Abstract
During immune-mediated death, death-inducing granzyme (Gzm) proteases concentrate in the nucleus of cells targeted for immune elimination, suggesting that nuclear processes are important targets. Here we used differential 2D proteomics of GzmA-treated nuclei to identify potential GzmA substrates. Of 44 candidates, 33 were RNA-binding proteins important in posttranscriptional RNA processing, including 14 heterogeneous nuclear ribonucleoproteins (hnRNP). Multiple hnRNPs were degraded in cells undergoing GzmA-, GzmB-, or caspase-mediated death. GzmA and caspase activation impaired nuclear export of newly synthesized RNA and disrupted pre-mRNA splicing. Expressing GzmA-resistant hnRNP A1 inhibited GzmA-mediated cell death and rescued pre-mRNA splicing, suggesting that hnRNP A1 is an important GzmA substrate. Cellular stresses are known to inhibit initiation of cap-dependent translation. Disrupting pre-mRNA processing should block further new protein synthesis and promote death by interfering with pathways induced to protect cells from death.
Keywords: apoptosis, cytotoxic T lymphocyte
Killer lymphocytes deploy cytotoxic granule serine proteases to activate programmed cell death in cells targeted for immune elimination (1). Humans express five granzymes (Gzms), of which GzmA and GzmB are the most abundant and best characterized. GzmB activates the caspases and also directly cleaves some caspase substrates (2). GzmA induces caspase-independent programmed cell death, characterized by ssDNA damage and a unique pathway of mitochondrial damage without mitochondrial outer membrane permeabilization (3). GzmA and GzmB share few substrates. GzmA, a homodimer with tryptase activity, binds its substrates through an extended exosite and does not recognize a predictable cleavage site peptide (4). Thus, in silico methods cannot predict its substrates.
A key mitochondrial substrate was identified by proteomics of GzmA-treated mitochondria (5). This approach succeeded because a minimal Gzm concentration and incubation time were used to treat intact organelles in which potential substrates are in their native state. Although ∼300 mitochondrial proteins were resolved, GzmA altered only a few. Thus, GzmA is a highly specific protease. Although hundreds of potential substrates have been identified by proteomics (6), only 14 GzmA substrates have been verified in GzmA-mediated cell death. Both Gzms concentrate in target-cell nuclei, suggesting that nuclear substrates are important. In fact, all but two validated intracellular GzmA substrates either are predominantly nuclear or move to the nucleus during oxidative stress [histones H1/H2B/H3, lamins A/B/C, PARP-1, Ku70, nucleolin (NUCL), SET, HMGB2, and APE1] (3).
To understand GzmA’s nuclear function, we compared the proteome of isolated nuclei before and after GzmA treatment. Forty-four candidate substrates were identified, of which 33 are RNA-binding proteins that regulate mRNA processing. These RNA-binding proteins included 14 heterogeneous nuclear ribonucleoproteins (hnRNP) that assemble on nascent transcripts and participate in nearly all steps of mRNA maturation (7). Many hnRNPs are both GzmA and caspase substrates, cleaved during death by cytotoxic granules, death receptors, and cancer drugs. Thus, inactivating hnRNPs is a shared feature of programmed cell death. GzmA treatment or caspase activation, but not nonlethal oxidative stress, disrupted splicing and export of newly synthesized RNAs.
Results
Identification of Candidate Nuclear GzmA Substrates.
We compared the proteome of isolated nuclei incubated with GzmA or buffer (Fig. 1 and Fig. S1). The GzmA concentration and incubation time used were the minimum needed to detect clearly cleavage of PARP-1 and lamins A/C, known substrates. Approximately 1,554 spots were resolved. Because the nuclear proteome comprises ∼1,200–2,500 proteins, most nuclear proteins were resolved. Ninety-six spots were reduced by ≥10-fold in intensity by GzmA. Spots of similar apparent mass that might represent posttranslational modifications were grouped into 42 samples and analyzed by in gel digestion and mass spectrometry. Forty-four nuclear proteins, identified by at least three peptides, that had predicted molecular weight and isoelectric point (pI) similar to their migration were selected as candidates (Fig. 1, Table S1, and Dataset S1). The hits included previously described GzmA substrates, lamins A/B/C, and NUCL.
Fig. 1.
Identification of candidate nuclear GzmA substrates. K562 nuclei were treated with 1 μM GzmA or buffer for 30 min at 37 °C, and nuclear proteins were resolved using 2D gel electrophoresis and visualized by silver staining. Spots that changed in intensity at least 10-fold after GzmA treatment were grouped into 42 spots that migrated with similar apparent molecular weight and were analyzed by mass spectrometry. Forty-four GzmA candidate nuclear substrates (Table S1 and Dataset S1) were analyzed by Ingenuity software for known protein–protein interactions. Proteins with similar functions that are not annotated as interacting were added. Previously unknown targets are in blue, previously validated substrates that scored as hits are indicated in green, and those not scored as hits are in yellow.
