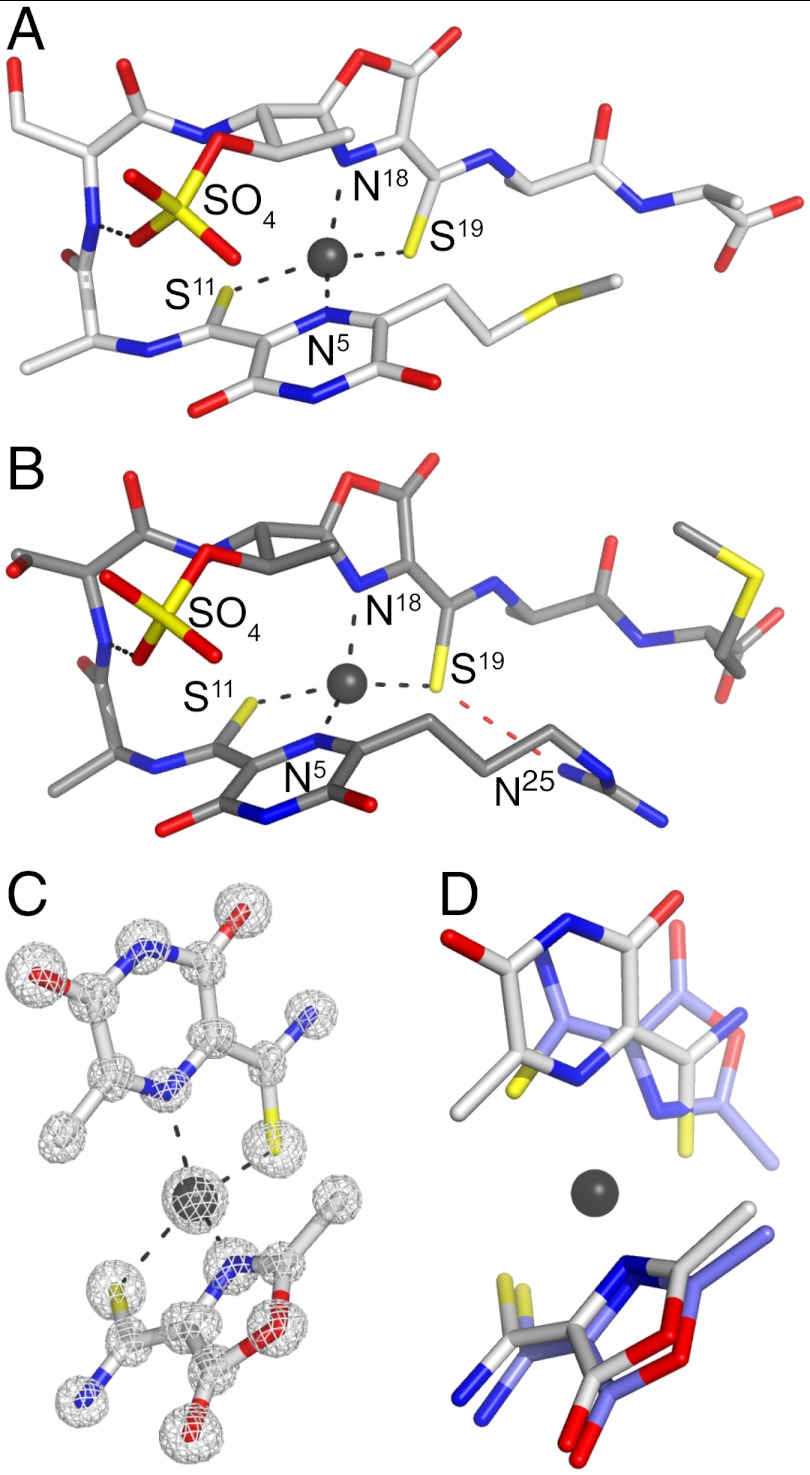
Attempts to crystallize M. hirsuta CSC1 Cu(I)-mb were unsuccessful, and very low yields of the full-length Methylocystis strain M mbtin precluded crystallization trials. However, we have been able to crystallize Cu(I)-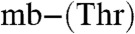 from Methylocystis strains M and hirsuta CSC1 (Table S3 and SI Text) (M. rosea mbtins were not crystallized due to their similarity to the mbtins from Methylocystis strain M). Both molecules possess very similar (root mean square deviation for the Cα atoms of 0.51 Å) hairpin-like structures (Fig. 3
A and B), which are very different from the more compact arrangement of the M. trichosporium OB3b mbtins (13, 17). NMR studies have indicated that the mbtin from Methylocystis SB2 (18) has a comparable overall fold to the structures we have determined. The Methylocystis mbtins (including M. rosea, SI Text) have similar peptide backbones composed of only four standard amino acids (Ala1, Ser2, Ala3, and either Met4 or Ala4 in Methylocystis strains M and hirsuta CSC1 mbtins, respectively) and two modified residues that include oxazolone and pyrazinedione rings (Fig. 3
A–C, Fig. S4, and SI Text). The pyrazinedione ring in the Methylocystis mbtins studied here replaces one of the oxazolone rings of the M. trichosporium OB3b molecules (an imidazole has been suggested in Methylocystis SB2 mbtin; ref. 18). Pyrazinediones have been found in very few naturally produced compounds, with the only examples being small molecules from certain fungi (35), and its involvement in coordinating a metal is, as far as we are aware, unprecedented. The structurally characterized mbtins have Cu(I) sites coordinated by a N2S2 ligand set with very similar distorted tetrahedral geometries (Fig. 3 and Table S4), although the C-terminal sulfur and nitrogen ligands in the Methylocystis mbtins are switched compared to their positions in the M. trichosporium OB3b molecules (Fig. 3D).
from Methylocystis strains M and hirsuta CSC1 (Table S3 and SI Text) (M. rosea mbtins were not crystallized due to their similarity to the mbtins from Methylocystis strain M). Both molecules possess very similar (root mean square deviation for the Cα atoms of 0.51 Å) hairpin-like structures (Fig. 3