Thirty-three of 44 candidate proteins—a significant overrepresentation—participate in posttranscriptional regulation of RNA (P = 1.8 × 10−14; Ingenuity). These proteins included 14 of 24 hnRNP proteins (A0, A1, A2/B1, A3, C1/C2, C-like 1, D, G, L, M, Q, and U). hnRNPs assemble on nascent transcripts to regulate RNA processing and nuclear export. The list also included other RNA-binding proteins, including the mRNA-splicing and -processing protein SF2/ASF-1 (SFRS1), proteins involved in biogenesis of nucleoli and ribosomes, including nucleolin (NUCL) and nucleophosmin (NPM1), and mRNA export proteins. A protein interaction network of the candidate substrates was constructed, which confirmed the enrichment for RNA-processing proteins and suggested that GzmA may target multiple proteins within some complexes (Fig. 1). Besides hnRNP complexes, multiple hits were in the Drosha complex (DDX5, DDX17, hnRNP M, and hnRNP D) (8) and in NUCL complexes with hnRNP D or NPM1. Thus, GzmA may interfere with RNA processing.
GzmA Cleaves Multiple hnRNPs.
We verified that GzmA cleaves multiple hnRNP family members. Nuclei treated with GzmA, catalytically inactive GzmA (S-AGzmA), or GzmB were analyzed by immunoblot for some hnRNP proteins (Fig. 2 A and B). hnRNP A1, A2/B1, C1/C2, and U were all cleaved by GzmA with kinetics similar to those of PARP-1 in a dose- and time-dependent manner, as indicated by either a stable cleavage product or decreased intensity of the full-length protein. S-AGzmA had no effect. hnRNPs C1/C2, and U, but not A1 and A2/B1, also were cleaved by GzmB. Another proteomics hit, Nono/p54nrb, was not a clear GzmA substrate; a slight mobility shift suggested it might be cleaved near one end, but this possibility was not pursued. GzmA also cleaved all analyzed hnRNPs within hnRNP complexes immunopurified from HeLa nuclei using hnRNP C1/C2 or A1 antibodies (Fig. S2A).
Fig. 2.
GzmA cleaves multiple hnRNP proteins. (A) K562 nuclei were treated with increasing GzmA concentrations or with 250 or 1,000 nM GzmA for indicated times, and hnRNP cleavage was assessed by immunoblot. PARP-1 is a known GzmA substrate; Ku-86 was a loading control. S-A, inactive GzmA. (B) GzmA treatment of isolated K562 nuclei compared with GzmB. Treatment was with 1 μM Gzm for 1 h. U, untreated control. (C) K562 cells were treated with PFN and/or GzmA or GzmB at the indicated dose and time. hnRNP cleavage was assessed by immunoblot. Control samples were treated with buffer or with PFN only, GzmA only, or GzmB only for 1 h. SET is a known GzmA substrate (29); β-actin was a loading control. Blots are representative of at least three independent experiments.
To determine whether hnRNPs are physiologically relevant targets, we evaluated their cleavage in K562 cells treated with perforin (PFN) and Gzms (Fig. 2C). hnRNPs A1 and A2/B1 were cleaved by GzmA in intact cells with dose and time dependency similar to that of the known substrate SET. However, the cleavage fragments were labile and were not detected in whole cells. In contrast to treatment of isolated nuclei (Fig. 2B), GzmB and PFN treatment of intact cells led to hnRNP A1 and A2/B1 cleavage, confirming previous proteomics studies (9–16) suggesting that these hnRNPs might be caspase targets. When cells were treated with PFN and GzmB in the presence of the pan-caspase inhibitor zVAD-fmk, hnRNP A1 remained unchanged, confirming that hnRNP A1 is not a direct GzmB target but is a caspase target (Fig. S2B). hnRNP A1 also was cleaved when NK-92 natural killer (NK) cells attacked 721.221 B cells (Fig. S2C). hnRNP A1 cleavage occurred, but was reduced, in the presence of zVAD. Thus, hnRNP A1 is a shared substrate of GzmA and activated caspases. In fact, all six validated hnRNPs (A1, A2/B1, C1/C2, and U) were cleaved in GzmB-treated cells. However, hnRNP C1/C2 and U were cleaved directly by GzmB and the caspases, whereas hnRNP A1 and A2/B1 were cleaved efficiently only via caspase activation (Fig. 2 B and C and Fig. S2B).
hnRNPs Are Degraded During Caspase-Dependent Death.
To investigate whether hnRNPs are degraded during caspase-mediated death, Jurkat cells were treated with anti-Fas or doxorubicin, and HeLa cells were treated with staurosporine (Fig. S3). A PARP-1 cleavage fragment was seen within 2 h after the addition of anti-Fas. hnRNP A1, C1/C2, and U levels also declined within 2–4 h of anti-Fas treatment. Cleavage was inhibited by zVAD-fmk. HMGB2, an abundant DNA-binding protein, was an uncleaved control. Doxorubicin, which induced apoptosis more slowly, based on PARP-1 cleavage and procaspase-3 disappearance, also led to caspase-dependent hnRNP A1 and C1/C2 cleavage and a decrease in hnRNP U. hnRNP A1, C1/C2, and U also were degraded in staurosporine-treated HeLa cells, roughly coincident with caspase activation. Thus, hnRNP degradation occurs in multiple apoptotic pathways.