A and B), which are very different from the more compact arrangement of the M. trichosporium OB3b mbtins (13, 17). NMR studies have indicated that the mbtin from Methylocystis SB2 (18) has a comparable overall fold to the structures we have determined. The Methylocystis mbtins (including M. rosea, SI Text) have similar peptide backbones composed of only four standard amino acids (Ala1, Ser2, Ala3, and either Met4 or Ala4 in Methylocystis strains M and hirsuta CSC1 mbtins, respectively) and two modified residues that include oxazolone and pyrazinedione rings (Fig. 3
A–C, Fig. S4, and SI Text). The pyrazinedione ring in the Methylocystis mbtins studied here replaces one of the oxazolone rings of the M. trichosporium OB3b molecules (an imidazole has been suggested in Methylocystis SB2 mbtin; ref. 18). Pyrazinediones have been found in very few naturally produced compounds, with the only examples being small molecules from certain fungi (35), and its involvement in coordinating a metal is, as far as we are aware, unprecedented. The structurally characterized mbtins have Cu(I) sites coordinated by a N2S2 ligand set with very similar distorted tetrahedral geometries (Fig. 3 and Table S4), although the C-terminal sulfur and nitrogen ligands in the Methylocystis mbtins are switched compared to their positions in the M. trichosporium OB3b molecules (Fig. 3D).
Fig. 3.
The crystal structures of the Methylocystis mbtins. The overall structures of Cu(I)-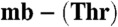 from M. hirsuta CSC1 (A) and Methylocystis strain M (B) with the hydrogen-bonding interaction made by the sulfate group and that between the S19 and N25 atoms (dashed red line, 3.2 Å) in the Methylocystis strain M molecule shown, and the copper ions represented as gray spheres. Chemical structures of the Methylocystis mbtins are shown in Fig. S4. (C) The Cu(I) binding site of M. hirsuta CSC1
from M. hirsuta CSC1 (A) and Methylocystis strain M (B) with the hydrogen-bonding interaction made by the sulfate group and that between the S19 and N25 atoms (dashed red line, 3.2 Å) in the Methylocystis strain M molecule shown, and the copper ions represented as gray spheres. Chemical structures of the Methylocystis mbtins are shown in Fig. S4. (C) The Cu(I) binding site of M. hirsuta CSC1 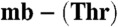 including the 2|Fobs| - |Fcalc| electron-density map calculated using the phases from the final model contoured at 2σ. (D) A superimposition of the copper sites of M. hirsuta CSC1 Cu(I)-
including the 2|Fobs| - |Fcalc| electron-density map calculated using the phases from the final model contoured at 2σ. (D) A superimposition of the copper sites of M. hirsuta CSC1 Cu(I)-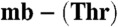 (light gray) and M. trichosporium OB3b Cu(I)-mb (slate blue, Protein Data Bank code 2xjh).