GzmA Disrupts Export of Newly Synthesized RNA.
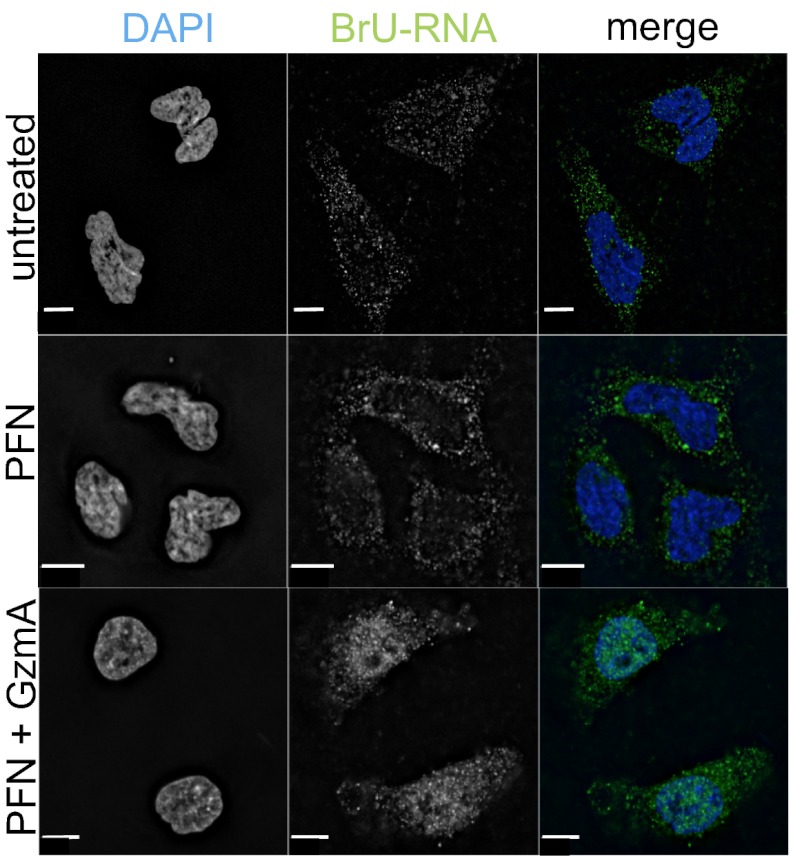
Because cleavage of hnRNPs and other RNA-binding proteins might disrupt mRNA processing and nuclear export, we used immunofluorescence microscopy and a BrdU antibody that cross-reacts with BrU to assess the localization of newly synthesized BrU-labeled RNA in HeLa cells treated for 1 h with PFN and GzmA (Fig. 3). In untreated cells or in cells treated only with PFN, BrU-labeled RNA was mostly cytoplasmic, suggesting efficient processing and export. However, in cells treated with GzmA and PFN newly synthesized RNA was retained largely in the nucleus.
Fig. 3.
GzmA causes nuclear retention of newly synthesized RNA. HeLa cells were labeled with BrU during treatment with PFN with or without 1 μM GzmA. Cells were fixed after 1 h and stained for BrdU, which also recognizes BrU (green), and DAPI (blue). Most newly synthesized RNA was retained in the nucleus after treatment with PFN and GzmA. Images are representative of three independent experiments. (Scale bars, 10 μm.)
GzmA Disrupts Pre-mRNA Splicing.
To investigate whether the RNA export defect was caused by deficient splicing and/or export, we designed primers to amplify spliced (primer pairs in adjacent exons), unspliced (primer pairs in adjacent exon and intron), and total (primer pairs within an exon) transcripts. We selected mRNAs (MYC, FOS, DUSP5, and E2F1) with short half-lives (T1/2 ∼10–90 min) to weight the analysis toward newly synthesized mRNAs. Nuclear and cytoplasmic RNA were isolated from untreated cells and from cells treated for 1 h with PFN and/or Gzms. Arsenite, which causes noncytotoxic oxidative stress, was a control noncytotoxic cellular stress that induced FOS, DUSP5, and E2F1. PFN- and GzmB-treated cells were analyzed with and without zVAD-fmk. Subcellular fractionation was verified by immunoblot for tubulin, which was not detected in the nuclear fractions. The ratio of spliced/unspliced RNA in each compartment was normalized to untreated cells. The ratios were not altered significantly by treatment with PFN alone or arsenite (Fig. 4A). However, splicing was sharply reduced (up to ∼10-fold) for all four genes after treatment with PFN and either Gzm. The reduction in splicing after treatment with GzmA/PFN was significant for MYC, FOS, and DUSP5; a possible explanation for the lack of a significant reduction in E2F1 may be that E2F1 mRNA has the longest half-life. Caspase inhibition during GzmB treatment largely restored splicing to control levels. Thus, both GzmA and GzmB, the latter in a caspase-dependent manner, disrupted splicing of newly synthesized mRNAs.
Fig. 4.