(light gray) and M. trichosporium OB3b Cu(I)-mb (slate blue, Protein Data Bank code 2xjh).
The sulfate group in the Methylocystis mbtins is attached to a Thr side chain (O-sulfonation), and makes stabilizing interactions (Fig. 3 A and B). A hydrogen bond is formed with the backbone amide of Ser2, and a π-anion interaction (36–39) occurs between the sulfate and the distal edge of the pyrazinedione ring. This interaction will be favored by copper coordination that will decrease electron density on the aromatic ring (36). The absence of the sulfonate group affects the affinity for Cu(I) more than that for Cu(II) and results in 40–50 mV decreases in Em (Table S2). In siderophores from marine bacteria, sulfonation of the Fe(III)-binding catechol rings has been observed, and it has been suggested that these forms may represent a distinct class of siderophores (31).
The large variation in Em between the Methylocystis strains M and hirsuta CSC1 mbtins is not due to Cu(I) site differences, as these have remarkably similar structures (Fig. 3 A and B and Table S4) that are equally solvent exposed. The main determinant of the Em is a second-coordination sphere change caused by alterations at the N termini (Fig. S4). The positively charged guanidine group in the mbtin from Methylocystis strain M (and presumably also M. rosea mbtin) hydrogen bonds via N25 to the coordinating S19 atom (Fig. 3B). This interaction is absent in the M. hirsuta CSC1 mbtins that have a thioether moiety at the N terminus (Fig. 3A). This hydrogen bond will decrease electron density on the coordinating sulfur, disfavoring the Cu(II) form over the Cu(I) form and increasing the Em value [this interaction can also be rationalized electrostatically, with the positively charged guanidine group destabilizing Cu(II)] (40). Because an N-terminal guanidine group is present in the Methylocystis SB2 mbtin (18) we would predict a high Em and a low Cu(II) affinity. It has been suggested that Cu(I)-mbtin acts as an electron donor to pMMO (27, 41, 42). If this is the case, alterations in the Em values of mbtins will have functional relevance in optimizing the rate of electron transfer to pMMO. Methanobactins are probably sufficiently flexible to allow structural changes upon redox interconversion, unlike copper sites that are involved in biological electron transfer (43, 44), which will hinder such reactivity. The observed Em differences of mbtins are therefore probably a consequence of modifications made primarily to alter the affinity for Cu(II).
Methanobactin-Mediated Copper Acquisition.
The large differences in the Cu(II) affinities of mbtins will influence their relative ability to sequester copper from natural sources, which will have functional relevance for methane-oxidizing bacteria, particularly in mixed populations. The mbtin from M. trichosporium OB3b can be internalized (30), consistent with this molecule being the initial source of copper for trafficking pathways to cellular destinations (mbtins may also help diminish copper toxicity, Fig. S5 and SI Text). It is not clear if the membranes that harbor pMMO are contiguous with the cytoplasmic membrane, and therefore whether pMMO may provide a rare example of a bacterial cytoplasmic protein that requires copper. The removal of copper from mbtins is not favored due to their high affinities for Cu(I). This may prevent the copper metallochaperone CopZ and the copper transporting ATPase CopA, which are involved in copper efflux from the prokaryotic cytosol (both are present in methane-oxidizing bacteria; refs. 45–47), from acquiring Cu(I) directly from mbtins as their affinities are at least three orders of magnitude weaker (8, 48–52). Homologues of the copper-dependent transcriptional activator CueR (53) are also present in methane-oxidizing bacteria (45–47) and probably have Cu(I) affinities closer to those of mbtins (53), making copper transfer to CueR thermodynamically more favorable than transfer to a metallochaperone.