GzmA interferes with mRNA splicing. (A–C) HeLa cells were untreated or treated with PFN and/or 0.5 μM GzmA or GzmB (± zVAD-fmk) or with arsenite (Ars) for 1 h before isolation of RNA from fractionated nuclei and cytoplasm. PCR primers were chosen to amplify unspliced, spliced, or total RNA. First, qRT-PCR results were normalized to GAPDH, and then the ratios were normalized to their value in untreated cells (ratio = 1). (A) Ratio of spliced:unspliced nuclear mRNA. (B) Ratio of cytoplasmic:nuclear total RNA for spliced genes (MYC and FOS) compared with intronless genes (JUN and IFNA1). Data are pooled from four independent experiments. Asterisks indicate a significant difference compared with untreated cells (P < 0.05). (C) FOS and JUN RNA was isolated from fractionated nuclei, and protein was extracted from whole-cell lysates. (Top) FOS and JUN transcripts (total RNA) were amplified by qRT-PCR normalized to GAPDH and then to expression in untreated cells. Both genes were significantly induced under all conditions. Data are mean ± SD of four independent experiments. (Middle) Protein expression was assessed by immunoblot with β-actin as loading control. (Bottom) Blots of four independent experiments were quantified by densitometry showing the intensity of the c-Fos or c-Jun band relative to the loading control (mean ± SEM percent of untreated cells; *P < 0.05).
Nuclear export of intron-containing transcripts requires splicing. The cytoplasm of untreated cells contained ∼10–20 times more spliced mRNA for these genes than did the nuclear fractions. Thus, once splicing occurs, nuclear export is efficient. To assess whether defective RNA export was caused mostly by inefficient splicing and/or by inhibition of export, we next compared the ratio of cytoplasmic:nuclear total and spliced RNA, normalized to untreated cells, for MYC and FOS with the ratio for two intronless genes, JUN and IFNA1. As expected, MYC and FOS mRNA export was impaired severely in Gzm-treated cells, as compared with untreated cells or cells treated only with PFN or arsenite, but the export of JUN and IFNA1 mRNA was not altered significantly (Fig. 4B). However, nuclear export of spliced MYC, FOS, DUSP5, and E2F1 mRNAs, assessed by the cytoplasmic:nuclear spliced mRNA ratio, was unchanged (Fig. S4). Thus, spliced mRNAs were exported efficiently, and nuclear retention was caused by defective splicing.
FOS and JUN are induced by cellular stress. In fact, total FOS and JUN RNA increased ∼eightfold within 1 h after the addition of GzmA and PFN and >30-fold with GzmB and PFN (Fig. 4C). FOS mRNA, which requires splicing, was not spliced or exported after treatment with PFN and either Gzm, whereas the export of intronless JUN mRNA was not impaired by the Gzms (Fig. 4B). Thus, we expected that c-FOS up-regulation would be blunted in cells undergoing programmed cell death but that c-JUN protein could be induced. Arsenite induced both c-JUN and c-FOS mRNA and protein (Fig. 4C). After treatment with Gzms and PFN, c-FOS protein declined in 1 h, as compared with levels in control cells, whereas c-JUN levels increased dramatically. Thus, induction of early-response proteins that orchestrate the cellular repair response probably is severely disrupted during programmed cell death only if their transcripts need splicing.
hnRNP A1 Cleavage Disrupts Its Nuclear Localization.
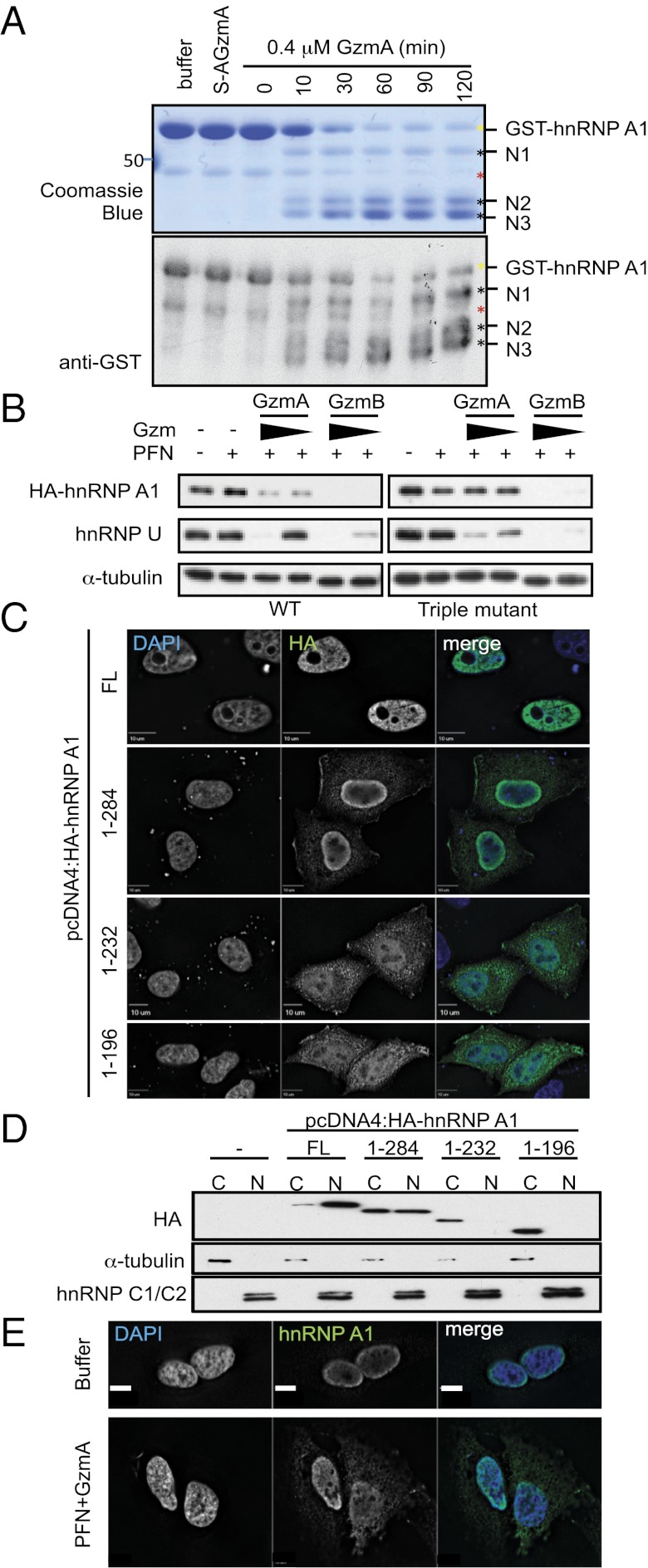
We next focused on hnRNP A1, the best-studied hnRNP. An N-terminal GST-hnRNP A1 fusion protein was generated to identify GzmA cleavage sites. After purified GST-hnRNP A1 was treated with GzmA for 10 min, three N-terminal cleavage products were seen that increased with time as the full-length protein decreased (Fig. 5A). In-gel chymotryptic digestion and mass spectrometry identified R196, R232, and R284 as the cleavage sites. Site-directed mutagenesis of the three putative cleavage sites (R196/232/284A) protected mutant hnRNP from GzmA cleavage in vitro (Fig. S5A) and in transfected target cells treated with GzmA and PFN (Fig. 5B) or NK cells, provided zVAD-fmk was present (Fig. S2C). Thus, these three arginines are functional GzmA cleavage sites. hnRNP A1 has three RNA-binding domains, followed by a C-terminal noncanonical nuclear localization sequence (Fig. S5B). It shuttles between the nucleus and cytoplasm but is mostly nuclear. Cleavage at these sites would separate or disrupt the nuclear localization signal from the RNA-binding domains, likely disrupting hnRNP A1 nuclear localization. In fact, immunofluorescence microscopy (Fig. 5C) and immunoblot of cell fractions (Fig. 5D) showed that successive C-terminal truncations of N-terminal HA-tagged hnRNP A1 that would be produced by GzmA cleavage accumulated increasingly in the cytoplasm. Full-length HA-hnRNP A1, like endogenous hnRNP A1, was mostly nuclear (Fig. 5E). Moreover, endogenous hnRNP A1 redistributed toward the cytoplasm after GzmA and PFN treatment for 1 h (Fig. 5E). To verify that hnRNP A1 cleavage is responsible for its mislocalization, HeLa cells expressing WT or GzmA-uncleavable triple-mutant HA-hnRNP A1 were treated with PFN and GzmA and examined by immunofluorescence microscopy (Fig. S5C). In untreated cells, HA staining for both WT and mutant hnRNP A1 was exclusively nuclear. WT hnRNP A1 partly stained in the cytoplasm after treatment with GzmA and PFN, but the GzmA-uncleavable mutant remained nuclear. Similar mislocalization of endogenous hnRNP A1 occurred within 2 h after the addition of staurosporine, increasing further after 3 h (Fig. S5D). Thus, both GzmA and caspases interfere with hnRNP A1 nuclear localization.
Fig. 5.
GzmA cleaves hnRNP A1 after R196, R232, and R284, resulting in cytoplasmic mislocalization. (A) Recombinant purified GST-hnRNP A1 was treated with 0.4 μM GzmA or S-AGzmA and examined by Coomassie blue (Upper) or GST immunoblot (Lower). The GST tag was at the N terminus. Three N-terminal cleavage products (black asterisks) appear within 10 min. Yellow asterisk indicates full-length GST-hnRNP A1; red asterisk indicates a contaminating band. Mass spectrometry of excised bands indicated cleavage after R196, R232, and R284. (B) HeLa cells, expressing WT (Left) or GzmA-uncleavable mutant (Right) HA-hnRNP A1, were treated with PFN plus GzmA or GzmB (0.5 and 0.16 μM, respectively) for 1 h. HA-hnRNP A1 cleavage was assessed by immunoblot probed for HA. α−Tubulin was the loading control, and endogenous hnRNP U served as a cleavage control. WT HA-hnRNP A1 and endogenous hnRNP U were cleaved by both Gzms; mutant HA-hnRNP A1 was resistant to cleavage by GzmA but not by GzmB. (C) HeLa cells expressing full-length (FL) HA-hnRNP A1 or indicated truncations were stained with anti-HA (green) and DAPI (blue). (D) HeLa cells expressing GzmA-generated HA-hnRNP A1 truncations were separated into cytoplasmic (C) and nuclear (N) fractions, and HA-hnRNP A1 localization was assessed by anti-HA immunoblot. Tubulin and hnRNP C1/C2 are fractionation controls for the cytoplasm and nucleus, respectively. (E) Immunofluorescence localization of endogenous hnRNP A1. Untransfected HeLa cells were treated with buffer or PFN plus 1 μM GzmA for 1 h before staining for hnRNP A1 (green) and DAPI (blue). Blots and images are representative of at least three independent experiments.