TonB-dependent transporters (45–47) appear to be responsible for the active uptake of mbtin (30). Analogous systems are responsible for siderophore uptake in bacteria (31, 32, 54, 55), and in the case of the mbtin-like pyoverdines, specificity for the host receptor is achieved by the peptidic part of the siderophore (56, 57). To gain insight into the physiological function of mbtins, we investigated the uptake of different Cu(I)-mbtins, and their ability to promote switchover from sMMO to pMMO in M. trichosporium OB3b and M. hirsuta CSC1 cells (Fig. 4). Nonnative Cu(I)-mbtins are internalized by both organisms, and the absence of the apo-form of the added mbtin in the media during these experiments demonstrates that the endogenous mbtin (also not observed) is not involved in this process (Fig. 4A). Copper accumulation, and switchover from sMMO to pMMO, however, is more efficient with the native Cu(I)-mbtin (Fig. 4
B and C). Furthermore, copper uptake by M. hirsuta CSC1 appears to be slower with the native Cu(I)-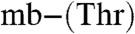 compared to the native full-length form (Fig. 4C), although sMMO activity decreases are similar (Fig. 4C and Fig. S5B). Shortened forms of both the Methylocystis and the M. trichosporium OB3b mbtins (17) are able to promote physiological responses. Cu(I)-mb from M. hirsuta CSC1 and M. trichosporium OB3b have very similar Cu(I) affinities yet mediate copper uptake and switchover at different rates, and we therefore propose that the amino acid sequence of a mbtin must be important for the interaction with the receptor. Copper-regulated transition from using sMMO to pMMO is not fully understood (15, 21). The observation that the Methylocystis strains M (switchover) and rosea (nonswitchover) mbtins have similar properties and structures would suggest that Cu-mbtin is not directly involved in the switchover mechanism.
compared to the native full-length form (Fig. 4C), although sMMO activity decreases are similar (Fig. 4C and Fig. S5B). Shortened forms of both the Methylocystis and the M. trichosporium OB3b mbtins (17) are able to promote physiological responses. Cu(I)-mb from M. hirsuta CSC1 and M. trichosporium OB3b have very similar Cu(I) affinities yet mediate copper uptake and switchover at different rates, and we therefore propose that the amino acid sequence of a mbtin must be important for the interaction with the receptor. Copper-regulated transition from using sMMO to pMMO is not fully understood (15, 21). The observation that the Methylocystis strains M (switchover) and rosea (nonswitchover) mbtins have similar properties and structures would suggest that Cu-mbtin is not directly involved in the switchover mechanism.
Fig. 4.
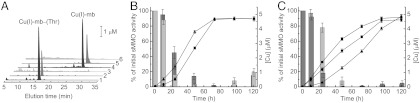
Methanobactin-mediated copper uptake in methane-oxidizing bacteria. (A) Analytical HPLC analysis of the extracellular medium of M. trichosporium OB3b (lines 1–3) and M. hirsuta CSC1 (lines 4–6) cells immediately after the addition of M. hirsuta CSC1 Cu(I)-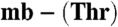 (line 1) and M. trichosporium OB3b Cu(I)-mb (line 4), and after incubation for 48 and 96 h (lines 2 and 3, respectively) and 72 and 120 h (lines 5 and 6, respectively). Copper uptake by (lines) and sMMO activity of (bars) M. trichosporium OB3b (B), and M. hirsuta CSC1 (C) cells after the addition of M. trichosporium OB3b Cu(I)-mb (5 μM, triangles and light gray bars) and M. hirsuta CSC1 Cu(I)-
(line 1) and M. trichosporium OB3b Cu(I)-mb (line 4), and after incubation for 48 and 96 h (lines 2 and 3, respectively) and 72 and 120 h (lines 5 and 6, respectively). Copper uptake by (lines) and sMMO activity of (bars) M. trichosporium OB3b (B), and M. hirsuta CSC1 (C) cells after the addition of M. trichosporium OB3b Cu(I)-mb (5 μM, triangles and light gray bars) and M. hirsuta CSC1 Cu(I)-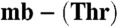 (5 μM, circles and dark gray bars). Also included in C is copper uptake after the addition of the M. hirsuta CSC1 Cu(I)-mb to M. hirsuta CSC1 cells (5 μM, squares) [the sMMO activities in this experiment (Fig. S5B) are the same within error as those obtained with Cu(I)-
(5 μM, circles and dark gray bars). Also included in C is copper uptake after the addition of the M. hirsuta CSC1 Cu(I)-mb to M. hirsuta CSC1 cells (5 μM, squares) [the sMMO activities in this experiment (Fig. S5B) are the same within error as those obtained with Cu(I)-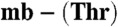 ].
].
The utilization of mbtin-bound copper, either by pMMO, other copper enzymes, or regulatory systems presumably requires copper removal, which our data indicate are thermodynamically unfavorable (release will also be kinetically hindered by the relatively buried nature of the coordinatively saturated metal site). The variation in Em values of mbtins, along with a comparison to iron removal from siderophores, provides insight into possible copper release mechanisms. The low Em value of enterobactin (-750 mV at pH 7; ref. 58), due to a strong preference for Fe(III) (Kb of 1049 M-1; ref. 59), is below that accessible by physiological reductants making release by conversion to Fe(II) alone impossible and such siderophores are enzymatically hydrolyzed prior to reduction (57, 58, 60). Iron release via reduction of the metal only (57, 58, 61) occurs for siderophores that possess higher Em values, for example ferrichrome A (Em approximately 300 mV higher than that of enterobactin; ref. 58). The mechanism of copper removal from mbtins may also depend on their Em value. Cu(I)-mbtins with lower Em values (approximately 500 mV), such as that from M. hirsuta CSC1, will be more readily converted to the Cu(II) forms by physiological oxidants, which would assist release as the affinity for Cu(II) is much lower than that for Cu(I). Cu(I)-mbtins with higher Em values (approximately 750 mV) may require enzymatic degradation to facilitate removal of the metal.
In summary, mbtins from switchover and nonswitchover Methylocystis strains have been characterized in detail and the features of these molecules that influence mbtin-mediated copper acquisition by methane-oxidizing bacteria identified. Although involving different modified amino acids, the ligand set and geometry of the copper sites of mbtins are highly conserved, as are their high affinities for Cu(I). The differences in Em values of mbtins most likely reflect alterations in their ability to sequester Cu(II) from natural sources that may influence in situ species selection and the intracellular copper release mechanism. We propose that the amino acid composition of mbtins plays an important role in their recognition and uptake. This analysis of the variations in the properties of mbtins and their physiological impact provides insights into the only known prokaryotic copper-acquisition system and has importance for understanding greenhouse gas suppression by these environmentally critical organisms.
Materials and Methods
Isolation and Purification of Methanobactins.
For mbtin isolation methane-oxidizing bacteria were grown either in the presence (1 μM) or absence of copper (17). Purification of crude extracts obtained after passing extracellular media through C18 cartridges (Sep-Pak Plus; Waters) was performed by reverse-phase HPLC using a previously described procedure (17) with some modifications (SI Text). The purity of the isolated mbtins was verified by analytical HPLC (SI Text). In the case of Methylocystis strain M, many cycles of growth were required before any mbtin could be detected in the media, and yields were considerably lower than those for all other mbtins.
Characterization of Methanobactins.
Mass determinations were performed by electrospray ionization mass spectrometry (17). UV-visible spectra (25 °C) were acquired on a PerkinElmer λ35 spectrophotometer, typically using sealed quartz cuvettes. Copper concentrations were measured by atomic absorption spectrometry and were used to determine molar absorption coefficients (ε values) of Cu(I)-mbtins (17). Concentrations of apo-mbtins were obtained from titrations with Cu(I) in the presence of bicinchoninic acid as described previously (17). Enzymatic digestion (22 °C) with carboxypeptidase Y from Saccharomyces cerevisiae (Sigma; EC 3.4.16.1) was carried out in 10 mM ammonium acetate pH 7 and the mixture analyzed by analytical HPLC (SI Text). The stability of mbtins in cell-free spent media was assessed as described previously (17).