GzmA-Resistant hnRNP A1 Inhibits Death and Rescues Splicing.
To determine whether hnRNP A1 cleavage is important during GzmA-mediated death, HeLa cells overexpressing WT or GzmA-resistant hnRNP A1 were treated with PFN and either Gzm and were evaluated by 51Cr release assay (Fig. 6A) and annexin V/propidium iodide staining (Fig. 6B). Expression of triple-mutant hnRNP A1 rendered target cells more resistant to GzmA but equally sensitive to GzmB as cells expressing WT protein. Furthermore, GzmA-uncleavable hnRNP-A1 expression significantly restored MYC, DUSP5, and FOS splicing after GzmA treatment but did not affect their splicing after GzmB treatment (Fig. 6C). Thus, hnRNP A1 is an important GzmA substrate, because blocking its cleavage inhibits GzmA-mediated death and rescues splicing.
Fig. 6.
Uncleavable hnRNP A1 rescues splicing and inhibits cell death. (A–C) HeLa cells stably expressing WT or GzmA-uncleavable mutant HA-hnRNP A1 were treated with PFN plus GzmA or GzmB. Death was monitored by a 4-h 51Cr release assay (A) and annexin V/propidium iodide staining 1 h later for flow cytometry (B). Data in A show mean ± SD of three independent experiments of percent specific lysis after subtraction of percent specific lysis of control cells treated with PFN alone. Data in B show mean ± SEM of pooled data from six independent experiments. (C) RNA isolated from whole cells 1 h after treatment was analyzed by qRT-PCR to compare spliced/unspliced total cellular RNA. The ratio was normalized to untreated cells. Data are mean ± SEM of five independent experiments. In all panels, statistical differences between WT and mutant hnRNP A1-expressing cells were calculated using a paired Student’s t test.
Discussion
The rapid concentration of Gzms in target cell nuclei motivated us to analyze changes in the proteome of GzmA-treated nuclei. Only 6% of protein spots changed after GzmA treatment, confirming GzmA’s specificity. Notably, some abundant nuclear protein spots were unchanged. Treating proteins in intact nuclei likely reduced background that might occur in treating cell lysates. We identified 44 potential nuclear GzmA substrates, which included four previously known GzmA substrates (lamins and NUCL), but others (PARP-1, Ku70, Ape1, and histones) were missed. By setting a stringent criterion (10-fold less protein), we biased our hits toward key substrates at the price of reduced sensitivity. In fact, we were able to validate all the hits we examined experimentally (with the possible exception of Nono). Seventy-five percent of the candidate substrates, including 14 hnRNPs, are RNA-binding proteins that orchestrate posttranscriptional RNA processing. We verified that six of six hnRNPs examined were cleaved during GzmA-mediated death and also confirmed previous proteomics studies that suggested that these hnRNPs also are caspase targets (9–16).
Because RNA-binding proteins dominated our screen, we examined the effect of Gzms on RNA processing. A key common and unrecognized feature of caspase-independent and caspase-dependent programmed cell death is disruption of pre-mRNA splicing and nuclear export of newly synthesized RNA. At least 11 of the 44 candidate GzmA substrates, including hnRNP A1 and ASF1, have important roles in pre-mRNA splicing. Pre-mRNA levels of the early-response genes investigated in this study (FOS, JUN, DUSP5, E2F1, MYC, and IFNA1) increased by 7–50 fold within 1 h of treatment with either Gzm. Thus, transcription is unimpaired during the early stages of programmed cell death. However, because of impaired splicing, early-response proteins, whose mRNAs (like most mRNAs) require splicing, are not up-regulated. Inhibiting synthesis of cellular stress-response proteins, many of which have tightly regulated transcripts with short half-lives, likely interferes with cellular repair.