Cu(I) Affinity Determinations.
Cu(I) affinities (Kb values) were mainly determined (25 °C) by competition experiments with BCS in 20 mM Hepes pH 7.5 plus 200 mM NaCl (17). The Cu(I) affinities of certain mbtins were measured (24 ± 2 °C) by incubating the apo-form of one mbtin and the copper form of a second mbtin for at least 6 h. Prior to HPLC analysis of this mixture, 10 equivalents of ethylenediaminetetraacetic acid were added. Peaks were integrated and compared to mbtin alone, run under the same conditions. The Cu(I) affinity was calculated using Eq. 1;
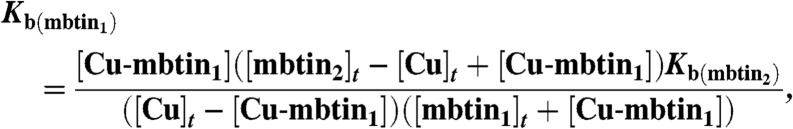 |
[1] |
where mbtin1 is the mbtin for which the affinity is being determined and mbtin2 is the mbtin of known Cu(I) affinity. [Cu]t and [mbtin]t represent the total concentrations of Cu(I) and mbtin (either 1 or 2), respectively, and [Cu-mbtin1] represents the concentration of the copper-bound form of mbtin1.
Electrochemistry.
The direct measurement of the Em values of the copper-loaded mbtins (approximately 500 μM) was carried out by CV as described previously (ref. 17 and SI Text). Measurements (24 ± 2 °C) were performed in 20 mM Hepes at pH 7.5 plus 90 mM NaCl typically at a scan rate of 20 mV/s. Em values are quoted referenced to the normal hydrogen electrode.
Crystallization and Structure Determinations.
Crystallization conditions are described in the SI Text along with data collection and refinement statistics (Table S3). Briefly, X-ray diffraction data were collected at the IO2 beamline at the Diamond Light Source. The datasets were integrated using XDS (62) and scaled with SCALA (63). The phases were solved by single-wavelength anomalous dispersion using the signal from copper with SHELXC/D/E pipeline (64). COOT (65) was used for model building and SHELXL (66) for refinement. The atomic coordinates have been deposited in the Protein Data Bank [accession codes 2ygj and 2ygi for Methylocystis strains M and hirsuta CSC1 Cu(I)-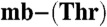 , respectively].
, respectively].
Physiological Activity of Methanobactins.
Growth, sMMO activity, and copper uptake by M. hirsuta CSC1 or M. trichosporium OB3b cells to which Cu(I)-mb, Cu(I)-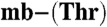 , or CuSO4 was added (5 μM) were monitored over 120 h as described in the SI Text.
, or CuSO4 was added (5 μM) were monitored over 120 h as described in the SI Text.
Supplementary Material
Acknowledgments.
We thank the staff of the Diamond Light Source synchrotron radiation source, Prof. Hiroo Uchiyama (University of Tsukuba) for providing Methylocystis strain M, Dr. Helen Talbot for preliminary LC-MS analyses, Drs. Adrianna Badarau, Kevin Waldron, and Charles Knapp for useful discussions, Semeli Platsaki for help with copper uptake studies, and a reviewer for suggesting the in vivo experiments. This work was funded by Natural Environment Research Council Grant NE/F00608X.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PBD ID codes 2ygj and 2ygi for Methylocystis strains M and hirsuta CSC1 Cu(I)-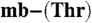 , respectively).
, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112921109/-/DCSupplemental.
References
- 1.Kim B, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 2.Turski ML, Thiele DJ. New roles for copper metabolism in cell proliferation, signaling, and disease. J Biol Chem. 2009;284:717–721. doi: 10.1074/jbc.R800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huffman DL, O’Halloran TV. Function, structure, and mechanism of intracellular copper trafficking proteins. Annu Rev Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 4.Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 5.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banci L, et al. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 8.Badarau A, Dennison C. Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803. Proc Natl Acad Sci USA. 2011;108:13007–13012. doi: 10.1073/pnas.1101448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Feo CJ, Aller SG, Unger VM. A structural perspective on copper uptake in eukaryotes. Biometals. 2007;20:705–716. doi: 10.1007/s10534-006-9054-7. [DOI] [PubMed] [Google Scholar]
- 10.Fitch MW, et al. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1993;59:2771–2776. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiSpirito AA, et al. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol. 1998;180:3606–3613. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Téllez CM, et al. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl Environ Microbiol. 1998;64:1115–1122. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HJ, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 14.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci USA. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 16.Balasubramanian R, Rosenzweig AC. Copper methanobactin: A molecule whose time has come. Curr Opin Chem Biol. 2008;12:245–249. doi: 10.1016/j.cbpa.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Ghazouani A, et al. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg Chem. 2011;50:1378–1391. doi: 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 18.Krentz BD, et al. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry. 2010;49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramanian R, et al. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murrell JC, McDonald IR, Gilbert B. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 2000;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 22.Zahn JA, DiSpirito AA. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon S, Kraemer SM, DiSpirito AA, Semrau JD. An assay for screening microbial cultures for chalkophore production. Environ Microbiol Rep. 2010;2:295–303. doi: 10.1111/j.1758-2229.2009.00125.x. [DOI] [PubMed] [Google Scholar]
- 24.Choi DW, et al. Spectral and thermodynamic properties of methanobactin from γ-proteobacterial methane oxidizing bacteria: A case for copper competition on a molecular level. J Inorg Biochem. 2010;104:1240–1247. doi: 10.1016/j.jinorgbio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, et al. Purification and physical-chemical properties of methanobactin: A chalkophore from Methylosinus trichosporium OB3b. Biochemistry. 2005;44:5140–5148. doi: 10.1021/bi047367r. [DOI] [PubMed] [Google Scholar]
- 26.Hakemian AS, et al. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I) J Am Chem Soc. 2005;127:17142–17143. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi DW, et al. Effect of methanobactin on the activity and electron paramagnetic resonance spectra of the membrane-associated methane monooxygenase in Methylococcus capsulatus Bath. Microbiology. 2005;151:3417–3426. doi: 10.1099/mic.0.28169-0. [DOI] [PubMed] [Google Scholar]
- 28.Choi DW, et al. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45:1442–1453. doi: 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 29.Behling LA, et al. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J Am Chem Soc. 2008;130:12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. J Biol Chem. 2011;286:37313–37319. doi: 10.1074/jbc.M111.284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandy M, Butler A. Microbial iron acquisition: Marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 33.Lindner AS, et al. Methylocystis hirsuta sp nov., a novel methanotroph isolated from a groundwater aquifer. Int J Syst Evol Microbiol. 2007;57:1891–1900. doi: 10.1099/ijs.0.64541-0. [DOI] [PubMed] [Google Scholar]
- 34.Wartiainen I, Grethe Hestnes A, McDonald IR, Svenning MM. Methylocystis rosea sp nov., a novel methanotrophic bacterium from Arctic wetland soil, Svalbard, Norway (78°N) Int J Syst Evol Microbiol. 2006;56:541–547. doi: 10.1099/ijs.0.63912-0. [DOI] [PubMed] [Google Scholar]
- 35.Savard ME, Melzer MS, Boland GJ, Bensimon C, Blackwell BA. A new 1-hydroxy-2,6-pyrazinedione associated with hypovirulent isolates of Sclerotinia minor. J Nat Prod. 2003;66:306–309. doi: 10.1021/np020445w. [DOI] [PubMed] [Google Scholar]