Disruption of pre-RNA splicing and RNA export is not a general feature of cellular stress, because, consistent with previous reports, it did not occur during nonapoptotic oxidative stress (17). Heat shock transiently interferes with pre-mRNA splicing of at least some genes (18). However, global changes in pre-RNA splicing and export of newly synthesized RNAs during apoptosis have not been reported previously. Caspase activation, and cellular stress more generally, disrupts translation initiation. This disruption is accomplished by caspase cleavage of eIF4G, eIF2α, and other initiation factors only during apoptosis and by phosphorylation of eIF2α by a variety of stress-induced kinases during both stress and apoptosis (19). Because translation uses about half of the cell’s energy consumption, global inhibition of translation husbands resources at times of stress. However, some mRNAs, especially cap-independent transcripts whose products may help the cell survive stress and apoptosis, overcome this block. During stress there are shifts from cap-dependent to internal ribosome entry site (IRES)-dependent translation and toward alternative splicing, especially of mRNAs for genes such as bcl-XL, whose splice variants often play opposing roles in cell survival (20). Alternative splicing is regulated by SFRS1/ASF1 and hnRNP A1 binding to exonic sites to enhance or inhibit, respectively, binding of U2AF and the U2 small nucleotide ribonucleoprotein to the 3′ splice site (21). Both ASF1 and hnRNP A1 were hits in our screen for GzmA nuclear substrates, although we did not confirm that ASF1 is a bona fide target, and ASF1 is not a known caspase substrate. Alternative pre-mRNA splicing occurs early in caspase-mediated death (22). Future studies are needed to compare splice-site selection during caspase-independent and -dependent programmed cell death and to examine the role of hnRNP A1 and ASF1 cleavage in this process. The relative efficiency of cleavage of hnRNP A1 and/or ASF1 may influence the ratio of the splice variants that get made. Changes in alternative splicing might play a more important role in more protracted forms of apoptosis (such as following UV irradiation or chemotherapy) than in the relatively rapid Gzm-induced cell death.
Impaired RNA processing should synergize with the inhibition of translation to block de novo protein synthesis even of mRNAs that bypass the stress-induced block in cap-dependent translation initiation. If a target cell is unable to synthesize new proteins, it will be crippled in repairing cellular damage instigated by death stimuli. In previous studies we significantly reduced GzmA-mediated death by overexpressing noncleavable forms of key substrates, including Ape1, Ndufs3, PARP1, and Ku70 (3). These experiments work—perhaps surprisingly—because inducing cell death is not an all-or-none process: Programmed cell death disrupts multiple cellular pathways that together overcome cellular repair. Indeed, GzmA-noncleavable hnRNP A1, which rescued splicing, rendered cells significantly more resistant to GzmA-mediated death. hnRNP A1, one of the most abundant nuclear proteins, binds to nascent transcripts as soon as they are transcribed and remains bound during their nuclear export (23). It regulates both cap-dependent and IRES-mediated translation. It also is implicated in ribosomal RNA (rRNA) processing, which potentially could interfere with the translation of unspliced mRNAs and those intron-containing mRNAs that succeed in getting spliced and exported. Because of its central role in RNA processing, it is perhaps not surprising that hnRNP A1 on its own is a physiologically important GzmA target.
In some stresses, hnRNP A1 is phosphorylated by MAPK p38 and translocates to the cytoplasm (24). The cytoplasmic translocation of hnRNP A1 we observed following GzmA and staurosporine treatment could result from hnRNP A1 cleavage and/or stress-induced phosphorylation. However, cleavage is likely the dominant mechanism behind mislocalization, at least for GzmA, because the noncleavable mutant retained its nuclear localization. Expression of a phosphomimetic mutant of hnRNPA1 that localizes to the cytoplasm activates caspase 3. The proapoptotic effect of hnRNP A1 cleavage may have been partly the result of its cytoplasmic mislocalization.
Although multiple proteins involved in mRNA export are candidate GzmA targets (Fig. 1), mRNA export or protein expression of an unspliced immediate early gene product (c-JUN) was not inhibited in Gzm-treated cells. Further study is needed to assess if and when candidate proteins involved in RNA export are cleaved during programmed cell death and whether their cleavage affects export. Export may be impaired at times later than we analyzed. The list of candidate GzmA substrates also included RNA-binding proteins that affect synthesis and processing of rRNAs (which make up ∼80% of cellular RNA) and microRNAs. Because newly synthesized RNA overall was retained globally in the nucleus after treatment with GzmA and PFN, the processing of other classes of RNAs, especially abundant rRNAs and tRNAs, is disturbed during apoptosis. Our identification of four proteins in the large Drosha complex as potential GzmA substrates suggests that microRNA processing is impaired also. High-throughput sequencing of nuclear and cytoplasmic RNAs or of newly synthesized RNAs in cells undergoing programmed cell death should help assess the effects of distinct programmed cell death pathways on all steps of RNA processing.
The SET complex, which contains three GzmA substrates (SET, HMGB2, and APE1) and is responsible for GzmA-mediated DNA damage, was the first recognized example of a programmed cell death protease targeting multiple substrates within the same complex (3). However, cleavage of multiple components of a multiprotein complex, as we found here for the hnRNPs, may be a more common phenomenon. Activated caspases also cleave multiple proteins in the N-CoR/SMRT complex (13). GzmA and the caspases may cleave multiple components of a complex, such as the hnRNP complexes, because they are dimers: While one monomer is attacking one component in the complex, the other monomer may be well positioned to attack another.