- 36.de Hoog P, Gamez P, Mutikainen I, Turpeinen U, Reedijk J. An aromatic anion receptor: Anion-π interactions do exist. Angew Chem Int Ed Engl. 2004;43:5815–5817. doi: 10.1002/anie.200460486. [DOI] [PubMed] [Google Scholar]
- 37.Jackson MR, et al. A preference for edgewise interactions between aromatic rings and carboxylate anions: The biological relevance of anion-quadrupole interactions. J Phys Chem B. 2007;111:8242–8249. doi: 10.1021/jp0661995. [DOI] [PubMed] [Google Scholar]
- 38.Schneider H. Binding mechanisms in supramolecular complexes. Angew Chem Int Ed Engl. 2009;48:3924–3977. doi: 10.1002/anie.200802947. [DOI] [PubMed] [Google Scholar]
- 39.Salonen LM, Ellermann M, Diederich F. Aromatic rings in chemical and biological recognition: Energetics and structures. Angew Chem Int Ed Engl. 2011;50:4808–4842. doi: 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- 40.Yanagisawa S, Banfield MJ, Dennison C. The role of hydrogen bonding at the active site of a cupredoxin: The Phe114Pro azurin variant. Biochemistry. 2006;45:8812–8822. doi: 10.1021/bi0606851. [DOI] [PubMed] [Google Scholar]
- 41.Choi DW, et al. Oxidase, superoxide dismutase, and hydrogen peroxide reductase activities of methanobactin from types I and II methanotrophs. J Inorg Biochem. 2008;102:1571–1580. doi: 10.1016/j.jinorgbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 43.Gray HB, Malmström BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 44.Dennison C. Investigating the structure and function of cupredoxins. Coord Chem Rev. 2005;249:3025–3054. [Google Scholar]
- 45.Ward N, et al. Genomic insights into methanotrophy: The complete genome sequence of Methylococcus capsulatus (Bath) PloS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein LY, et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J Bacteriol. 2010;192:6497–6498. doi: 10.1128/JB.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein LY, et al. Genome sequence of the methanotrophic alphaproteobacterium Methylocystis sp strain Rockwell (ATCC 49242) J Bacteriol. 2011;193:2668–2669. doi: 10.1128/JB.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Singleton C, Le Brun NE. High Cu(I) and low proton affinities of the CXXC motif of Bacillus subtilis CopZ. Biochem J. 2008;413:459–465. doi: 10.1042/BJ20080467. [DOI] [PubMed] [Google Scholar]
- 49.Xiao Z, Loughlin F, George GN, Howlett GJ, Wedd AG. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: Sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J Am Chem Soc. 2004;126:3081–3090. doi: 10.1021/ja0390350. [DOI] [PubMed] [Google Scholar]
- 50.Xiao Z, Wedd AG. The challenges of determining metal-protein affinities. Nat Prod Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 51.Badarau A, Dennison C. Copper trafficking mechanism of CXXC-containing domains: Insight from the pH-dependence of their Cu(I) affinities. J Am Chem Soc. 2011;133:2983–2988. doi: 10.1021/ja1091547. [DOI] [PubMed] [Google Scholar]
- 52.Xiao Z, et al. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: Detection probes and affinity standards. J Biol Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Changela A, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 54.Faraldo-Gómez JD, Sansom MS. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 55.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer JM. Pyoverdines: Pigments, siderophores and potential taxonomic markers of fluorescent Pseudomomas species. Arch Microbiol. 2000;174:135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 57.Schalk I. Metal trafficking via siderophores in gram-negative bacteria: Specificities and characteristics of the pyoverdine pathway. J Inorg Biochem. 2008;102:1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 58.Cooper SR, McArdle JV, Raymond KN. Siderophore electrochemistry: Relation to intracellular iron release mechanism. Proc Natl Acad Sci USA. 1978;75:3551–3554. doi: 10.1073/pnas.75.8.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loomis LD, Raymond KN. Solution equilibria of enterobactin and metal-enterobactin complexes. Inorg Chem. 1991;30:906–911. [Google Scholar]
- 60.Langman L, Young IG, Frost GE, Rosenberg H, Gibson F. Enterochelin system of iron transport in Escherichia coli: Mutations affecting ferric-enterochelin esterase. J Bacteriol. 1972;112:1142–1149. doi: 10.1128/jb.112.3.1142-1149.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matzanke BF, Anemüller S, Schünemann V, Trautwein AX, Hantke K. FhuF, part of a siderophore-reductase system. Biochemistry. 2004;43:1386–1392. doi: 10.1021/bi0357661. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 64.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheldrick GM, Schneider TR. SHELXL: High-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.