Most previously identified GzmA substrates are unique to caspase-independent death. The two substrates shared with the caspases, PARP-1 and lamin B, illustrate processes that might need to be disrupted for programmed cell death and for avoiding necrosis. For example, failure to inactivate PARP-1 depletes cellular ATP, needed for programmed death (25, 26). Lamin cleavage may be needed to disrupt the nuclear envelope. In some cases the same pathways are disrupted but by targeting different proteins. For example, the repair of dsDNA breaks is inhibited by GzmA by targeting Ku70 and by the caspases by cleaving DNA-PKcs (27, 28). The identification of multiple hnRNP proteins as shared targets of both GzmA and the caspases suggests that inhibiting mRNA processing is another critical feature of programmed cell death. It will be of interest to identify how many of the GzmA candidate targets are shared targets with GzmB and/or the caspases and whether other posttranscriptional RNA processing proteins are targeted during apoptosis.
Materials and Methods
Isolated nuclei were treated with GzmA or buffer and were analyzed by 2D SDS/PAGE isoelectric focusing gels, and candidate GzmA substrates were identified by mass spectrometry. Candidates were validated by immunoblot and shown to be cleaved during immune-mediated death. Changes in RNA splicing and cellular localization were followed by immunofluorescence microscopy and quantitative RT-PCR (qRT-PCR). Details are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Gideon Dreyfuss for hnRNP antibodies and Arlene Sharpe, Paul Anderson, Tom Kirchhausen, Melissa Moore and members of the J.L. laboratory for helpful discussions. This work was supported by National Science Foundation predoctoral fellowships (to D.K.R. and M.P.T.), a Kurt und Senta Herrmann-Foundation fellowship (to M.W.), and National Institutes of Health Grant AI45587 (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201327109/-/DCSupplemental.
References
- 1.Chowdhury D, Lieberman J. Death by a thousand cuts: Granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lord SJ, Rajotte RV, Korbutt GS, Bleackley RC. Granzyme B: A natural born killer. Immunol Rev. 2003;193:31–38. doi: 10.1034/j.1600-065x.2003.00044.x. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman J. Granzyme A activates another way to die. Immunol Rev. 2010;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hink-Schauer C, Estébanez-Perpiñá E, Kurschus FC, Bode W, Jenne DE. Crystal structure of the apoptosis-inducing human granzyme A dimer. Nat Struct Biol. 2003;10:535–540. doi: 10.1038/nsb945. [DOI] [PubMed] [Google Scholar]
- 5.Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme P, et al. The substrate specificity profile of human granzyme A. Biol Chem. 2010;391:983–997. doi: 10.1515/BC.2010.096. [DOI] [PubMed] [Google Scholar]
- 7.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 9.Waterhouse N, et al. Heteronuclear ribonucleoproteins C1 and C2, components of the spliceosome, are specific targets of interleukin 1beta-converting enzyme-like proteases in apoptosis. J Biol Chem. 1996;271:29335–29341. doi: 10.1074/jbc.271.46.29335. [DOI] [PubMed] [Google Scholar]
- 10.Brockstedt E, et al. Identification of apoptosis-associated proteins in a human Burkitt lymphoma cell line. Cleavage of heterogeneous nuclear ribonucleoprotein A1 by caspase 3. J Biol Chem. 1998;273:28057–28064. doi: 10.1074/jbc.273.43.28057. [DOI] [PubMed] [Google Scholar]
- 11.Thiede B, Dimmler C, Siejak F, Rudel T. Predominant identification of RNA-binding proteins in Fas-induced apoptosis by proteome analysis. J Biol Chem. 2001;276:26044–26050. doi: 10.1074/jbc.M101062200. [DOI] [PubMed] [Google Scholar]
- 12.Thiede B, Siejak F, Dimmler C, Rudel T. Prediction of translocation and cleavage of heterogeneous ribonuclear proteins and Rho guanine nucleotide dissociation inhibitor 2 during apoptosis by subcellular proteome analysis. Proteomics. 2002;2:996–1006. doi: 10.1002/1615-9861(200208)2:8<996::AID-PROT996>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Damme P, et al. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- 16.Van Damme P, et al. Complementary positional proteomics for screening substrates of endo- and exoproteases. Nat Methods. 2010;7:512–515. doi: 10.1038/nmeth.1469. [DOI] [PubMed] [Google Scholar]
- 17.Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- 19.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 20.Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 22.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 24.Lewis SM, et al. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu P, et al. The cytotoxic T lymphocyte protease granzyme A cleaves and inactivates poly(adenosine 5′-diphosphate-ribose) polymerase-1. Blood. 2009;114:1205–1216. doi: 10.1182/blood-2008-12-195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herceg Z, Wang ZQ. Failure of poly(ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol Cell Biol. 1999;19:5124–5133. doi: 10.1128/mcb.19.7.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu P, et al. Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku70. EMBO Rep. 2006;7:431–437. doi: 10.1038/sj.embor.7400622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casciola-Rosen L, et al. Apopain/CPP32 cleaves proteins that are essential for cellular repair: A fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beresford PJ, Kam CM, Powers JC, Lieberman J. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc Natl Acad Sci USA. 1997;94:9285–9290. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.